Background: Non-native intermediates are preferred substrates for GroEL.
Results: GroEL binds with the burst phase intermediate of malate synthase G (MSG), subsequently eliminating the slowest kinetic phase of refolding with the assistance of GroES and ATP.
Conclusion: GroEL modulates the folding trajectory of MSG, thereby accelerating its refolding.
Significance: The study provides much needed insight into GroEL/GroES-assisted folding of large, multidomain proteins.
Keywords: Aggregation, Molecular Chaperone, Protein Conformation, Protein Dynamics, Protein Folding, GroEL, Malate Synthase G, Folding Intermediates, Large Protein Folding, Refolding Kinetics
Abstract
Despite a vast amount information on the interplay of GroEL, GroES, and ATP in chaperone-assisted folding, the molecular details on the conformational dynamics of folding polypeptide during its GroEL/GroES-assisted folding cycle is quite limited. Practically no such studies have been reported to date on large proteins, which often have difficulty folding in vitro. The effect of the GroEL/GroES chaperonin system on the folding pathway of an 82-kDa slow folding protein, malate synthase G (MSG), was investigated. GroEL bound to the burst phase intermediate of MSG and accelerated the slowest kinetic phase associated with the formation of native topology in the spontaneous folding pathway. GroEL slowly induced conformational changes on the bound burst phase intermediate, which was then transformed into a more folding-compatible form. Subsequent addition of ATP or GroES/ATP to the GroEL-MSG complex led to the formation of the native state via a compact intermediate with the rate several times faster than that of spontaneous refolding. The presence of GroES doubled the ATP-dependent reactivation rate of bound MSG by preventing multiple cycles of its GroEL binding and release. Because GroES bound to the trans side of GroEL-MSG complex, it may be anticipated that confinement of the substrate underneath the co-chaperone is not required for accelerating the rate in the assisted folding pathway. The potential role of GroEL/GroES in assisted folding is most likely to modulate the conformation of MSG intermediates that can fold faster and thereby eliminate the possibility of partial aggregation caused by the slow folding intermediates during its spontaneous refolding pathway.
Introduction
In the cell, proper folding of a majority of proteins is assisted by a class of proteins called chaperones. GroEL and GroES, both from the bacterium Escherichia coli, are the best characterized chaperones. GroEL along with its co-chaperonin GroES facilitates folding of many proteins via ATP-dependent release, preventing their misfolding and aggregation and channeling them on the correct folding pathway (1–5). GroEL binds to a wide range of proteins ranging from 2 kDa to more than 100 kDa both in vivo and in vitro (6–10). Based on the size of the protein, two types of mechanisms for GroEL-assisted folding, cis and trans, have been delineated in the past. Proteins of up to ∼ 60 kDa fold via the cis mechanism where polypeptide and GroES bind to the same GroEL ring (11), but large proteins (>60 kDa) that cannot be pushed inside the GroEL cavity fold via the trans mechanism where polypeptide and GroES bind to the opposite GroEL rings (12, 13). The interplay of GroEL, GroES, and ATP in chaperone-assisted folding has been well delineated, and their effect on the refolding of polypeptides has been thoroughly demonstrated (14–17). However, the kinetics and molecular events associated with the refolding polypeptide during its GroEL/GroES-assisted folding cycle are still poorly understood.
Binding of GroEL to the substrate proteins modulates the energy landscape for their folding process by lowering the kinetic barriers that separate various states on the folding pathway (1–3). Such kinetic barriers are responsible for slow folding of proteins in the absence of chaperones, which often results in their aggregation. Modulation is also caused by binding and hydrolysis of ATP to GroEL (18, 19) due to which folding can occur preferably through a single route when many routes are otherwise available (20). The binding energy of GroEL may be utilized to unfold kinetically trapped or misfolded conformers, allowing their productive folding (21–23). However, GroEL binding may not always be associated with unfolding of bound polypeptide. The unfolding action of GroEL is usually observed when it binds to early unstructured or collapsed intermediates (20, 24, 25), whereas no unfolding has been reported for proteins where late structured intermediates bind to GroEL (26, 27). This implies that GroEL binding may result in folding or unfolding or in no structural change depending on the conformation of protein that binds to GroEL. Following binding, proteins may require both GroES and ATP or only ATP for their release from the GroEL complex. This is usually dictated by the ease with which protein is able to fold spontaneously. Poor refolders generally require both GroES and ATP (28–31), whereas relatively easy refolders require only ATP (32, 33) for the GroEL-assisted folding.
Over the last decade, it has been well established that GroEL/GroES are involved in driving protein folding under conditions where spontaneous folding simply does not occur (1, 11, 34–37); however, how they facilitate the process is still not completely understood. To address this question, it is important to know the nature of conformational changes taking place for the protein in solution and when it transits through the GroEL/GroES folding cycle. It is difficult to carry out such studies with the natural substrate proteins of GroEL as their folding mechanisms are poorly understood, and partly folded forms of these proteins that bind to GroEL are aggregation-prone. These constraints make them poor folding models in vitro. Thus, such studies aimed at understanding the events experienced by the polypeptides undergoing GroEL-assisted folding utilize proteins either that are slow folding or whose folding mechanisms are well characterized. Few such studies have been reported on small proteins where GroEL enhanced their refolding without changing their respective folding pathways (38, 39).
Here, we attempted to address these questions by using an aggregation-prone, large (82-kDa), and multidomain protein malate synthase G (MSG).3 Its folding pathway has recently been characterized (40) and was found to populate both early collapsed and late structured intermediates. Although its folding is reversible in vitro, it is a slow folding protein where the native state is formed very slowly from stable native-like intermediate, thus making the process aggregation-prone at higher protein concentrations (40). Considering the nature of the substrate binding property of GroEL, which preferentially binds to the partially folded intermediates (1–3), we decided to explore the chaperone binding ability of non-native MSG, prevention of its aggregation, and the mechanism of assisted folding. Furthermore, MSG was chosen as a model protein for studying the mode of modulation of the folding pathway of a multidomain protein by GroEL.
EXPERIMENTAL PROCEDURES
Materials
DH5α, BL21, and BL21-DE3 Escherichia coli strains were used for cloning, expression, and purification of MSG, GroEL, and GroES. MSG was cloned in pET28b vector at NcoI and XhoI sites containing His6 at the C terminus. pACYCEL containing GroEL gene was a generous gift from Prof. Hideki Taguchi (Tokyo Institute of Technology, Yokohama, Japan). pET11a containing GroES gene was a generous gift from Prof. A. L. Horwich (Yale University, New Haven, CT). Guanidine hydrochloride was purchased from USB Corp. All other chemicals were of ultrapure grade.
Purification of MSG, GroEL, and GroES
MSG and GroES were overexpressed in BL21-DE3 E. coli cells, whereas GroEL was overexpressed in BL21 E. coli cells. Plasmids pET28b, pACYCEL, and pET11a were used for overexpression of MSG, GroEL, and GroES, respectively. Chaperones were purified as described previously (14–16) with necessary modifications. Cells were lysed in a French press, and lysates were centrifuged at 18,000 rpm for 1.5 h. Supernatant was collected and filtered through a 0.45-μm filter before applying to chromatographic columns in an ÄKTA FPLC system. GroEL was purified using Fast Flow Q anion exchange and 15ISO hydrophobic interaction chromatography. After the FPLC purification process, GroEL was treated with Affi-Gel Blue to remove bound substrate proteins. MSG purification was a single step process using a nickel-chelating column (HisTrap HP, GE Healthcare) as described previously (40). For GroES purification, first, differential precipitation of protein mixture was done by lowering the pH, which allowed elimination of other proteins, and then GroES was purified using Fast Flow SP cation exchange chromatography.
Aggregation Kinetics
MSG was incubated with and without GroEL at a 1:2 molar ratio in 20 mm Tris buffer, pH 7.8, 10 mm MgCl2, 10 mm KCl at 55 °C, and its aggregation kinetics was monitored by light scattering at 500 nm in a Cary Varian Eclipse fluorometer using a quartz cuvette with a path length of 1 cm. Solutions were constantly stirred to avoid formation of bubbles. Excitation and emission wavelengths were set at 500 nm with 5 and 2.5 slit widths each. Final MSG concentration was 1 μm.
Identification of GroEL-MSG Binary Complex
GroEL-MSG complex was formed by incubating 1 μm MSG with 2 μm GroEL at 55 °C for 10 min after which the mixture was centrifuged at 13,000 rpm for 20 min to remove any aggregated protein. Supernatant was collected, vigorously vortexed, and cooled to 4 °C before running through a 14-ml prepacked size exclusion column (TSKgelG4000SWXL, Tosoh, Japan) in a Waters HPLC system. The peak top of the GroEL fraction was collected, lyophilized, and run on a 10% SDS gel followed by Coomassie Blue staining to detect the presence of any MSG band. 20 mm Tris buffer, pH 7.8, 10 mm MgCl2, 10 mm KCl was used as the mobile phase at a 1 ml/min flow rate. Samples were injected into the column using a 200-μl loop.
GroEL/GroES-assisted in Vitro Refolding of GdnHCl-denatured MSG
MSG refolding was monitored by measuring activities at different time intervals in a Beckman Coulter DU800 spectrophotometer. MSG was denatured in 3 m GdnHCl for 2 h at 25 °C, and unfolding was confirmed by loss of enzymatic activity. To monitor spontaneous refolding, denatured MSG was diluted directly into refolding buffer containing 20 mm Tris, 300 mm NaCl, 10 mm MgSO4, 10 mm KCl, 1 mm tris(2-carboxyethyl)phosphine hydrochloride, 10% glycerol, pH 7.9. The refolding buffer composition was optimized previously for spontaneous refolding of MSG (40). Because we wanted to compare the kinetics, the same buffer was used for spontaneous and chaperone-assisted folding. Chaperone-assisted refolding was carried out by diluting denatured MSG in refolding buffer containing GroEL such that the final MSG and GroEL concentrations were 0.25 and 0.5 μm, respectively. Reactivation of GroEL-MSG complex was performed by adding 2.5 mm ATP 10 min after complex formation. When indicated, GroES was added at a concentration of 1 μm before ATP. The final GdnHCl concentration was kept at 0.1 m, and buffer containing 0.25 μm native MSG in 0.1 m GdnHCl was used as a control. The activity of MSG was measured by determining the rate of glyoxylate-dependent release of free CoA from acetyl-CoA at 25 °C. Each assay mixture contained 100 μl of 1 m Tris buffer, pH 7.9; 10 μl of 1 m MgCl2; 10 μl of 0.1 m glyoxylate; 40 μl of 10 mm acetyl coenzyme A; and protein in a total volume of 900 μl. The reaction was initiated by adding refolded protein solutions at the indicated time intervals to the activity assay mixture. The amount of protein used in the assay was always 12 μg. The reaction was stopped after 5 min by adding 2 ml of 6 m GdnHCl and 100 μl of 10 mm 5,5′-dithiobis(2-nitrobenzoic acid) for color development, and absorbance was measured at 412 nm. For the above activity assay, a 1-ml quartz cuvette with a light path of 1 cm was used. The percentage of recovery of MSG activity in different samples was calculated after considering an equivalent amount of native MSG activity as 100%. Activity curves were plotted by fitting the data points into a best fit equation, and corresponding refolding rate constants were calculated.
Refolding Kinetics Experiments
Refolding kinetics of MSG was monitored using intrinsic tryptophan and ANS fluorescence of protein as probes. Refolding was initiated by manual mixing by rapid dilution of denatured protein in refolding buffer such that the final GdnHCl concentration was 0.1 m, and its refolding kinetics was followed. For refolding in the presence of GroEL, unfolded MSG was diluted in GroEL-containing buffer. The MSG and GroEL concentrations used were 0.25 and 1 μm, respectively, and the dead time of mixing was 10 s. Kinetics experiments were done in a Biologic MOS-450 optical system using a 1-cm-path length cuvette. Refolding kinetic traces were monitored by measurement of the change in tryptophan fluorescence at 340 ± 10 nm using a band pass filter (Asahi Spectra). The excitation wavelength was 295 nm. For ANS-monitored kinetics, refolding kinetic traces were monitored by measurement of the decrease in ANS fluorescence at 450 nm using a band pass filter (Asahi Spectra). The excitation wavelength was 350 nm. Folding kinetics of MSG in the absence and presence of GroEL was followed by diluting denatured MSG in refolding buffer containing ANS alone and both ANS and GroEL, respectively. Reactivation kinetics of GroEL-bound MSG was followed by addition of GroES and ATP in ANS-containing buffer. The dead time was 10 s. The final ANS concentration was 50 μm. MSG, GroEL, and GroES concentrations were 0.25, 1, and 2 μm, respectively. ATP was added at 5 mm concentration. Correction for background fluorescence caused by ANS in reactions lacking MSG was carried out.
Fluorescence spectra were recorded in a PerkinElmer Life Sciences LS 55 luminescence spectrometer using a 120-μl quartz cuvette with a path length of 1 cm. MSG-GroEL complex (I-G; see Fig. 9 for a description of the different intermediates) was formed as described above (MSG/GroEL, 0.25:1), and its tryptophan fluorescence spectra were recorded 10 min after GroEL addition at 25 °C. Reactivation of MSG was performed by GroES and ATP addition to GroEL-MSG complex. Intermediate IC-G was generated by stopping the reactivation of MSG 60 s after ATP addition by adding 10 mm EDTA. Unfolding of IC-G was carried out by incubating it with EDTA for 30 min at 30 °C. For tryptophan fluorescence, samples were excited at 295 nm with excitation and emission slit widths of 5 nm each. Background fluorescence of chemically identical samples lacking substrate protein was subtracted. The ratios of MSG/GroEL used in all experiments refer to 14-mers.
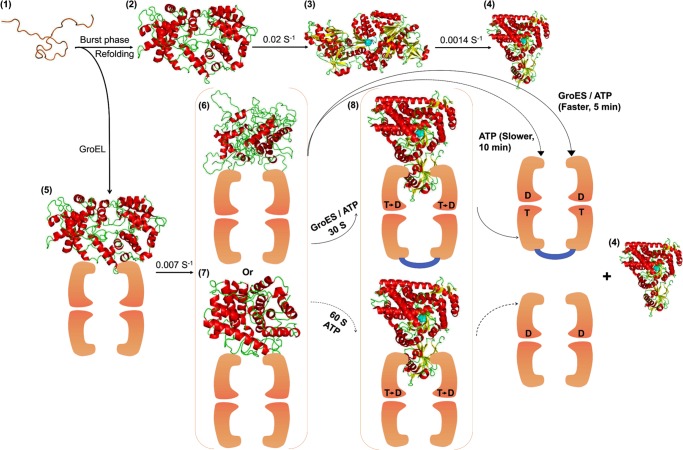
FIGURE 9.
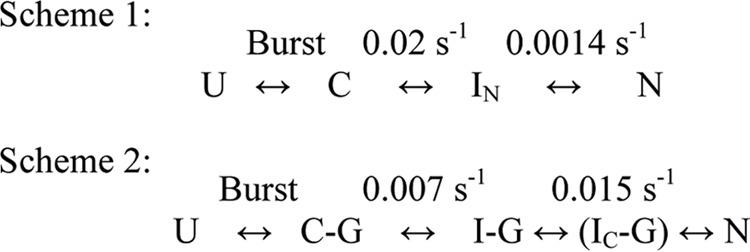
Folding mechanisms of MSG in the absence (Scheme 1) and presence (Scheme 2) of GroEL. Scheme 1 represents the mechanism of refolding of 0.25 μm MSG in 0.1 m GdnHCl and 20 mm Tris, pH 7.9, 300 mm NaCl, 10 mm MgSO4, 10 mm KCl, 1 mm tris(2-carboxyethyl)phosphine hydrochloride, 10% glycerol and is reproduced from previous work (40). Scheme 2 represents the mechanism of refolding of 0.25 μm MSG in the same buffer in the presence of 1 μm GroEL. U, unfolded MSG; C, burst phase intermediate; C-G, burst phase intermediate of MSG bound to GroEL; IN, functional intermediate of MSG having the same enzymatic activity and tryptophan fluorescence as native protein formed in spontaneous refolding; I-G, steady state GroEL-bound MSG; IC-G, compact intermediate bound to GroEL formed 30 s after GroES/ATP addition and 60 s after ATP addition to GroEL-MSG complex (I-G); N, native MSG.
Trypsin Digestion Experiments to Monitor Conformational Changes of MSG
Trypsin digestion assay was carried out to monitor conformational changes in MSG during chaperone-assisted refolding. GroEL-MSG complex was formed as above and divided into two parts. One part was treated with trypsin 60 s after ATP addition, whereas in the second part, no ATP was added before trypsin addition. ATP-mediated reactivation of MSG was stopped after 60 s by addition of 10 mm EDTA followed by its trypsin digestion. For sample without GroEL, trypsin was added 5 min after initiation of MSG refolding. Trypsin treatment was performed at 37 °C for 15 min using a 1:100 (w/w) ratio of trypsin to MSG. Native MSG was used as a control and treated with trypsin in the same way. The reaction was stopped by boiling the samples in SDS loading dye for 5 min. Band densities of MSG in different reactions were compared with each other using the band quantitation tool of the gel documentation unit taking native MSG band as reference. The relative quantity of MSG left after trypsin digestion was taken as a measure of proteolytic resistance to trypsin. Trypsin treatment was applied for 15 min as after that native MSG itself becomes sensitive to trypsin. The same amount of MSG was loaded in all the lanes so its corresponding band intensities in each of the lanes can be directly compared with each other.
RESULTS
GroEL Prevents Thermal Aggregation of MSG
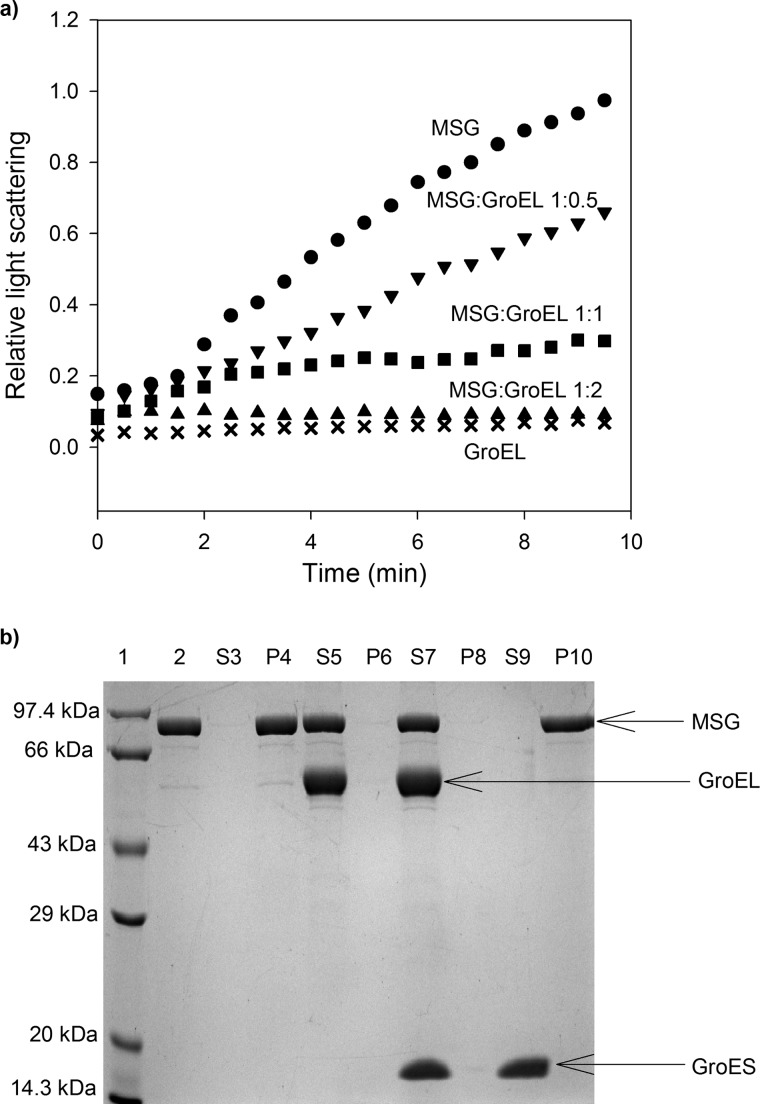
The effect of the presence of GroEL on the prevention of aggregation of MSG is shown in Fig. 1a. We observed that the thermal aggregation of MSG decreased with the addition of increasing amounts of GroEL, and there was complete suppression of aggregation in the presence of a 2-fold molar excess of GroEL over MSG. In a control experiment, GroEL did not show any increase in light scattering intensity when incubated at 55 °C, showing that GroEL is stable at this temperature (Fig. 1a). In Fig. 1b, supernatant and pellet fractions of heat-treated samples of MSG on SDS-PAGE are presented. It can be seen that when GroEL was present in a 2:1 molar ratio over the protein aggregation of MSG was completely prevented as it appears only in the supernatant (Fig. 1b, lanes 5 and 7). In the absence of GroEL, MSG was found exclusively in the pellet (Fig. 1b, lanes 3, 4, 9, and 10). GroES was not required to prevent aggregation of MSG (Fig. 1b, lanes 5–8), and it could not prevent protein aggregation (Fig. 1b, lanes 9 and 10) alone.
FIGURE 1.
Prevention of thermal aggregation of MSG by GroEL. Aggregation of MSG was monitored by light scattering and SDS-PAGE analysis of supernatant-pellet fractions. a, native MSG at a concentration of 1 μm was incubated in the absence (●) and in the presence of different molar ratios of GroEL in 20 mm Tris buffer, pH 7.8, 10 mm MgCl2, 10 mm KCl at 55 °C. ▾, 0.5:1 GroEL/MSG; ■, 1:1 GroEL/MSG; ▴, 2:1 GroEL/MSG; ×, GroEL only. Aggregation kinetics was monitored by light scattering at 500 nm and normalized with the highest value. b, SDS-PAGE showing supernatant (S)-pellet (P) fractions of heat-treated MSG with and without chaperones. MSG at a concentration of 1 μm was incubated with 2 μm GroEL at 55 °C for 10 min and centrifuged, and supernatant-pellet fractions were run on a 12% SDS gel after normalization. When present, GroES was added in a 2:1 molar ratio to GroEL. Lane 1, protein molecular mass marker; lane 2, native MSG; lane 3, supernatant fraction of heat-treated MSG in the absence of GroEL (S3); lane 4, pellet fraction of S3; lane 5, supernatant fraction of heat-treated MSG in the presence of GroEL (S5); lane 6, pellet fraction of S5; lane 7, supernatant fraction of heat-treated MSG in the presence of GroEL/ES (S7); lane 8, pellet fraction of S7; lane 9, supernatant fraction of heat-treated MSG in the presence of only GroES (S9); lane 10, pellet fraction of S9.
GroEL Forms Stable Complex with Non-native MSG
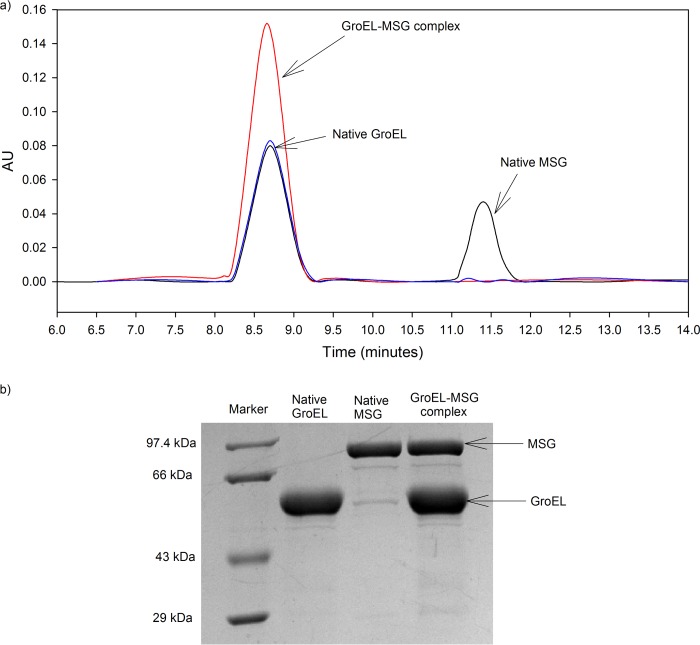
Native GroEL and native MSG were mixed in a 2:1 molar ratio (GroEL/MSG, 2:1) and run on a size exclusion column. Both eluted as separate peaks (Fig. 2a, shown in black) with native GroEL eluting at 8.70 min and native MSG eluting at 11.45 min. This shows that native MSG does not bind to GroEL. GroEL-MSG complex was generated by incubating native protein with GroEL in a 1:2 molar ratio at 55 °C for 10 min. The mixture was centrifuged at high speed to remove any aggregates present. The supernatant thus collected was vortexed vigorously and cooled to 4 °C before running on a size exclusion column in HPLC. If MSG does not form a stable complex with GroEL, it will be released from the complex upon vigorous shaking and cooling and elute as a separate peak. The elution profile of the supernatant after heating is shown in red in Fig. 2a. It can be seen that a single peak eluted at the native GroEL position, whereas no peak corresponding to native MSG was found. The GroEL peak fraction was collected, lyophilized, and run on a 10% SDS-polyacrylamide gel after resuspending it in SDS loading dye. Both MSG and GroEL were identified in the same lane (Fig. 2b, lane 4), indicating that non-native MSG forms a stable complex with GroEL. A slight shift in the elution time was observed from GroEL to GroEL-MSG complex, which elutes at 8.67 min. This could be due to a slight increase in the molecular mass of GroEL due to the presence of MSG. GroEL heated at 55 °C for 10 min was used as a control. It was found to elute at the same position as native GroEL and displayed a similar absorbance, showing that it was structurally stable at 55 °C (Fig. 2a, blue peak). It should be mentioned here that during thermal aggregation experiments the temperature varied from 55 to 4 °C, and as a result, the pH of the Tris buffer changed from 7 to 8.5. However, both MSG and GroEL were found to be structurally stable within this pH range at 25 °C (data not shown). Thus, pH changes do not contribute to the results obtained.
FIGURE 2.
GroEL forms stable binary complex with MSG. A binary complex of MSG and GroEL was formed as described under “Experimental Procedures.” a, gel filtration profiles monitored by absorbance measurements at 280 nm. AU, absorbance units. Native GroEL and native MSG elution peaks (black), GroEL preheated at 55 °C for 10 min (blue peak), and GroEL-MSG complex (red peak) are shown. GroEL and MSG concentration used were 2 and 1 μm, respectively. b, SDS-PAGE analysis of complex formation. Lane 1, medium range protein molecular mass marker (14.3–97.4 kDa); lane 2, native GroEL fraction; lane 3, native MSG fraction; lane 4, GroEL peak fraction (red peak).
GroEL/GroES Increase Rate of Recovery of Functional MSG
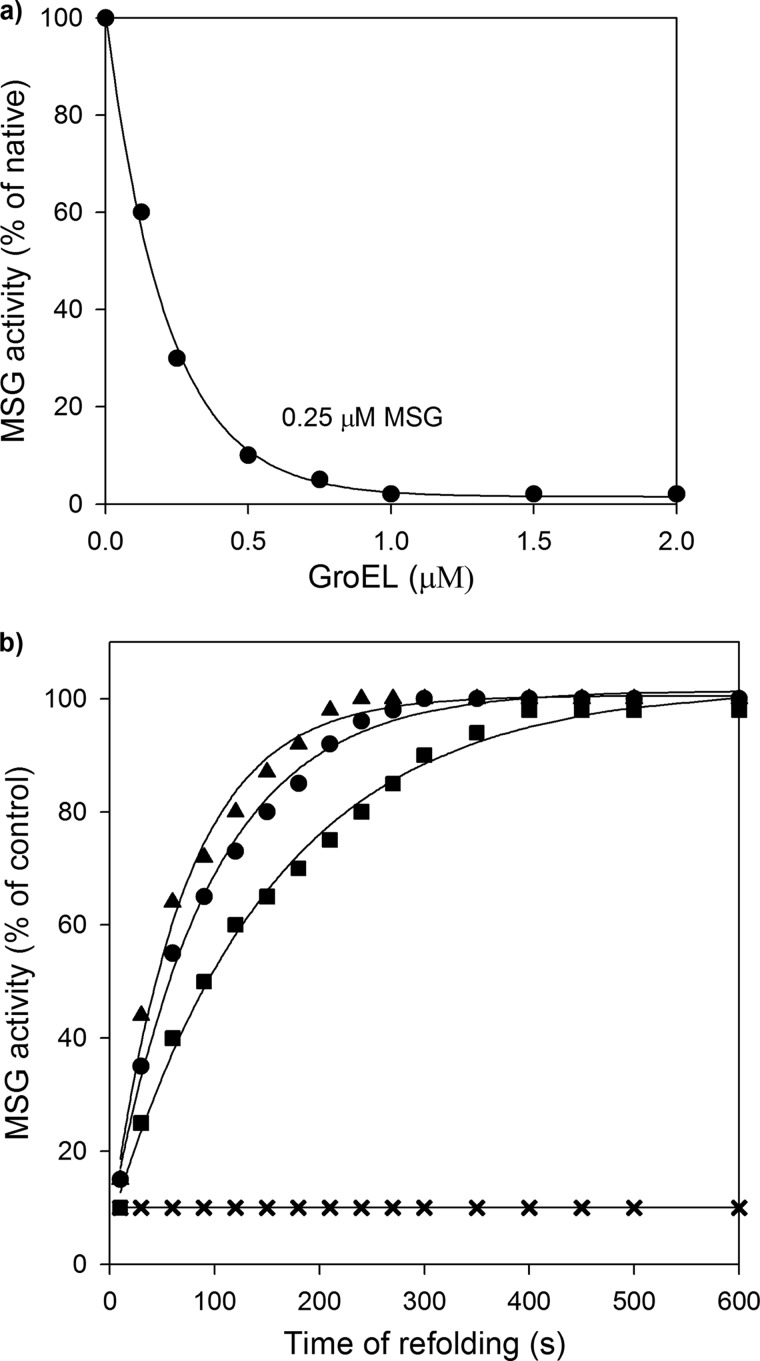
Our previous finding on the refolding reaction of MSG shows that GdnHCl-denatured MSG spontaneously refolds to 100% upon removal of denaturant (40). However, during the process, MSG accumulates slow folding intermediates, which make the process aggregation-prone at higher protein concentrations (40). Based on these observations, a possible role of GroEL was speculated in modulating the kinetics of MSG refolding. Hence, GroEL-assisted refolding of MSG was attempted, and GroEL-MSG binding was established by a simultaneous loss of enzymatic activity of MSG upon complex formation. MSG was unfolded in 3 m GdnHCl at 25 °C until it completely lost its activity. It was then diluted into GroEL-containing buffer such that the final concentration of MSG was 0.25 μm. The protein to GroEL ratio of 1:2 was maintained, which reduced MSG activity by 90% (Fig. 3a), indicating binding of MSG to GroEL during the refolding process. Unfolded MSG stably bound to GroEL and was released from the GroEL complex only after addition of ATP, which allowed refolding to occur with a rate of 0.006 s−1 (Fig. 3b). This was slower than the spontaneous refolding rate of MSG (k = 0.01 s−1). Addition of GroES to GroEL-MSG complex in a 2:1 molar ratio over GroEL accelerated MSG refolding (k = 0.015 s−1), making it slightly faster than spontaneous refolding. It can be seen that the GroES requirement for achieving complete reactivation of MSG from GroEL-MSG complex was not mandatory per se, but its presence doubled ATP-dependent refolding reaction rate (Fig. 3b). It is worth mentioning here that the 100% activity that was observed after 300 s in the case of spontaneous refolding corresponds to the functional intermediate of MSG (IN) and not to the native protein (40). As can be seen in Fig. 4, in a comparison of processes of formation of native MSG, GroEL/ES/ATP-assisted refolding was several times faster than spontaneous refolding.
FIGURE 3.
GroEL dependent reactivation of GdnHCl denatured MSG. MSG denatured in 3 m GdnHCl was diluted in refolding buffer containing 20 mm Tris, pH 7.9, 300 mm NaCl, 10 mm MgSO4, 10 mm KCl, 1 mm tris(2-carboxyethyl)phosphine hydrochloride, 10% glycerol in the presence and absence of GroEL such that the final GdnHCl concentration was 0.1 m. a, inhibition of spontaneous refolding by GroEL when MSG is refolded in the presence of different molar ratios of GroEL. b, time course of GroEL-, GroEL/GroES-, and GroEL/ATP-mediated refolding of MSG in vitro. MSG in 3 m GdnHCl was diluted in refolding buffer containing the indicated additions: GroEL (×), GroEL/ATP (■), GroEL/GroES/ATP (▴), and MSG only (●). At the indicated time intervals, aliquots from the refolding mixture were withdrawn and added into the activity assay mixture. Enzymatic activity was expressed as a percentage of what was obtained with an equivalent amount of native MSG in 0.1 m GdnHCl. Final MSG, GroEL, and GroES concentrations in the refolding buffer were 0.25, 0.5, and 1 μm, respectively. ATP was added at 2.5 mm. Refolding rate constants were calculated by fitting activity curves to the best fit equation. Enzymatic activity values shown are the average values from three separate experiments.
FIGURE 4.
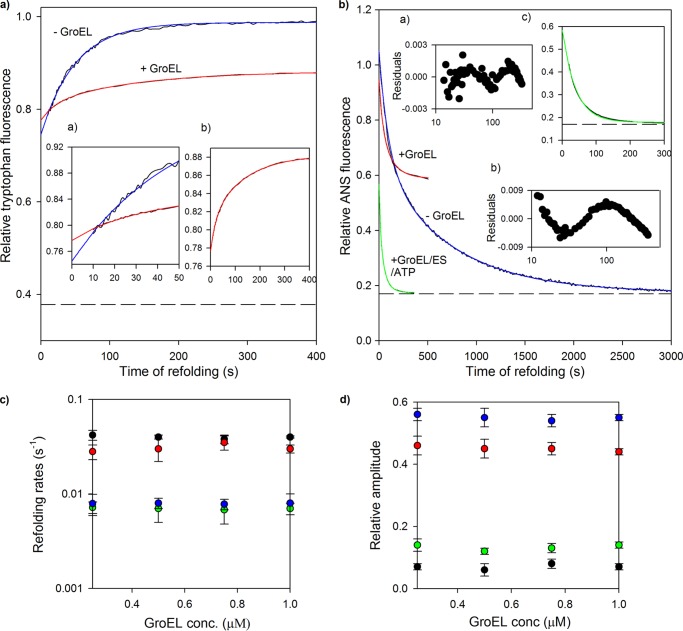
Folding kinetics of MSG in presence of GroEL. Refolding of MSG was monitored in the presence and absence of GroEL by intrinsic tryptophan and ANS fluorescence. MSG (0.25 μm) was refolded in the presence of 0 and 1 μm GroEL in the refolding buffer containing residual GdnHCl (0.1 m). a, refolding kinetic traces monitored by tryptophan fluorescence at 340 nm when protein was excited at 295 nm. Blue and red continuous lines represent single and double exponential fits of kinetic refolding traces of MSG in the absence and presence of GroEL, respectively. Inset a shows the first 50 s of refolding, and inset b shows the refolding kinetic trace of GroEL-bound MSG. The broken line represents fluorescence of unfolded MSG in 3 m GdnHCl. The fluorescence of the relevant concentration of GroEL was subtracted from the GroEL-containing trace, and all fluorescence values were then normalized to a value of 1 for the fluorescence of MSG at 400 s of refolding in the absence of GroEL after which no further change in tryptophan fluorescence of MSG takes place. b, ANS fluorescence-monitored refolding kinetics of MSG at 450 nm. Blue and red continuous lines represent double exponential fits of kinetic refolding traces of MSG in the absence and presence of GroEL, respectively. The blue curve was derived by diluting denatured MSG in buffer containing ANS, whereas for the red curve, ANS fluorescence was monitored with time after diluting denatured MSG in buffer containing both GroEL and ANS. The green curve depicts a double exponential fit of the reactivation of GroEL-bound MSG. Insets a and b show the residuals of the double and single exponential fits of the green curve. Reactivation kinetics of GroEL-bound MSG was followed by addition of GroES and ATP in GroEL- and ANS-containing buffer. Inset c shows the first 300 s of the reactivation kinetics of GroEL-bound MSG. The t = 0 value of the green trace represents the ANS fluorescence value of GroEL-bound MSG, which is the same as the final steady state value obtained from the red trace. The final ANS concentration was 50 μm. Fluorescence values were corrected for background fluorescence caused by ANS in reactions lacking MSG and normalized to a value of 1 for ANS fluorescence of MSG at t = 0 of refolding in the absence of GroEL. The dashed line represents ANS fluorescence of native MSG. Refolding was performed by manual mixing with a dead time of 10 s. Data for refolding of MSG in the absence of GroEL were reproduced from previous work (40) for comparison. c, tryptophan fluorescence-monitored refolding rate constants of fast phase (black circles) and slow phase (green circles) and ANS fluorescence-monitored refolding rate constants of fast phase (red circles) and slow phase (blue circles) of GroEL-assisted refolding of MSG obtained from the respective double exponential fits of refolding at different GroEL concentrations (conc.). d, effect of GroEL concentration on their corresponding relative amplitudes. Relative amplitudes of the fast and slow phases monitored by ANS fluorescence were calculated with respect to the total ANS fluorescence change observed for the red curve (i.e. relative to the sum of both fast and slow phases). For the tryptophan fluorescence change, relative amplitudes were plotted using the linearly extrapolated unfolding base line in a. Nearly 70% of the tryptophan fluorescence change occurs in the burst phase. In c and d, the error bars represent the spread of measurements made in three separate experiments.
Modulation of Refolding Kinetics of MSG by GroEL
Fig. 4, a and b, show the kinetic traces of folding of 0.25 μm MSG in the presence and absence of 1 μm GroEL (a 1:4 molar ratio of protein to GroEL was used to ensure that no MSG molecule escapes binding) monitored by tryptophan and ANS fluorescence spectroscopy. Previous studies on the refolding kinetics of MSG in the absence of GroEL have shown that the tryptophan fluorescence change accompanying refolding in the presence of 0.1 m residual GdnHCl occurs in two phases: a burst phase (∼70%) and a slow phase (30%) with an apparent rate constant of 0.02 s−1 (Fig. 4a, blue curve). The burst phase intermediate (C) leads to the formation of a functional intermediate (IN) in the slow phase that gives rise to native MSG in the very slow phase (0.0014 s−1) that has no detectable tryptophan fluorescence change (40). When MSG refolding was carried out in the presence of 1 μm GroEL, the following observations were made (Fig. 4a, red curve): 1) a decrease in the total amplitude of observable tryptophan fluorescence change, 2) a 10% increase in burst phase amplitude, 3) a decrease in the apparent rate constant of the slow phase (0.007 s−1), and 4) the appearance of a fast phase (0.04 s−1).
Fig. 4b shows the effect of GroEL on ANS fluorescence-monitored refolding kinetics of MSG. The hydrophobic molecule ANS binds to hydrophobic clusters on non-native protein species that are hydrated and therefore accessible (41), and such binding is accompanied by a large change in fluorescence emission of protein-bound ANS molecules. Folding of protein to the native state can be monitored by following ANS fluorescence, which decays as the protein folds due to release of bound ANS, with time. ANS did not interfere with the folding of MSG (data not shown) and did not bind to its native (N) or unfolded (U) states (Fig. 5b). Hence, ANS was used as a probe to investigate the native topology formation of MSG along the refolding time scale that is actually the rate-limiting step of the MSG folding pathway (40). ANS fluorescence-monitored refolding of MSG in the absence of GroEL consists of two kinetic phases: slow and very slow with corresponding rate constants of 0.012 and 0.0014 s−1, respectively (Fig. 4b, blue curve). When denatured MSG was diluted in the refolding buffer containing GroEL and ANS, ANS fluorescence of MSG was found to decay with time in two kinetic phases with rate constants of 0.03 and 0.008 s−1, respectively (Fig. 4b, red curve). Because the ANS-monitored rates in the presence of GroEL are similar to the tryptophan-monitored rates (0.04 and 0.007 s−1; Fig. 4a, red curve), both the probes are probably monitoring the same kinetic processes. The formation of the native topology of MSG after GroES and ATP addition to GroEL-MSG complex was further monitored and compared with the spontaneous refolding process. Reactivation of GroEL-bound MSG (I-G) occurred in two kinetic phases with rate constants of 0.06 and 0.018 s−1 (Fig. 4b, green curve), respectively. A double exponential equation was found to be a better fit of the GroEL/ES/ATP kinetic trace than a single exponential equation as shown by the residuals (Fig. 4b, insets a and b). This might represent two transitions of I-G to N. These results clearly shows that GroEL-bound MSG acquires native topology upon its release from the complex considerably faster than when MSG refolds free in solution. Quite remarkably, the rate of recovery of the native conformation of MSG in GroEL/GroES/ATP-assisted refolding was about 10 times faster than in unassisted refolding (0.0014 versus 0.018 s−1). ANS fluorescence in each case was normalized with the highest ANS fluorescence value of MSG, i.e. the value at t = 0 in spontaneous refolding, which represents the burst phase intermediate (C).
FIGURE 5.
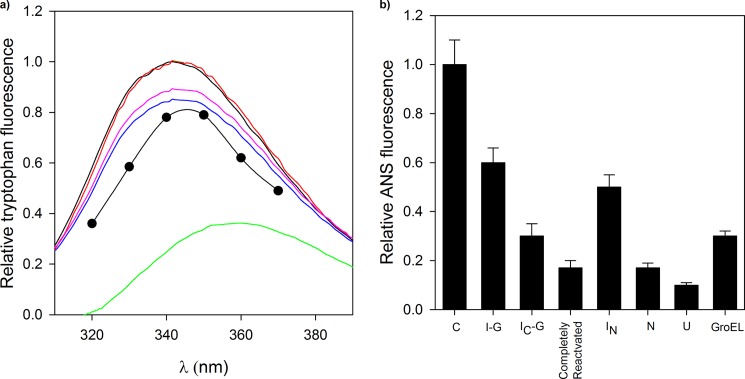
Monitoring conformational changes of MSG during refolding. Different intermediates of MSG formed in the presence and absence of GroEL were characterized by recording their intrinsic tryptophan and ANS fluorescence. a, tryptophan fluorescence spectra of different conformations of MSG. Unfolded MSG (U; green curve), GroEL-bound MSG (I-G; blue curve), reactivated MSG 30 s after GroES and ATP addition (IC-G; red curve), IC-G incubated with 10 mm EDTA for 30 min at 30 °C (pink curve), and native MSG (N; black curve) are shown. The fluorescence spectra of the burst phase intermediate (C; ●) is reproduced from previous work (40). I-G represents the steady state complex of MSG with GroEL. Background fluorescence of chemically identical reactions lacking substrate proteins (due to minor impurities of GroE preparation) was subtracted. b, histogram showing ANS fluorescence of different conformations of MSG. C and IN refer to the burst phase and functional intermediates, respectively, of MSG formed in spontaneous refolding as described in the text. Completely reactivated MSG is formed 5 min after GroES and ATP addition. The rest of the conformations are described in a. ANS fluorescence values of transient intermediates of MSG (C, IN, and IC-G) are derived from the refolding kinetic curves in Fig. 4b as described in the text. ANS fluorescence of GroEL-bound MSG (I-G), native (N) and unfolded MSG (U), and completely reactivated MSG were recorded by incubating them with 50 μm ANS for 5 min at 25 °C. Error bars represent the spread of measurements from three separate experiments.
GroEL Binds to the Burst Phase Intermediate of MSG and Alters Its Conformation
C appears to be a potential candidate for GroEL binding as it possesses high ANS binding capacity (Fig. 4b, blue curve). The tryptophan and ANS fluorescence changes with time that we observed in Fig. 4, a and b, suggest conformational changes in C upon GroEL binding because had it been only a GroEL binding effect the kinetics of overall tryptophan and ANS fluorescence change should not be so slow (∼0.007 s−1). However, it is possible that GroEL is not binding to the burst phase intermediate of MSG as speculated but to some of its late refolding intermediates, and the fluorescence rates we derived from Fig. 4, a and b (red curves), are actually rates of binding instead of rates of refolding. To exclude this possibility, tryptophan and ANS fluorescence kinetics of MSG were monitored in the presence of different GroEL concentrations. At all GroEL concentrations tested, the kinetic curves fitted to the double exponential equation (data not shown), and the respective rates were calculated. Fig. 4c shows that both the tryptophan- and ANS fluorescence-monitored rates of the fast and slow phases did not depend on GroEL concentration. Moreover, their corresponding relative amplitudes were also found to be independent of GroEL concentration (Fig. 4d), which means that there does not appear to be any fluorescence change associated with the binding of GroEL to MSG. These observations suggest that what we detected through fluorescence changes in Fig. 4, a and b, was not a GroEL-MSG binding event but a GroEL-assisted refolding event, implying conformational changes in the GroEL-bound form of MSG, i.e. the burst phase intermediate. Here, the experiments could be carried out only at stoichiometric or higher ratios of GroEL to ensure that the majority of the MSG molecules were in the GroEL-bound state. It can be seen that when MSG was refolded in the presence of a substoichiometric ratio of GroEL (MSG/GroEL, 0.25:0.125 μm) nearly 60% of its enzymatic activity persisted (Fig. 3a), which implies that the majority of the MSG molecules remain in the unbound state at lower GroEL concentrations. Complete activity of MSG disappeared only at higher GroEL concentrations (Fig. 3a). Thus, to ensure that unbound MSG molecules did not interfere with the refolding kinetics of GroEL-bound MSG and with the interpretation of kinetics results, higher GroEL concentrations were required in Fig. 4, c and d.
GroEL Assists MSG Refolding through Formation of a Compact Intermediate
To further explore whether the increase in refolding rate constants observed for chaperone-assisted refolding reflects a different folding trajectory adopted by MSG in the presence of GroEL, folding intermediates on the two pathways were characterized by tryptophan and ANS fluorescence, enzymatic activity, and trypsin digestion. Fig. 5a shows tryptophan fluorescence spectra of various conformations of MSG. GroEL-bound MSG (I-G; blue curve) has a tryptophan fluorescence intensity quite close to that of native (black curve) with its λmax emission the same as that of native MSG. The spectrum was taken after 10 min of dilution of denatured MSG in GroEL-containing buffer. However, I-G possessed strong ANS fluorescence as compared with native (N) or unfolded MSG (U) (Fig. 5b). The finding that the λmax emission for tryptophan fluorescence of C (Fig. 5a, black circles) shifted from 345 to 340 nm upon GroEL binding (I-G; Fig. 5a, blue curve) further supports our claim that the conformational state of the initial and final GroEL-bound forms of MSG are different. Reactivation of I-G was initiated by either ATP or GroES/ATP addition. Within 30 s of GroES/ATP addition to GroEL-MSG complex, the level of fluorescence intensity reached that of the native protein (Fig. 5a, red curve), and ANS fluorescence decreased by 50%, defining the folding intermediate IC-G (Fig. 5b). In the case of ATP, the above event happened within 60 s of ATP addition. Complete reactivation of GroEL-bound MSG happened only about 5 min after GroES/ATP addition and 10 min after ATP addition, respectively, and was accompanied by the recovery of native-like ANS fluorescence (Fig. 5b) and enzymatic activity (Fig. 3b). ANS fluorescence values of transient intermediates in Fig. 5b were derived from the ANS fluorescence-monitored refolding kinetic traces of MSG in Fig. 4b. The t = 0 and t = 300 s points of the kinetic curve obtained in the absence of GroEL (Fig. 4b, blue curve) represent the ANS fluorescence values of C and IN, respectively, in Fig. 5b. Similarly, the t = 30 s point of the kinetic curve obtained in the presence of GroEL/GroES/ATP (Fig. 4b, green curve) represents the ANS fluorescence value of IC-G.
Unlike the completely reactivated form, IC-G was still associated with GroEL. IC-G was generated by adding ATP to GroEL-MSG complex, and the reaction was stopped after 60 s by adding the Mg2+ chelator EDTA. When the reaction containing IC-G and EDTA was maintained at 30 °C for 30 min, there was partial unfolding as indicated by the decrease in tryptophan fluorescence intensity (Fig. 5a, pink curve). Addition of sufficient Mg2+ to compensate for the EDTA present reinitiated the folding reaction (data not shown). This indicates that IC-G was still bound to GroEL as no unfolding was observed under the same conditions for the reaction without EDTA that was used as a control.
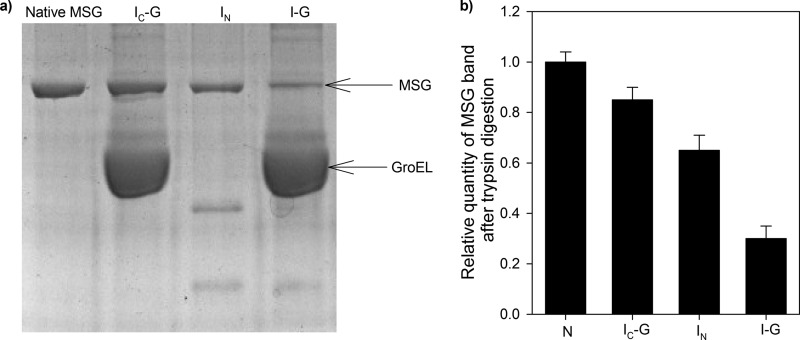
The observation that formation of IC-G from I-G was accompanied by an increase in tryptophan fluorescence (Fig. 5a) and a decrease in ANS fluorescence (Fig. 5b) indicates compaction of I-G to IC-G. This was confirmed by improved resistance of IC-G to trypsin (Fig. 6). IC-G was generated in the same way as described in the previous section. Quite remarkably, IC-G (Fig. 6a, lane 2) was the most resistant toward trypsin digestion among all conformations, whereas I-G was found to be the most sensitive (Fig. 6a, lane 4), followed by IN (Fig. 6a, lane 3). Fig. 6b shows the relative quantities of MSG left after trypsin digestion in each case. Resistance to proteolytic digestion was measured in terms of density of the MSG band in a trypsin-treated sample. Equal amounts of MSG were loaded in the wells to enable direct comparison in each case. Here, extreme care was taken to avoid any error in the loading amounts. Besides differences in the relative quantities of MSG, differences in the digestion pattern of various intermediates were also observed (Fig. 6a). Apparently, folding of GroEL-bound MSG (I-G) to the native state occurs via formation of a compact intermediate (IC-G) on the GroEL surface as also predicted by ANS fluorescence-monitored reactivation kinetics (Fig. 4b, green curve) of GroEL-bound MSG that suggests that there might be two transitions from I-G to native MSG formation.
FIGURE 6.
Trypsin digestion assay. Various conformations of MSG were tested by trypsin digestion assay. GroEL-MSG complex was prepared by diluting denatured MSG in GroEL-containing buffer in a 1:4 ratio to GroEL and incubating for 10 min at 25 °C and then dividing it into two parts. The first sample was treated with trypsin 60 s after ATP addition, and in the second sample, no ATP was added before trypsin addition. ATP-mediated reactivation of MSG was stopped after 60 s (IC-G) by addition of 10 mm EDTA following which trypsin was added. For sample without GroEL, trypsin was added 5 min after initiation of refolding (IN). Trypsin treatment was performed at 37 °C for 15 min using a 1:100 (w/w) ratio of trypsin to MSG. Native MSG was used as a control and was treated with trypsin in the same way. Trypsin digestion was stopped by boiling the samples in SDS loading dye for 5 min. a, 10% SDS-PAGE gel showing trypsin-digested mixtures. Lane 1, native MSG; lane 2, IC-G; lane 3, IN; lane 4, GroEL-bound MSG (I-G). b, histogram showing relative quantities of MSG band in each case. Band density was taken as a measure of proteolytic resistance of MSG. The same amount of MSG was loaded in all the lanes so its corresponding band intensities in each of the lanes can be directly compared with each other. Error bars represent the spread of measurements from three separate experiments.
GroES Binds in trans to MSG and Accelerates Its Refolding
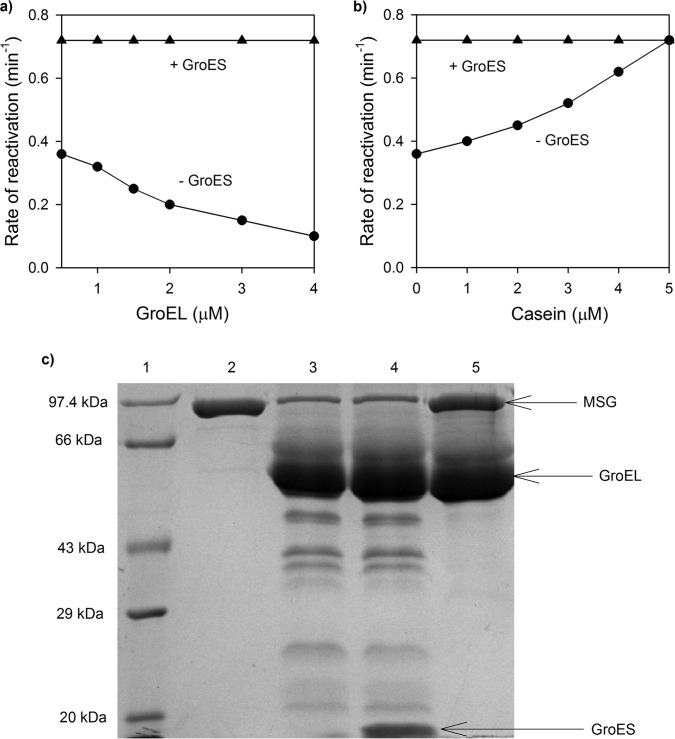
To test whether the slow rate of ATP-dependent reactivation observed in Fig. 3b is due to rebinding of non-native MSG to GroEL after ATP-dependent release, the concentration of GroEL in the refolding mixture was increased while keeping the MSG concentration the same (i.e. at 0.25 μm). The rate of ATP-dependent reactivation of MSG was retarded (Fig. 7a). This indicates that, after release, MSG could rebind to GroEL before folding is complete. To confirm this, αS1-casein was used as a competitor of protein binding by GroEL. Casein has certain properties of partially denatured protein. In its native form, it exposes a considerable portion of hydrophobic residues to the solvent and contains a high amount of disordered structure (42, 43). The rate of reactivation of MSG increased with an increase in casein concentration (Fig. 7b). Casein bound to GroEL when present in excess and thus prevented MSG rebinding to it. Hence, the ATP-dependent reactivation rate of MSG was increased. Interestingly, retardation of the reactivation rate of MSG due to rebinding was not observed when GroES was present in a 2:1 molar ratio to GroEL (Fig. 7a). Also, there was no effect of casein on the MSG reactivation rate in the presence of GroES (Fig. 7b). Thus, it appears that only when GroES is absent does MSG undergo multiple cycles of GroEL binding and ATP-dependent release before its folding is finally complete. GroES binding couples ATP hydrolysis with folding at GroEL by coordinating among protein binding regions of different subunits of GroEL. This action of GroES prevents release of complete protein in a single step and allows it to organize to a more compact structure before rebinding occurs (43). Hence, in the presence of GroES, MSG is forced into a folding pathway that involves staged release during a single round of GroEL interaction, which did not occur in the presence of only ATP. Moreover, addition of GroES and ADP to GroEL-MSG complex was unable to protect MSG from proteolytic digestion, confirming that GroES binds in trans to MSG (Fig. 7c). This shows that GroES accelerates refolding of MSG without encapsulating it inside the GroEL cavity.
FIGURE 7.
Effect of GroES on reactivation rate of GroEL bound MSG. MSG reactivation was monitored by measuring regain of enzymatic activity by MSG upon GroES/ATP addition. a, influence of increasing GroEL concentration on the MSG reactivation rate in the presence and absence of GroES. GroEL-MSG complex was generated as described previously (GroEL/MSG, 0.5:0.25 μm). After 10 min, increasing amounts of GroEL (total concentrations of GroEL are indicated) were added in the presence (▴) or absence (●) of GroES. GroES was always added in a 2:1 molar ratio to GroEL. Reactivation was initiated by adding ATP. b, effect of casein on MSG reactivation rates. Casein (0–5 μm) was added 5 min after GroES addition (1 μm) in GroES-containing reactions. After further incubation for 5 min, ATP (2.5 mm) was added. Rates in each case were calculated from their corresponding activity curves just like rates in Fig. 3b. c, SDS-PAGE showing that GroES does not encapsulate GroEL-bound MSG. GroES was added in a 2-fold molar excess to GroEL-MSG complex (MSG/GroEL/GroES, 1:2:4 μm) in the presence of ADP followed by trypsin digestion after which samples were run on a 10% SDS-polyacrylamide gel. Trypsin treatment was performed in the same way as described previously. Lane 1, medium range protein molecular mass marker; lane 2, trypsin-treated native MSG used as control; lane 3, GroEL-MSG complex treated with trypsin; lane 4, GroEL-MSG-GroES and ADP treated with trypsin; lane 5, GroEL-MSG complex without trypsin.
DISCUSSION
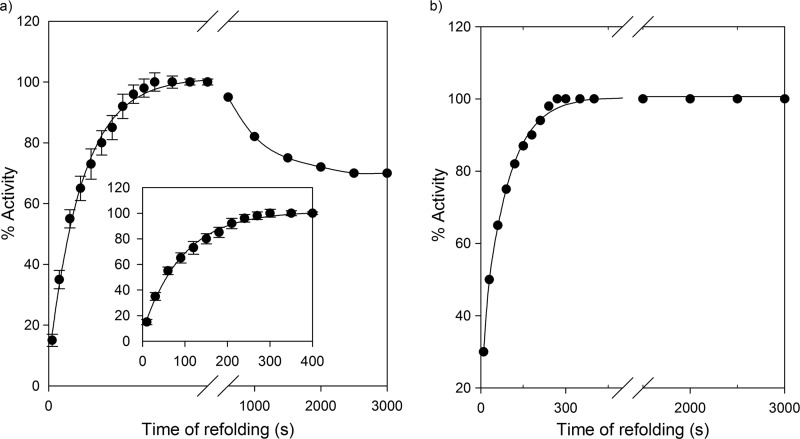
Despite the fact that MSG displays reversible unfolding in vitro in the absence of chaperones (40), its refolding is prone to aggregation at higher protein concentrations (i.e. > 0.5 μm) (40). Fig. 8 shows a comparison of spontaneous and GroEL-assisted refolding of MSG at a final MSG concentration of 1 μm. A notable phenomenon observed in the GroEL-assisted refolding of MSG is that the loss of 25–30% of the enzymatic activity with time observed in spontaneous refolding (Fig. 8a) did not occur in the presence of GroEL, which completely prevented MSG aggregation even at higher protein concentrations (Fig. 8b). This effect was specifically due to elimination of the slowest step of MSG refolding in the presence of GroEL/GroES (Fig. 9, Scheme 1 versus Scheme 2).
FIGURE 8.
GroEL/ES inhibit aggregation during refolding of GdnHCl denatured MSG. Refolding of MSG was carried out such that its final concentration was 1 μm in the presence and absence of 2 μm GroEL and monitored by enzymatic activity as described under “Experimental Procedures.” The final GdnHCl concentration in refolding buffer was kept at 0.1 m. The indicated time points correspond to the time after which aliquots from the refolding mixture were added to the activity assay mixture. a, spontaneous refolding of MSG. Data were reproduced from previous work (40) for comparison. Error bars represent measurements from three separate experiments. b, GroEL/ES-assisted refolding showing no loss in enzymatic activity with time. The black continuous line is the single exponential fit to data points.
The conformational change of GroEL-bound MSG observed in Fig. 4 could be a consequence of either folding or unfolding of MSG on the GroEL surface. With the current set of experiments, however, the two events could not be distinguished. Still, the observations of enhanced sensitivity toward trypsin digestion (Fig. 6) and enzymatically inactive state of I-G (Fig. 3b) favor the unfolding event more than the folding event. In this context, it would have been helpful if trypsin sensitivities of C and I-G could be compared, but the transient nature of C precluded this comparison. It may be speculated that because MSG is a multidomain protein only a very small portion of it would have actually bound to the GroEL cavity with most of it lying in the bulk solution. This could have resulted in localized unfolding in the GroEL-bound regions of the protein. However, additional experiments such as FRET are required to validate this. Nevertheless, it was assumed that GroEL binding led to the unfolding of C as this unfolding step may be important in transforming the protein for the subsequent accelerated folding process.
The burst phase intermediate of MSG (C) consists of some non-native secondary structure, the rearrangement of which continues until the formation of the native state (40). Binding to GroEL helps the slow folding collapsed intermediate C to adopt a more folding-compatible state (I-G) probably by causing disruption of non-native interactions in C that might slow down its folding to the native state (Fig. 9, Scheme 1). Once I-G is formed, it folds easily to the native state N (Fig. 9, Scheme 1 versus Scheme 2). It thus appears that GroEL is playing an active role in the folding of MSG by binding to its on-pathway intermediate C and causing reorganization of non-native interactions, which dramatically improved its rate of folding upon GroES/ATP addition. Another prominent effect of GroEL-assisted refolding of MSG is that the GroEL/GroES/ATP-dependent rate of reactivation of MSG obtained from the activity assay (0.015 s−1; Fig. 3b) and the ANS-monitored refolding rate that is reflective of native topology formation of MSG (0.018 s−1; Fig. 4b, green curve) are nearly the same. Based on the similar rates of the two processes, it can be inferred that the complete recovery of enzymatic activity occurs simultaneously with the native topology formation of MSG in the presence of the GroEL/GroES/ATP system in contrast to spontaneous refolding where the two processes are not simultaneous (Figs. 3b and 4b, blue curve).
The difference in spontaneous and GroEL/ES-assisted rates of refolding of MSG can be attributed to different folding trajectories adopted by MSG in the presence and absence of GroEL. Table 1 shows a comparison between different folding intermediates of MSG formed during its spontaneous and GroEL-assisted refolding based on intrinsic tryptophan and ANS fluorescence measurements, enzymatic activity, and trypsin sensitivity. The I-G and IC-G type intermediates are not formed when MSG is folded free in solution. Similarly, the IN type intermediate is not formed in the GroEL-assisted folding pathway of MSG. The observation that 40% of MSG activity was recovered within 30 s of GroES and ATP addition (Fig. 3b) suggests that IC-G must be enzymatically active. It can be seen from Table 1 that IC-G is structurally quite close to the native state of MSG and is the most compact among all the intermediates. Such intermediates are quite significant and have been reported earlier in the GroEL/ES-assisted folding pathways of dihydrofolate reductase and rhodanese (43). During protein folding, non-local interactions are important in stabilizing native structures of proteins and in making important energetic decisions (44). Hence, formation of a compact, highly restricted ensemble will drive the chain to form favorable non-local interactions with minimum energetic barriers, thus making the process of attainment of native structure faster. Because the entropic penalty for establishing long range interactions is large, the acceleration of the folding rate is predicted to be more pronounced for proteins with a high proportion of long range tertiary contacts (45) such as the GroEL-dependent proteins with complex α/β or α + β domain topologies (46). The rate-limiting step in the MSG folding pathway was predicted to be the result of folding of its two domains (α + β; which do not form the active site), besides cis/trans proline isomerization (40). Most likely, chaperone action might have facilitated both domain folding and proline isomerization events in MSG through formation of more folding-compatible intermediates.
TABLE 1.
Comparison of different intermediates formed during spontaneous and GroEL-assisted refolding
Refolding of MSG was carried out in 20 mm Tris, pH 7.9, 300 mm NaCl, 10 mm MgSO4, 10 mm KCl, 1 mm TCEP HCl, 10% glycerol in the absence and presence of 1 μm GroEL such that final GdnHCl and MSG concentrations were 0.1 m and 0.25 μm, respectively. A description of the different intermediates is given in Fig. 9.
| Properties | I-G | IC-G | IN | N |
|---|---|---|---|---|
| Tryptophan fluorescence | 0.88 | 1 | 1 | 1 |
| ANS fluorescence | 0.6 | 0.3 | 0.5 | 0.17 |
| Enzymatic activity | Not active | Active | Active | Active |
| Trypsin resistance (%) | 30 | 85 | 65 | 100 |
One of the important questions here is whether the concepts established by our study also apply to other large, multidomain proteins. To gain further insights into that question, a large protein is needed for which the spontaneous folding pathway is well characterized so that it is possible to monitor the effect of GroEL on the conformations of its intermediates and the folding process as such. However, because of the lack of information on multidomain protein folding and the unavailability of their folding pathways, a comparison of assisted and unassisted folding processes of large proteins has not been explored in details so far. Moreover, non-native intermediates of most of the large proteins are aggregation-prone and difficult to accumulate, making it extremely difficult to perform such studies. Few such studies have been performed with proteins that are either not authentic GroEL substrates (38), do not have an intrinsic tendency to aggregate (38, 47, 48), or are small sized proteins undergoing the cis mechanism of folding (39, 47, 48). In this context, the present study perhaps provides a novel comparison between the spontaneous and chaperone-assisted folding of a truly aggregation-sensitive multidomain protein that is unique in its category. To date, the only significant information we have on the GroEL-assisted folding of large proteins is that they undergo the trans mechanism of folding to reach to their native state (12, 13). However, what transpires to the substrate protein during this process is unprecedented. Thus, the present study on the changing conformational properties of a large protein along the chaperone-assisted folding pathway advances our current knowledge of GroEL-mediated folding of large proteins. Moreover, this type of study can perhaps open the window to explore interactions between folding intermediates and GroEL for elucidating the chaperone-assisted folding pathways of multidomain proteins. In conclusion, GroEL/GroES accelerate refolding of MSG without encapsulation by apparently changing the folding mechanism, which is reflected through modification of its folding intermediates. To summarize our findings, we propose a model for GroEL/GroES-assisted folding of MSG in Fig. 10.
FIGURE 10.
GroEL/GroES-assisted folding model of MSG. The burst phase intermediate of MSG, C (2), is captured by GroEL (orange colored) upon dilution from GdnHCl-denatured MSG (1) to form GroEL-MSG complex, C-G (5). This binding induces minor structural rearrangements in C-G at a slow rate to give rise to a more folding-compatible state, I-G (6 and 7); I-G could be more unfolded (6) or more folded (7) than C-G. Further addition of GroES/ATP or ATP releases the GroEL-bound form of MSG (I-G), which folds to the native state (4) via formation of a compact intermediate, IC-G (8), that is structurally quite close to the native MSG. GroES (shown in blue) binds in trans to the folding polypeptide and doubles the ATP-dependent reactivation rate. Spontaneous refolding (1–4) proceeds through the functional intermediate, IN (3), of which conversion to the native MSG (4) is the slowest step in the MSG refolding pathway. GroEL-mediated folding averts this slowest kinetic phase by channeling the burst phase intermediate of MSG, C (2), to a different folding route (5–8). T and D represent ATP and ADP, respectively. The active site is depicted in cyan. The intermediate IC-G (8) is the most compact of all the intermediates.
Acknowledgments
We are extremely grateful to Prof. Jayant B. Udgaonkar, National Centre for Biological Sciences, Bangalore, India for helping with fluorescence kinetics experiments on the Biologic MOS-450 optical system and for invaluable assistance and advice. We gratefully acknowledge Aditya Padhi, School of Biological Sciences, Indian Institute of Technology Delhi for help in giving the beautiful shape to Fig. 10. Jon Tally, Department of Biochemistry and Molecular Biology, Kansas University Medical Center is also acknowledged for reading the manuscript carefully and providing valuable comments.
Footnotes
- MSG
- malate synthase G
- ANS
- 1-anilinonaphthalene sulfonate
- GdnHCl
- guanidine hydrochloride.
REFERENCES
- 1. Fenton W. A., Horwich A. L. (1997) GroEL-mediated protein folding. Protein Sci. 6, 743–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horwich A. L., Farr G. W., Fenton W. A. (2006) GroEL-GroES-mediated protein folding. Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 3. Lin Z., Rye H. S. (2006) GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 41, 211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 5. Noivirt-Brik O., Unger R., Horovitz A. (2007) Low folding propensity and high translation efficiency distinguish in vivo substrates of GroEL from other Escherichia coli protein. Bioinformatics 23, 3276–3279 [DOI] [PubMed] [Google Scholar]
- 6. Houry W. A., Frishman D., Eckerskorn C., Lottspeich F., Hartl F. U. (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402, 147–154 [DOI] [PubMed] [Google Scholar]
- 7. Viitanen P. V., Gatenby A. A., Lorimer G. H. (1992) Purified chaperonin 60 (groEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci. 1, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen L., Sigler P. B. (1999) The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell 99, 757–768 [DOI] [PubMed] [Google Scholar]
- 9. Ewalt K. L., Hendrick J. P., Houry W. A., Hartl F. U. (1997) In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90, 491–500 [DOI] [PubMed] [Google Scholar]
- 10. Lau C. K., Churchich J. E. (1999) Binding of polylysine to GroEL. Inhibition of the refolding of mMDH. Biochim. Biophys. Acta 1431, 282–289 [DOI] [PubMed] [Google Scholar]
- 11. Sigler P. B., Xu Z., Rye H. S., Burston S. G., Fenton W. A., Horwich A. L. (1998) Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 67, 581–608 [DOI] [PubMed] [Google Scholar]
- 12. Chaudhuri T. K., Farr G. W., Fenton W. A., Rospert S., Horwich A. L. (2001) GroEL/GroES-mediated folding of a protein too large to be encapsulated. Cell 107, 235–246 [DOI] [PubMed] [Google Scholar]
- 13. Paul S., Singh C., Mishra S., Chaudhuri T. K. (2007) The 69 kDa Escherichia coli maltodextrin glucosidase does not get encapsulated underneath GroES and folds through trans mechanism during GroEL/GroES-assisted folding. FASEB J. 21, 2874–2885 [DOI] [PubMed] [Google Scholar]
- 14. Weissman J. S., Rye H. S., Fenton W. A., Beechem J. M., Horwich A. L. (1996) Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell 84, 481–490 [DOI] [PubMed] [Google Scholar]
- 15. Hayer-Hartl M. K., Weber F., Hartl F. U. (1996) Mechanism of chaperonin action: GroES binding and release can drive GroEL-mediated protein folding in the absence of ATP-hydrolysis. EMBO J. 15, 6111–6121 [PMC free article] [PubMed] [Google Scholar]
- 16. Rye H. S., Burston S. G., Fenton W. A., Beechem J. M., Xu Z., Sigler P. B., Horwich A. L. (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 388, 792–798 [DOI] [PubMed] [Google Scholar]
- 17. Rye H. S., Roseman A. M., Chen S., Furtak K., Fenton W. A., Saibil H. R., Horwich A. L. (1999) GroEL-GroES cycling: ATP and non-native polypeptide direct alternation of folding-active rings. Cell 97, 325–338 [DOI] [PubMed] [Google Scholar]
- 18. Todd M. J., Lorimer G. H., Thirumalai D. (1996) Chaperonin-facilitated protein folding: optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 4030–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gulukota K., Wolynes P. G. (1994) Statistical mechanics of kinetic proofreading in protein folding in vivo. Proc. Natl. Acad. Sci. U.S.A. 91, 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bhutani N., Udgaonkar J. B. (2001) GroEL channels the folding of thioredoxin along one kinetic route. J. Mol. Biol. 314, 1167–1179 [DOI] [PubMed] [Google Scholar]
- 21. Weissman J. S., Kashi Y., Fenton W. A., Horwich A. L. (1994) GroEL-mediated protein folding proceeds by multiple rounds of binding and release. Cell 78, 693–702 [DOI] [PubMed] [Google Scholar]
- 22. Zahn R., Perrett S., Fersht A. R. (1996) Conformational states bound by the molecular chaperones GroEL and SecB: a hidden unfolding (annealing) activity. J. Mol. Biol. 261, 43–61 [DOI] [PubMed] [Google Scholar]
- 23. Zahn R., Axmann S. E., Rücknagel K. P., Jaeger E., Laminet A. A., Plückthun A. (1994) Thermodynamic partitioning model for hydrophobic binding of polypeptides by GroEL: I. GroEL recognizes the signal sequences of β-lactamase precursor. J. Mol. Biol. 242, 150–164 [DOI] [PubMed] [Google Scholar]
- 24. Walter S., Lorimer G. H., Schmid F. X. (1996) A thermodynamic coupling mechanism for GroEL mediated unfolding. Proc. Natl. Acad. Sci. U.S.A. 93, 9425–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhutani N., Udgaonkar J. B. (2000) A thermodynamic coupling mechanism can explain the GroEL-mediated acceleration of the folding of barstar. J. Mol. Biol. 297, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 26. Lilie H., Buchner J. (1995) Interaction of GroEL with a highly structured folding intermediate: iterative binding cycles do not involve unfolding. Proc. Natl. Acad. Sci. U.S.A. 92, 8100–8104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gervasoni P., Staudenmann W., James P., Gehrig P., Plückthun A. (1996) β-Lactamase binds to GroEL in a conformation highly protected against hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. U.S.A. 93, 12189–12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chuang J. L., Wynn R. M., Song J. L., Chuang D. T. (1999) GroEL/GroES-dependent reconstitution of α2β2 tetramers of human mitochondrial branched chain α-ketoacid decarboxylase. J. Biol. Chem. 274, 10395–10404 [DOI] [PubMed] [Google Scholar]
- 29. Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. (1989) Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature 342, 884–889 [DOI] [PubMed] [Google Scholar]
- 30. Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F. X., Kiefhaber T. (1991) GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry 30, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 31. Ranson N. A., Burston S. G., Clarke A. R. (1997) Binding, encapsulation and ejection: substrate dynamics during a chaperonin-assisted folding reaction. J. Mol. Biol. 266, 656–664 [DOI] [PubMed] [Google Scholar]
- 32. Brunschier R., Danner M., Seckler R. (1993) Interactions of phage P22 tailspike protein with GroE molecular chaperones during refolding in vitro. J. Biol. Chem. 268, 2767–2772 [PubMed] [Google Scholar]
- 33. Mattingly J. R., Jr., Iriarte A., Martinez-Carrion M. (1995) Homologous proteins with different affinities for groEL. J. Biol. Chem. 270, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 34. Thirumalai D., Lorimer G. H. (2001) Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct. 30, 245–269 [DOI] [PubMed] [Google Scholar]
- 35. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 36. Saibil H. R., Ranson N. A. (2002) The chaperonin folding machine. Trends Biochem. Sci. 27, 627–632 [DOI] [PubMed] [Google Scholar]
- 37. Fenton W. A., Horwich A. L. (2003) Chaperonin-mediated protein folding: fate of substrate polypeptide. Q. Rev. Biophys. 36, 229–256 [DOI] [PubMed] [Google Scholar]
- 38. Coyle J. E., Texter F. L., Ashcroft A. E., Masselos D., Robinson C. V., Radford S. E. (1999) GroEL accelerates the refolding of hen lysozyme without changing its folding mechanism. Nat. Struct. Biol. 6, 683–690 [DOI] [PubMed] [Google Scholar]
- 39. Horst R., Fenton W. A., Englander S. W., Wüthrich K., Horwich A. L. (2007) Folding trajectories of human dihydrofolate reductase inside the GroEL-GroES chaperonin cavity and free in solution. Proc. Natl. Acad. Sci. U.S.A. 104, 20788–20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dahiya V., Chaudhuri T. K. (2013) Functional intermediate in the refolding pathway of a large and multi-domain protein malate synthase G. Biochemistry 52, 4517–4530 [DOI] [PubMed] [Google Scholar]
- 41. Stryer L. (1965) The interaction of naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 42. Waxman L., Goldberg A. L. (1986) Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La). Science 232, 500–503 [DOI] [PubMed] [Google Scholar]
- 43. Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. (1991) Chaperonin-mediated protein folding occurs at the surface of groEL via a molten globule-like intermediate. Nature 352, 36–42 [DOI] [PubMed] [Google Scholar]
- 44. Dill K. A. (1990) Dominant forces in protein folding. Biochemistry 29, 7133–7155 [DOI] [PubMed] [Google Scholar]
- 45. Takagi F., Koga N., Takada S. (2003) How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: molecular simulations. Proc. Natl. Acad. Sci. U.S.A. 100, 11367–11372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 47. Tang Y. C., Chang H. C., Roeben A., Wischnewski D., Wischnewski N., Kerner M. J., Hartl F. U., Hayer-Hartl M. (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 [DOI] [PubMed] [Google Scholar]
- 48. Sharma S., Chakraborty K., Müller B. K., Astola N., Tang Y. C., Lamb D. C., Hayer-Hartl M., Hartl F. U. (2008) Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133, 142–153 [DOI] [PubMed] [Google Scholar]