Abstract
Conotoxins are peptide neurotoxins produced by predatory cone snails. They are mostly cysteine-rich short peptides with remarkable structural diversity. The conserved signal peptide sequences of their mRNA-encoded precursors have enabled the grouping of known conotoxins into a limited number of superfamilies. However, the conotoxins within each superfamily often present variable sequences, cysteine frameworks, and post-translational modifications. To understand better how conotoxins are diversified, we performed a venomic study with C. flavidus, an uninvestigated vermivorous Conus species, by combining transcriptomic and proteomic analyses. In order to obtain the full-length conotoxin sequences, protease digestion was not performed with the venom extraction prior to spectra acquisition via tandem mass spectrometry (MS/MS). Because conotoxins are produced from mRNA-encoded precursors by means of proteolytic cleavage, nonspecific digestion of precursors was applied during the database search. Special attention was also paid in interpreting the MS/MS spectra. All together, these analyses identified 69 nonredundant cDNA sequences and 31 conotoxin components with confident MS/MS spectra. A new Q-superfamily was also identified. More importantly, this study revealed that conotoxin-encoding transcripts are diversified by hypermutation, fragment insertion/deletion, and mutation-induced premature termination, and that a single mRNA species can produce multiple toxin products through alternative post-translational modifications and alternative cleavages of the translated precursor. These multiple diversification strategies at different levels may explain, at least in part, the diversity of conotoxins, and provide the basis for further investigation.
Cone snails (Conus species) are predatory mollusks that inhabit tropical and subtropical shallow seawater. Their venom ducts produce a mixture of peptides, generally known as conotoxins, that have exquisite specificity for different ion channels, receptors, and transporters. These toxins are used to immobilize or paralyze prey, as well as for defense (1). Because of their specificity, conotoxin peptides are considered the ideal neurobiological tools for investigating specific receptors and channels. Furthermore, they are valuable lead compounds for clinical therapy. The conotoxin ω-MVIIA, isolated from Conus magus, is known commercially as Prialt (ziconotide) and is already in clinical use for the treatment of chronic pain (2, 3).
Apart from their great potential for neurobiological and therapeutic applications, an emerging enigma concerning conotoxins is their striking diversity. Conotoxins are generally short disulfide-rich peptides of 10 to 40 amino acids. Every Conus species can produce more than 1000 different conotoxins (4, 5), and the number of Conus species with morphological differences is currently estimated as 800 (6). Hence, the total number of conotoxins is thought to be over 1 million. Because fewer than 2000 conotoxins have been conclusively identified in 92 different Conus species (7) and 99.8% of conotoxins are yet to be discovered, the mechanisms of conotoxin diversity are far from clear.
Despite this conundrum, it has been established that conotoxin peptides are produced from corresponding mRNA-encoded precursors that typically contain a highly conserved N-terminal signal peptide, a more variable pro-peptide, and a hypervariable mature peptide (8, 9). Based on the conserved signal peptide sequence, the disulfide-rich conotoxins are grouped into at least 17 superfamilies: the A-, D-, I1-, I2-, I3-, J-, L-, M-, O1-, O2-, O3-, P-, S-, T-, V-, Y-, and K-conotoxin superfamilies (7, 10). The non-disulfide-rich conotoxins are grouped into different families; these include conantokin, contulakin, conorfamide, conomap, conophan, contryphan, conkunitzin, conolysin, and conopressin, with conantokin and contulakin being recently categorized as B- and C-superfamily toxins, respectively (11).
In the mature peptides of conotoxins, the high content of cysteine residues and the number of post-translational modifications (PTMs)1 are also remarkable. Thus far, 23 different cysteine frameworks, numbered I to XXIII, have been identified (7, 10). The list of PTMs found in conotoxins includes hydroxylation of proline, valine, and lysine; carboxylation of glutamate; C-terminal amidation; cyclization of N-terminal glutamine; glycosylation; sulfation; bromination; residue epimerization; and so on. In fact, conotoxins are considered to be one of the most highly post-translationally modified classes of gene products known (12).
In order for the diversification of conotoxins to be better understood, a greater number of conotoxin sequences, at both the nucleotide and the amino acid sequence level, need to be identified. The conserved signal peptide sequence of conotoxin precursors has facilitated the large-scale identification of conotoxin cDNA sequences (13–16). However, it remains impossible to predict the sequences of mature conotoxins, in particular the termini of the mature peptides and their PTMs, based only on cDNA sequences. As a result, most information regarding conotoxin primary sequences has previously been obtained via traditional peptide chemistry techniques, which require relatively large amounts of the precious native conotoxin material.
Mass spectrometry (MS) techniques have been applied for conotoxin identification for more than two decades (17, 18). Some PTMs have been clearly identified with MS (19–22), which is a definite advantage of MS over cDNA sequencing. Recently, MS-based proteomics techniques have come into favor for the large-scale study of conotoxin sequences. For instance, proteomic analysis of different sections of the Conus textile venom duct has been performed to explore the venom processing (23). Some elegant de novo sequencing works have also been done with conotoxins, but specialized MS methodologies were required (24, 25). Integrating the data from the proteomic and transcriptomic analyses would facilitate the interpretation of MS results. Such a strategy was attempted in two recent studies, but high-accuracy tandem MS (MS/MS) was not applied to identify the conotoxin components (26, 27).
In this work, we studied the venom from Conus flavidus, a previously uninvestigated vermivorous Conus snail collected in the South China Sea, using a combined transcriptomic and proteomic approach. The analysis identified 69 nonredundant conotoxin cDNA sequences, as well as 31 conotoxins with confident MS/MS evidence. A new superfamily of conotoxins, the Q-superfamily, was also identified. Moreover, our results revealed various strategies of conotoxin diversification at different levels.
EXPERIMENTAL PROCEDURES
cDNA Library Construction
C. flavidus specimens were collected from the South China Sea near Hainan, China. The venom ducts were excised from live snails and immediately frozen in liquid nitrogen. One gram of frozen venom duct (from >10 specimens) was used for mRNA extraction, and mRNAs shorter than 1000 base pairs (bps) were used to construct the cDNA library (Invitrogen, Shanghai, China).
Analysis of cDNA Sequences
From this cDNA library, 1032 clones were sequenced and analyzed according to the following procedure. Vector sequences were detected using ClustalX 2.1 software for multiple sequence alignment (28) and were deleted until the 3′ end of a 74-bp common adaptor sequence (CTGATAGTGACCTGTTCGTTGCAACAAATTGATGAGCAATGCTTTTTTATAATGCCAACTTTGTACAAAAAAGT) was reached. The sequences at the 3′ end to the poly-A tag (>20 As; 414 of 1032 clones) or at the 5′ end to the poly-T tag (>20 Ts; in 5 of 1032 clones) were deleted. The cDNA sequences were then translated into protein sequences in six reading frames. All resulting sequences were extracted from start codon to stop codon, and sequences shorter than 50 amino acids were discarded. All peptides that passed through these filters were treated as valid peptides, and BLAST searches were conducted for each peptide sequence against the National Center for Biotechnology Information (NCBI) nonredundant protein database (downloaded on July 7, 2011) by using the BLASTP algorithm in the ncbi-blast-2.2.25+-ia32-linux program. The top 10 matched hits with e-values lower than 10−6 were returned for each query. Hits with the minimum e-value among all six queries derived from the same clone were used for the appropriate protein annotation of each clone. All sequence alignments were performed using the ClustalW alignment tool in the MEGA5.0 software package with default parameters.
Gene Ontology Annotation Analysis
Identified peptide sequences were blasted against the Gene Ontology (GO) sequence database (go_20110716-seqdb) by using the BLASTP algorithm compiled in the ncbi-blast-2.2.25+-ia32-linux software. The e-value cutoff was set at less than 0.01. GO IDs were uniquely extracted for each peptide, and the alternative IDs or obsolete IDs were removed. Every GO ID was replaced by its corresponding parent GO ID according to the hierarchy-resembled graph structure described in gene_ontology_ext.obo, until their parent GO IDs were among the three root terms: GO:0003674 (molecular_function), GO:0005575 (cellular_component), and GO:0008150 (biological_process).
Crude Venom Extraction
The venom ducts of 10 C. flavidus specimens were cut into short fragments with scissors. The fragments were then extracted successively with 10 ml 0.1% trifluoroacetic acid three times, 10 ml 20% acetonitrile twice, 10 ml 40% acetonitrile twice, and 10 ml 50% acetonitrile once. All venom duct extraction solutions were pooled together and lyophilized for future use.
Mass Spectrometry Analysis
Crude venom extract (∼300 μg) was dissolved in 50 mm NH4HCO3 (300 μl), reduced with 1 mm dithiothreitol at room temperature for 1 h, and then alkylated with 5.5 mm iodoacetamide at room temperature in the dark for 45 min. To remove hydrophobic proteins, lipids, salts, and tissue debris, the reaction solution was passed through an EmporeTM C18-SD 1-ml cartridge (3M, St. Paul, MN), and the peptide components were eluted from the cartridge with 90% acetonitrile and vacuum dried.
LC-MS analysis of the alkylated venom sample was performed on an LTQ-Orbitrap Velos coupled with a Surveyor HPLC (Thermo Scientific, Waltham, MA). One-third of the venom peptide mixture was loaded onto a C18 column, and a 60-min gradient was applied for washing and elution. Ions in the m/z range of 300–1800 were acquired. The automatic gain control target values were 1,000,000 for the MS spectra and 200,000 for the MS/MS spectra; the maximum injection times were 1000 ms for the MS spectra and 500 ms for the MS/MS spectra; and the resolution was 60,000 for the MS spectra and 15,000 for the MS/MS spectra. The top three peaks with a minimum signal above 5000 were activated by higher-energy C-trap dissociation (HCD), and their MS/MS spectra were acquired in the Orbitrap analyzer. The normalized collision energy was 48, and the activation time was 30.
MS Data Analysis
The software package pFind Studio 2.8 (29, 30) was applied to analyze the MS data. Raw files were first converted to MGF files by pXtract and searched with pFind against the decoy database constructed with 69 cDNA-encoded nonredundant precursor sequences obtained from the cDNA library of C. flavidus in this work, as well as all 4572 protein sequences downloaded from the ConoServer database (on December 11, 2012). The maximum mass deviation was set as 10 ppm for both MS and MS/MS ions. Carbamidomethylation of Cys was a fixed modification, and the variable modifications were oxidation of Met; hydroxylation of Pro, Val, and Lys; cyclization of the N-terminal glutamine; bromination of Trp; sulfation of Tyr; carboxylation of Glu; and C-terminal amidation. A nonspecific enzyme was used for the theoretical digestion. Identified results were compiled by pBuild, and peptides with false discovery rates greater than 0.01 were discarded. The MS/MS spectra interpretation was done automatically by pLabel, and then the charge states were manually checked and the interfragmented ions were manually labeled. To extract deconvoluted MS peaks, DeconTools AutoPrecessor_v1.0.4672 was applied (31).
cDNA Cloning of Q-superfamily Conotoxins with Rapid Amplification of cDNA Ends
To obtain more cDNA sequences of Q-superfamily conotoxins, gene-specific primers were designed according to the conserved sequence in the 5′ untranslated region and the 5′ end of the coding region of the Q-conotoxin cDNAs identified in the C. flavidus cDNA library in this work. These primers were then used for PCR amplification. The sequences of the gene-specific primers were as follows: P1, 5′-AGTCCTGTCTCCTGCCCGCTG-3′; P2: 5′-GCCATCGGTTCAAGACAGACT-3′; and P3: 5′-TGCACACACTGGAAATGCTGCTGC -3′. Kits for 3′ rapid amplification of cDNA ends (RACE) from Invitrogen or Takara (Dalian, China) were used to clone Q-superfamily conotoxin cDNAs from C. flavidus, C. quercinus, and C. caracteristicus as described previously (32). The cDNAs amplified via PCR were cloned into the pGEM-T easy vector (Promega, Madison, WI) for sequencing.
RESULTS
Data Processing of the cDNA Library
From the cDNA library of C. flavidus, a total of 1032 clones were sequenced. The cDNA lengths ranged from 96 bp to 1279 bp, with an average length of 982 bp. Among these clones, 414 sequences were cloned with cis mRNA sequences marked with poly-A tags, and five sequences were cloned with trans mRNA sequences marked with poly-T tags. The sequence direction of the other 613 clones could not be determined. The translation of all six reading frames generated 3312 peptides longer than 50 amino acids from 787 clones. All 3312 sequences had start and stop codons, and BLAST was used to search the sequences against the NCBI nonredundant database. The peptides were relatively short (51 to 346 amino acids; median = 74 amino acids), and therefore a stringent e-value cutoff (<10−6) was used to filter matched hits with high confidence. Hits with the minimum e-value for the same clone were taken as the final protein annotation of each clone. As a result, 353 clones with 259 nonredundant peptides had matched hits with an e-value of less than 10−6. No trans translational products were identified among the cis mRNA clones, and vice versa.
In order to elucidate the possible biological functions of the proteins encoded by these cDNA clones, these sequences were also searched against the GO sequence database. A total of 209 nonredundant peptides from 287 clones had matched hits with an e-value of less than 0.01. The molecular function distribution (supplemental Fig. S1) showed that the most populated categories of the translated products of the short mRNAs in the venom duct were enzymes and structural molecules such as ribosomal proteins. Conotoxins, which are grouped with channel regulator activity and receptor regulator activity, were the third major functional category. In addition, transporters, enzyme regulators, transcription factors, electron carriers, molecular transducers, antioxidants, and nutrient reservoirs were also present in the transcriptomic sequences of the C. flavidus venom duct.
Conotoxins Identified in the C. flavidus cDNA Library
Conotoxin precursors encoded by these cDNA sequences were first identified by NCBI BLAST. Peptides annotated with “conotoxin/conantokin/contulakin/conorfamide/conomap/conophan/contryphan/conkunitzin/conolysin/conopressin” were extracted from the NCBI BLAST results, which yielded 63 nonredundant precursor sequences from 96 clones (supplemental Table S1). For precursors without significant BLAST results, cysteine patterns were extracted and empirically examined. In this way, six nonredundant precursor sequences from eight clones were identified as conotoxins (supplemental Table S1), yielding 69 nonredundant precursor sequences in total. Because these six precursors without BLAST results were similar to one another and their signal peptide sequences were different from the other known signal peptide sequences (supplemental Fig. S2), we proposed grouping them into a new superfamily, the Q-superfamily.
The nomenclature of these 69 cDNAs and the corresponding precursor sequences identified in this study had three components: the Conus species identifier, “Fla”; the cysteine framework of the predicted mature peptide (determined by using the ConoPrec tools at the Conoserver website (7)); and a serial number (supplemental Table S1). If zero, two, or an odd number of cysteine residues were present in any given mature peptide, the format “Fla-serial number” was used. The 69 nonredundant cDNA sequences gave rise to 57 nonredundant mature conotoxins. The cleavage sites were predicted by using the ConoPrec tools at the ConoServer website (7) or, when not applicable, were determined empirically.
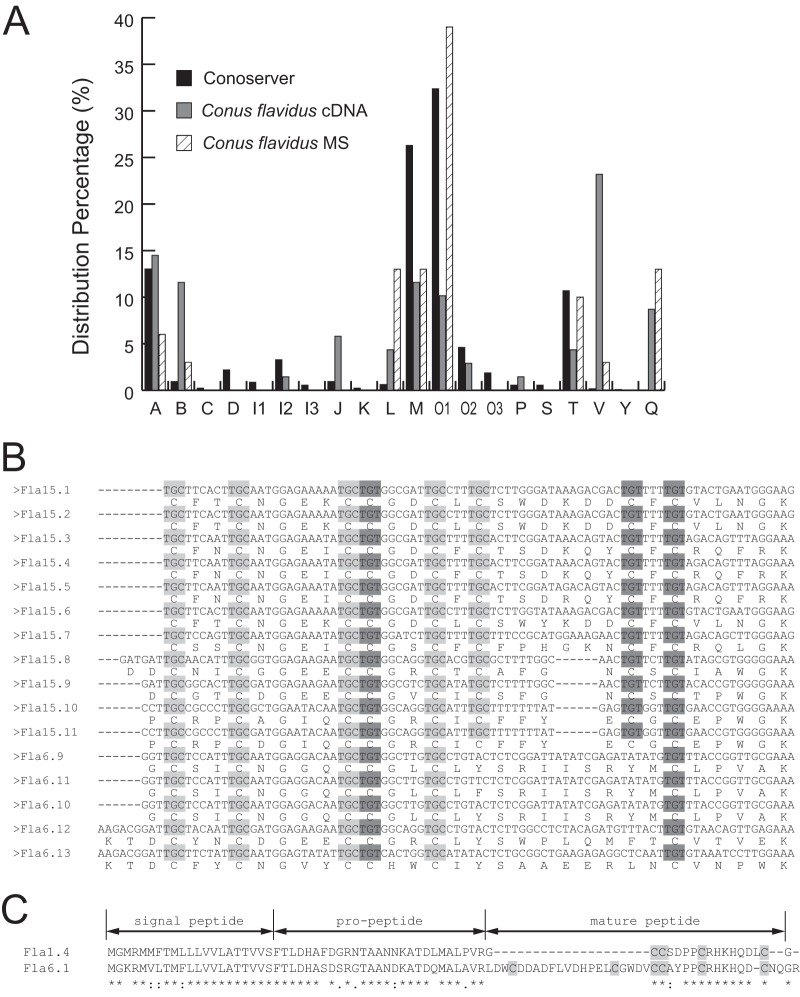
These 69 conotoxin cDNAs from C. flavidus belonged to 11 superfamilies, including the newly identified Q-superfamily (Fig. 1A). The most significant difference from the statistical distribution of the current data pool in the ConoServer database was that the C. flavidus transcriptome is rich in V-superfamily conotoxins, with 16 similar cDNA sequences identified in this study (Fig. 1B). Only two members of the V-superfamily, vi15.1 and vt15.1, were previously known from other Conus species (32). In particular, in addition to the cysteine framework XV (C-C-CC-C-C-C-C) previously identified in V-conotoxins, the cysteine framework VI/VII (C-C-CC-C-C), which is representative of the O-superfamily conotoxins, was also found in the V-conotoxins produced by C. flavidus. The alignment of the cDNA sequences corresponding to the mature peptides indicated that C. flavidus V-conotoxins with XV and VI/VII frameworks shared the same cysteine codons, the same intervals between two adjacent cysteine residues, and very similar lengths and sequences (Fig. 1B). This strongly suggests that these V-conotoxins evolved from the same ancestor.
Fig. 1.
Conotoxin cDNAs identified in the cDNA library of C. flavidus. A, distribution percentage of different conotoxin superfamilies in the ConoServer database, the conotoxins identified in the C. flavidus cDNA library, and the conotoxins identified via MS/MS in the venom of C. flavidus. B, comparison of the cysteine codon usage of V-conotoxins from C. flavidus with cysteine framework XV or VI/VII. The TGC cysteine codons are shown in light gray, and the TGT codons are shown in dark gray. C, comparison of the precursor sequences of two A-superfamily conotoxins from C. flavidus demonstrates that the alteration in the cysteine framework in Fla6.1 is caused by a fragment insertion.
The C. flavidus conotoxins belonging to other superfamilies also presented some unusual cysteine frameworks. For example, the A-superfamily conotoxins typically have cysteine framework I (CC-C-C) (33), which is shared by six of the seven A-conotoxins identified in this C. flavidus cDNA library. However, one predicted mature conotoxin of the A-superfamily (encoded by two cDNA sequences, Fla6.1 and Fla6.2) instead had cysteine framework VI/VII. Sequence alignment of these A-conotoxins indicated that this unusual cysteine framework was produced by a fragment insertion in the N terminus of the mature peptide of Fla6.1 (Fig. 1C). In addition, most of the M-superfamily conotoxins have cysteine framework III (CC-C-C-CC) (11). But three M-conotoxins from C. flavidus (Fla-13, Fla-14, and Fla-15) had two, five, and seven cysteine residues, respectively. Furthermore, four of the seven O1-conotoxins identified in the C. flavidus cDNA library shared the most common cysteine framework for this superfamily, VI/VII, whereas three other O1-conotoxins (Fla-16, Fla-17, and Fla-18) were cysteine-free. These results indicate that the mRNA sequences of conotoxins produced by C. flavidus are highly diversified.
MS/MS Analysis of the Venom Components of C. flavidus
In order to obtain further insight into C. flavidus conotoxins at the protein level, the crude venom extract was reduced, alkylated, and analyzed with LC-MS/MS on the LTQ-Orbitrap Velos. HCD fragmentation was applied so that highly accurate MS and MS/MS spectra were both acquired in the Orbitrap analyzer. Then, the deconvoluted molecular weights of all multiply charged peaks were extracted from the MS spectra with scan numbers smaller than 4000 (the remaining MS spectra were of too low quality), leading to the identification of more than 80,000 masses. The number of masses was reduced to 8604 when redundant peaks within a 5-ppm mass deviation were removed. These masses probably corresponded to venom components, but the mass of a molecule alone is not sufficient for it to be assigned to a given component.
To acquire confident sequence information regarding the conotoxin components, MS/MS data were searched against the 69 precursor sequences obtained from the C. flavidus cDNA library, as well as all 4572 protein sequences downloaded from ConoServer, with very stringent criteria (maximum mass deviation within 10 ppm for both MS and MS/MS data). In total, 31 different conotoxins were confidently identified (false discovery rates less than 0.01, score < 10−10 in pFind), and each of these was manually examined (Table I, Figs. 2–4, and supplemental Figs. S3–S26). Manual examination of the MS/MS analysis proved necessary, as two automatically assigned sequences (conantokin-fla and fla6e) were corrected in this step (Fig. 2). Both corrected sequences yielded significantly better coverage of fragment ions and a higher percentage of assigned peaks, and thus much better interpretation of the spectra.
Table I. Conotoxins identified via confident MS/MS analysis.
| Superfamily | cDNA | Precursor sequence | Conotoxin sequence | Conotoxin name | Monoisotopic mass (MH+) | Mass deviation (ppm) | MS/MS spectra |
|---|---|---|---|---|---|---|---|
| A | Fla1.3/1.4/1.5 | MGMRMMFTMLLLVVLATTVVSFTLDHAFDGRN(I/T)AANNKATDLMALPVRGCCSDPPCRHKHQDLCG | ↓CCSDPPCRHKHQDLC* | fla1a | 1968.7996 | 2.44 | Supplemental Fig. S3 |
| A | GCCSDPPCRHKHQDLC* | fla1b | 2025.8185 | 3.65 | Supplemental Fig. S4 | ||
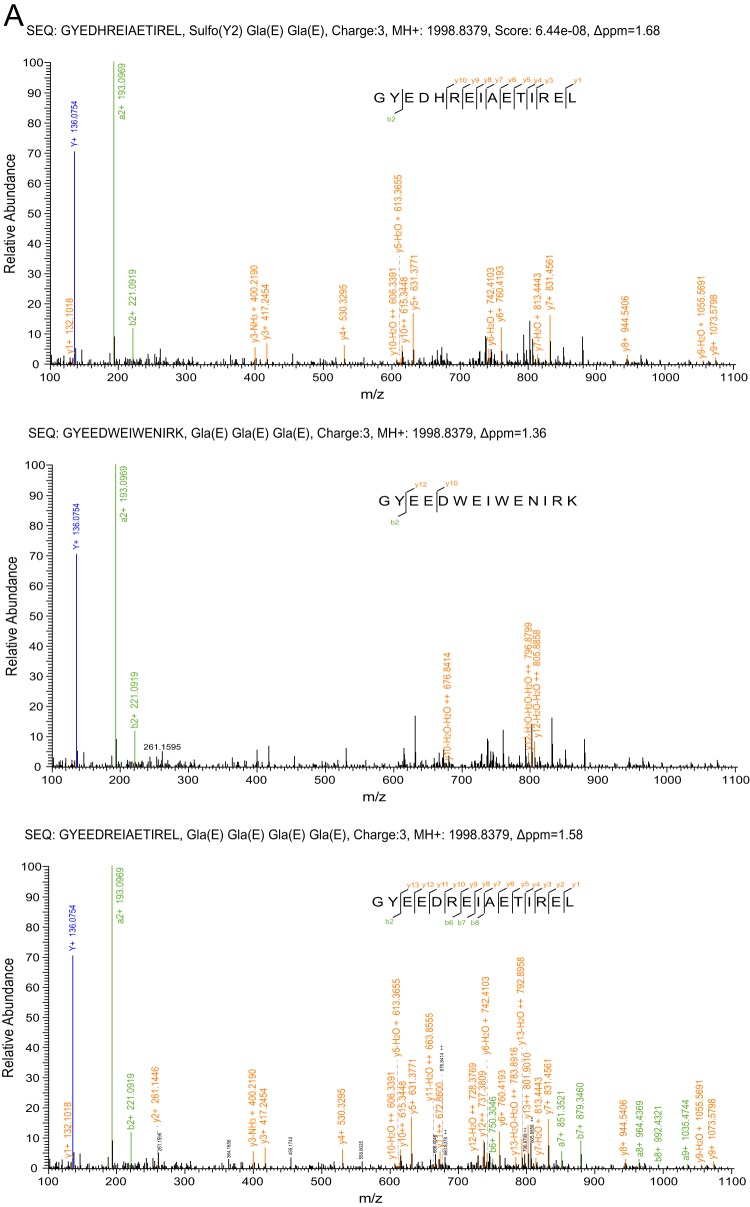
| B | Fla-5 | MQLYTHLYLLVPLVAFHLILGTGTLAHGDALTERRSADATALKPEPVLLQKSAARSTDDNDRDRLTQRKRILKKQGNMARGYEEDREIAETIRELNEAGK | GYEEDREIAETIREL↓ (four of these five Glu residues are carboxylated, the positions of which are not identified) | Conantokin-fla | 1998.8379 | 1.58 | Supplemental Fig. 2A |
| L | Fla14.3 | MKLSVTFIVVLMLTTSLTFGFSLSSNNGERAYGSHHSYVADQLVRRERASRACNPPCSDILTCLHGTCKHLGIG | ACNPPCSDILTCLHGTCKHLGI* | fla14a | 2523.1657 | 2.64 | Supplemental Fig. S5 |
| L | ACNPOCSDILTCLHGTCKHLGI* | fla14b | 2539.1588 | 3.31 | Supplemental Fig. S6 | ||
| L | ACNOOCSDILTCLHGTCKHLGI* | fla14c | 2555.1532 | 3.50 | Supplemental Fig. S7 | ||
| L | Fla14.2 | MKLSVTFIVVLMLTTSLTFGFSLSSNNGERAYGPRHSNVADQLVPREQASRTCYPPCIGYTYCKSGTCEYRQGRASTVFLKRQ | TCYPPCIGYTYCKSGTCEYRQ* | fla14d | 2663.1040 | 3.94 | Supplemental Fig. S8 |
| M | Fla3.4 | MMSKLGVLLTISLLLFPLTAVQLDGEQPVDLLALRTQDFAPEQSPWFDPVKRCCSKYCWECTPCCPYSSG | CCSKYCWECTOCCPYSS* | fla3a | 2319.7952 | 4.41 | Supplemental Fig. S9 |
| M | Lv3-V04 | MLKMGVVLFTFLVLFPLATLQLDADQPRARYAENKQDFNRNERTKMILSAVSASMGRQRRCCLWPACWGCVCCY | ↓RCCLWPACWGCVCCY | fla3b | 2017.7830 | 2.66 | Supplemental Fig. S10 |
| M | Fla3.2/3.3 | MLKMRVVLFTFLVLFPLATLQLDADQPRARYAENKQDFNRNERTKMILSAVSASMGRQRRCCLWPECGGCVCCY | ↓RCCLWPECGGCVCCY | fla3c | 2036.7294 | 3.47 | Supplemental Fig. S11 |
| M | ↓RCCLWPγCGGCVCCY | fla3d | 2080.7210 | 2.44 | Supplemental Fig. S12 | ||
| O1 | Fla-17 | MKLTCILIVAVLFSTAWTFVTVDDSINHLKYREWTDFRPWGKRIPEARDKMKNLKFSKLDNKKRRL | EWTDFRPW* | fla01 | 1135.5289 | 2.71 | Supplemental Fig. S13 |
| O1 | γWTDFRPW* | fla02 | 1179.5192 | 2.16 | Supplemental Fig. S14 | ||
| O1 | Fla-18 | MKLTCVMIVAALFLTAWTFVTADNSKYELENRGGLGHAGGWVKAGALGKDPGWGKRFLMARDEMKNPKASKLVP | GGLGHAGGWVKAGALG↓ | fla03 | 1407.7446 | 3.22 | Supplemental Fig. S15 |
| O1 | GGLGHAGGWVKAGALGKDPGW* | fla04 | 1990.0303 | 5.13 | Supplemental Fig. S16 | ||
| O1 | GGLGHAGGWVKAGALGKDPGW | fla05 | 1991.0250 | 0.26 | Supplemental Fig. S17 | ||
| O1 | ↓LGHAGGWVKAGALGKDPGW* | fla06 | 1875.9901 | 3.97 | Supplemental Fig. S18 | ||
| O1 | ↓HAGGWVKAGALGKDPGW* | fla07 | 1705.8867 | 3.15 | Supplemental Fig. S19 | ||
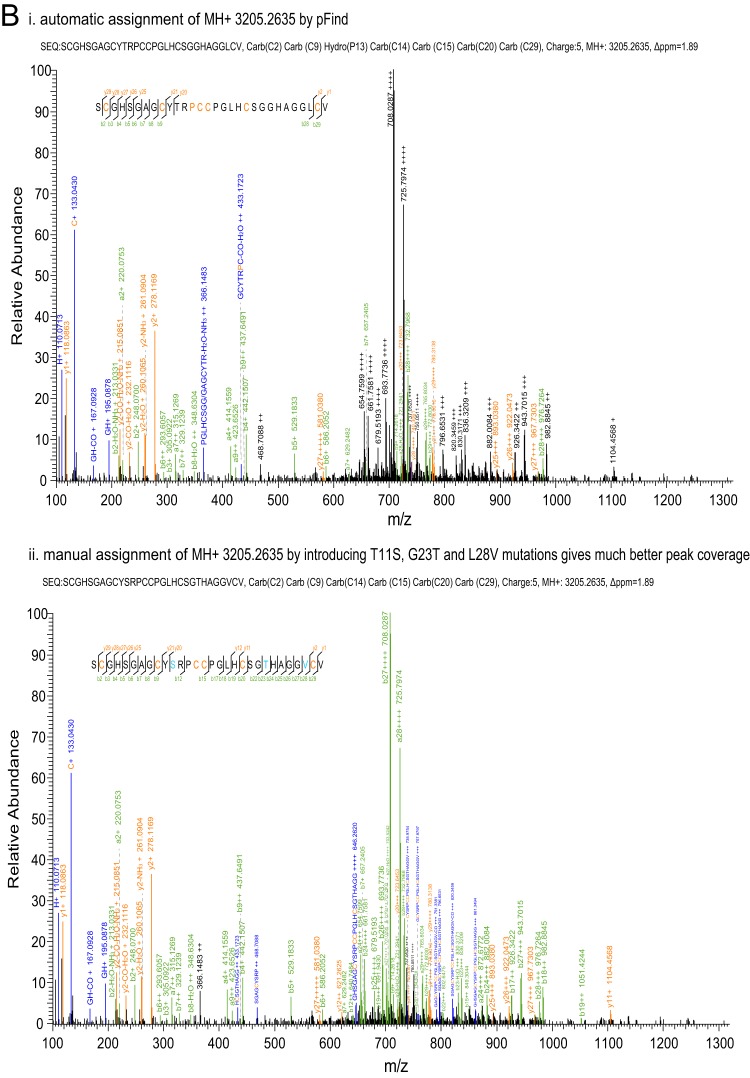
| O1 | / | / | SCGHSGAGCYSRPCCPGLHCSGTHAGGVCV | fla6e | 3205.2635 | 1.89 | Fig. 2B |
| O1 | Fla6.4 | MKLMCVLIIAVLFLTASQLITADYSRDKQEYRAERLRDAMGKFKGSRSCGHSGAGCYTRPCCPGLHCSGGQAGGLCV | SCGHSGAGCYTRPCCPGLHCSGGQAGGLCV | fla6a | 3180.2563 | 5.66 | Supplemental Fig. S20 |
| O1 | SCGHSGAGCYTROCCPGLHCSGGQAGGLCV | fla6b | 3196.2557 | 4.23 | Supplemental Fig. S21 | ||
| O1 | SCGHSGAGCYTROCCOGLHCSGGQAGGLCV | fla6c | 3212.2467 | 5.44 | Supplemental Fig. S22 | ||
| O1 | Fla6.5 | MKLTCALIVAMLFLTACQLTTTDDSRGRQKYPTERLRVKMRNPKLSKLTKTCDPPGDSCSRWYNHCCSKLCTSRNSGPTCSRP | TCDPOGDSCSRWYNHCCSKLCTSRNSGOTCSRP | fla6d | 3992.5991 | 1.33 | Supplemental Fig. S23 |
| T | Fla5.2 | MRCVPVFIILLLLSPSAPSVDAHPKTKDDVPLASFHDDAKRTLQRLWIKALCCYGYAFCCRLGR | ALCCYGYAFCCRL* | fla5a | 1712.7104 | 3.52 | Supplemental Fig. S24 |
| T | ↓LCCYGYAFCCRL* | fla5b | 1641.6746 | 2.85 | Supplemental Fig. S25 | ||
| T | LiC121 (P01681) | MRCVPVFIILLLLSPSAPSVDAHPKTKDDVPLASFHDDAKRTLQRLWIKALCCYGYRFCCPNFR | ALCCYGYRFCCPN↓ | fla5c | 1740.6666 | 4.82 | Supplemental Fig. S26 |
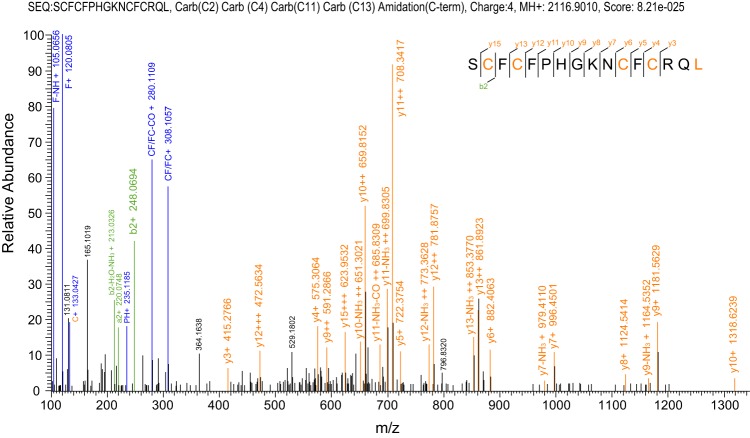
| V | Fla15.7 | MSTPKMTPFILLLLFSLTIRCGDGKAIQEDRDPSTRLLTGDKNRDLSVNRRCSSCNGEICCGSCFCFPHGKNCFCRQLGK | ↓SCFCFPHGKNCFCRQL* | fla15a | 2116.9010 | 3.52 | Fig. 3 |
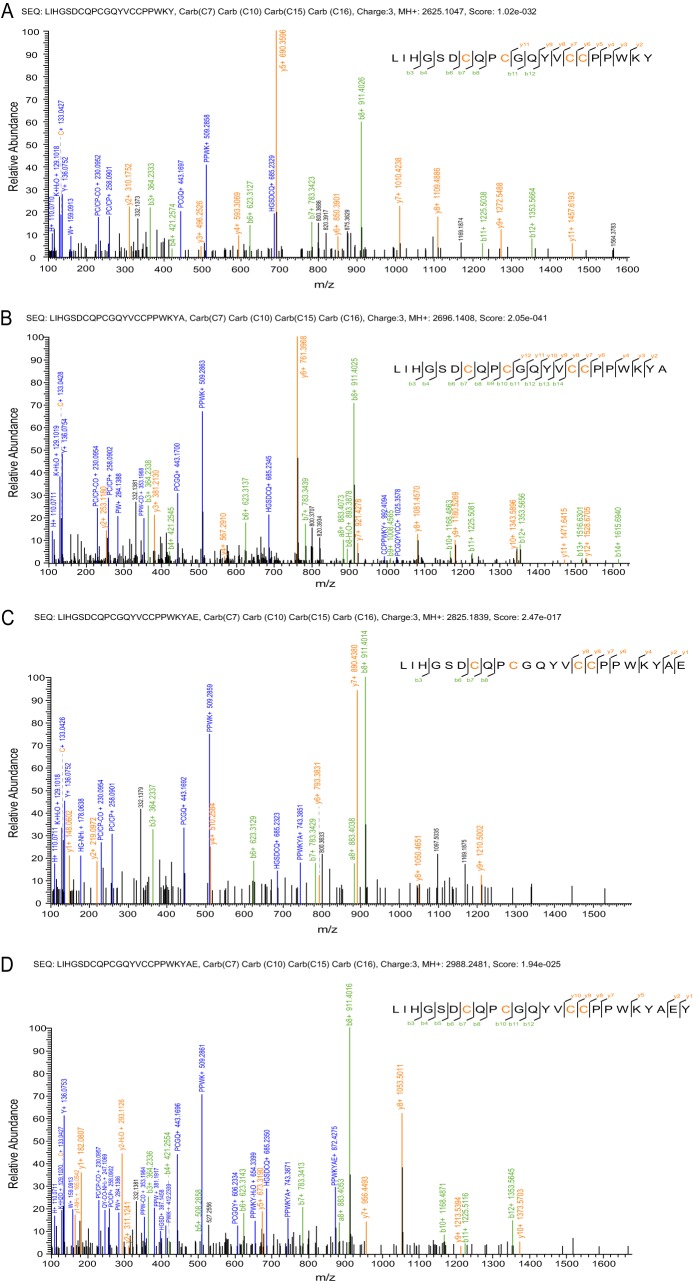
| Q | Fla16.1/16.2 | MHTLEMLLLLLLLLPLAPGEGDGQAVAGDRNPSEARSTHEHFLQRLIRLIHGSDCQPCGQYVCCPPWKYAEYRRFT | LIHGSDCQPCGQYVCCPPWKY↓ | fla16a | 2625.1047 | 3.59 | Fig. 4A |
| Q | LIHGSDCQPCGQYVCCPPWKYA↓ | fla16b | 2696.1408 | 3.86 | Fig. 4B | ||
| Q | LIHGSDCQPCGQYVCCPPWKYAE↓ | fla16c | 2825.1839 | 3.53 | Fig. 4C | ||
| Q | LIHGSDCQPCGQYVCCPPWKYAEY | fla16d | 2988.2481 | 3.04 | Fig. 4D |
Notes: O represents hydroxylated proline, and γ is the carboxylated glutamate residue. C-terminal amidation is denoted by an asterisk. Arrows indicate the unexpected proteolytic cleavage.
Fig. 2.
Manual examination of MS/MS spectra. A, the MS/MS spectrum of conantokin-fla was manually examined against three possible conantokin sequences without the PTMs that are present in the precursor ion. The mass accuracy of each spectrum was calculated with the mass of the precursor ion with corresponding PTMs shown in each spectrum heading. The third sequence was taken as the most confident result. B, the MS/MS spectrum of fla6e was automatically annotated (panel i) and then manually assigned (panel ii) by introducing point mutations of T11S, G23T, and L28V (shown in cyan in the inset sequences). Interfragmented ions were also manually labeled. In this figure, as well as in Fig. 3 and Fig. 4, the orange letters in the inset sequences indicate the residues with modifications listed in each spectrum heading.
Fig. 3.
MS/MS spectrum analysis of fla15a. The spectrum interpretation gives a confident sequence with only four cysteines, although its cDNA-encoded precursor has eight cysteines in the mature peptide region.
Fig. 4.
MS/MS spectra of Q-conotoxins fla16a (A), fla6b (B), fla6c (C), and fla6d (D). These four Q-conotoxins are derived from the same precursor, but with alternative cleavages at the C terminus.
These 31 conotoxin components identified from the venom belonged to the A-, B- (formerly conantokin), L-, M-, O1-, T-, V-, and Q-superfamilies, suggesting that these eight superfamilies are relatively dominant in the venom of C. flavidus (Fig. 1A). Most identified conotoxins (28 of 31) corresponded to 14 cDNA-encoded precursors obtained from the C. flavidus cDNA library, often with a high frequency of occurrence (supplemental Fig. S27). This implies consistency between the mRNA and protein components of the venom duct. In addition, two conotoxins identified via MS/MS analysis corresponded to two cDNA-encoded precursor sequences previously obtained from other Conus species, and one component did not correspond to any previously reported cDNA. Among the 31 identified conotoxins, only 3 (fla1b, fla6a, and fla5a) possessed sequences identical to those predicted from the precursors, underscoring the importance of characterizing conotoxins at the protein level.
Multiple Peptide Products from One Conotoxin Precursor
One remarkable phenomenon revealed by the MS/MS results is that conotoxin precursors often give rise to more than one mature peptide, with various numbers of PTMs or alternative cleavages; the most extreme case was that of five different mature peptides derived from the Fla-18 precursor (supplemental Figs. S15–S19). Consistent with the results for conotoxins from other Conus species, the most common PTMs observed in the C. flavidus conotoxins were C-terminal amidation, hydroxylation of proline, and carboxylation of glutamate, occurring in 15, 6, and 3 of 31 conotoxins, respectively.
Hydroxylation of proline causes a 16-Da increase in mass and is rather stable against HCD fragmentation; thus, this modification is clearly identified in MS/MS spectra. The mature peptide region of Fla14.3 has two proline residues. One, two, or none of these residues may be hydroxylated, giving rise to three different conotoxins in the venom (fla14a, fla14b, and fla14c; supplemental Figs. S5–S7). A similar situation exists for the Fla6.4 precursor, leading to three mature toxins with various numbers of hydroxylated proline residues (supplemental Figs. S20 and S21).
Carboxylation of glutamate gives rise to a residue termed Gla and increases the mass by 44 Da, which is seen in MS spectra. However, this modification is labile during HCD fragmentation so that no carboxylation remains in the MS/MS spectra (22, 34). As a result, the position of the carboxylation can be identified only when all of the Glu residues are carboxylated. In the cases of fla01 versus fla02 and fla3c versus fla3d, the MS/MS spectra of each pair were rather similar (supplemental Figs. S11–S14), but the difference between the masses of each pair (44 Da) clearly showed that there is only one Gla residue in fla02 and one Gla residue fla3d. Because only one Glu residue is present in each of these two sequences, the position of carboxylation is undoubtedly determined. In contrast, the mass of conantokin-fla shows that only four of five Glu residues in this toxin are carboxylated, but determining the positions of the resultant Gla residues is beyond the capability of MS/MS with the HCD fragmentation technique.
Apart from PTMs, alternative cleavage is another strategy by which multiple peptide products can be generated from one conotoxin precursor, as shown for the precursors Fla1.3/1.4/1.5, Fla-18, Fla5.2, and Fla16.1/16.2. The alternative cleavage sites occurred at either the N terminus (as in Fla1.3/1.4/1.5 and Fla5.2) or the C terminus (in Fla16.1/16.2), or at both termini (in Fla-18). Furthermore, the cleavage sites at the N terminus often occurred after a Gly residue, with one exception being cleavage after an Ala residue in fla5b. However, no clear sequence preference was observed for the alternative cleavage sites at the C terminus. The underlying mechanism for the different cleaving preferences at different termini certainly deserves further exploration.
Unexpected Cysteine Framework in a V-superfamily Conotoxin
Among all the cleavage sites identified in this work, the most unanticipated site was observed in a peptide produced from a precursor of Fla15.7. This cDNA sequence belongs to the V-superfamily, because it shares a homologous signal peptide sequence and cysteine framework XV (C-C-CC-CXC-CXC) in the mature toxin region with the other V-superfamily conotoxins. To our great surprise, a peptide corresponding to the C-terminal half of the predicted mature toxin was identified in the venom of C. flavidus (Fig. 3). This peptide had only four cysteine residues, although it is still named fla15a according to the classification of its cDNA. Intriguingly, the N-terminal cleavage site for the production of this peptide also occurred after a Gly residue. It is unknown whether the normal V-conotoxins with eight cysteine residues exist in the venom of C. flavidus, but they were not identified at the MS/MS level in the current study. Therefore, it is not known whether this truncated peptide is the only mature product of the precursor of Fla15.7. However, this is the first result to demonstrate that the cysteine framework in the precursor may be altered during the maturation process of conotoxins.
The Newly Identified Q-superfamily Conotoxins
A new Q-superfamily of conotoxins was identified from the cDNA library of C. flavidus (see above). Five of the six peptide precursors of this superfamily had cysteine framework XVI (C-C-CC), whereas the sixth precursor had cysteine framework VI/VII (supplemental Table S1). MS/MS analysis of the venom identified four peptides derived from precursor Fla16.1/Fla16.2, but with alternative cleavages (Fig. 4). The lengths of these four peptides (fla16a, fla16b, fla16c, and fla16d) differed from one other by one residue at the C terminus. The cleavages occurred at the carboxyl side of Tyr, Ala, or Glu, showing no obvious sequence preference. This implies that alternative cleavage at the C terminus of conotoxins may be not sequence dependent but position-dependent, and that it is probably mediated by carboxypeptidase-like enzymes.
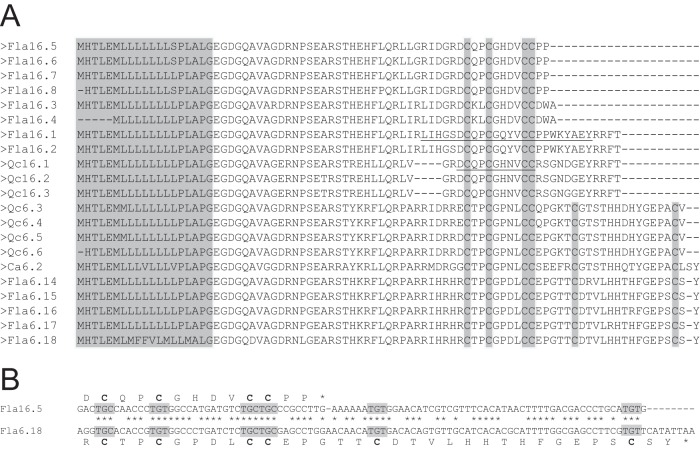
Among these Q-superfamily sequences, the predicted mature peptide of Fla16.5 is highly similar to qc16a, a conotoxin identified from C. quercinus (35). Therefore, qc16a probably belongs to the same Q-superfamily, indicating that C. flavidus is not the only Conus species that produces Q-superfamily conotoxins. Indeed, using the RACE method, we obtained a few Q-superfamily cDNA sequences not only from C. flavidus, but also from C. quercinus and C. caracteristicus (Fig. 5A). Among these, Qc16.1, Qc16.2, and Qc16.3 are highly similar to one another, and are probably the cDNAs corresponding to qc16a, if the 12 residues at the C terminus are cleaved.
Fig. 5.
cDNA-encoded precursor sequences of Q-superfamily conotoxins identified in this work. A, alignment of the precursor sequences of the Q-conotoxins shows that their mature toxins may have different lengths and different cysteine frameworks. The conserved signal peptide is shaded in gray. The mature peptide regions of Fla16.1 and Qc16.1 are underlined. B, comparison of the cDNA and protein sequences of Fla16.5 and Fla6.18 shows that the alteration in the length and cysteine framework of the Q-conotoxins is caused by a single nucleotide change in the cDNA sequence, which leads to the premature termination of the encoding region.
As shown by the results from the C. flavidus cDNA library (supplemental Table S1), the Q-superfamily contains sequences with not only cysteine framework XVI, but also framework VI/VII (Fig. 5A). Interestingly, alignment of the cDNA sequences with different lengths and different cysteine frameworks in the corresponding mature peptide regions (Fig. 5B) makes it obvious that the alteration in peptide length at the C terminus is due to a single nucleotide mutation that results in earlier termination of the encoding region. This is probably another strategy employed by Conus species to expand the diversity of conotoxins at the gene level.
DISCUSSION
Conotoxins are a huge reservoir of gene-encoded short peptides with remarkable structural diversity. The biological activities and applications of selected conotoxins have been studied, but the mechanism of conotoxin diversity is still far from being understood. To gain more insight into the structural diversity of conotoxins, we performed a comprehensive venomic analysis of a previously uninvestigated Conus species, C. flavidus. This analysis revealed various conotoxin diversifications at different levels.
Important Considerations Concerning Experimental Procedures
In this study, transcriptomic and proteomic analyses were combined in order to obtain information at both mRNA and protein levels. The conserved signal peptide sequence of each superfamily of conotoxins provides a convenient basis for cDNA identification, which contributes significantly to the data pool of conotoxins. However, knowledge of the cDNA-deduced precursor sequences is often not sufficient to anticipate the sequences of their toxin products, because most conotoxins undergo complicated maturation processes involving currently unpredictable proteolytic cleavages and PTMs. This demands the characterization of conotoxins at the protein level, and ideally on a large scale.
Modern MS technology is a powerful method for protein characterization, providing not only information regarding the mass of a molecule, but also sequence information via the fragmentation of peptide ions in MS/MS setups. As conotoxins have highly variable sequences and various PTMs, a mass value can be attributed to many different sequences. Similarly, a precursor sequence can be subject to different cleavages and PTMs so as to artificially match many masses. For example, if the precursor sequence of Fla15.7 were given all possible nonspecific cleavages to generate products longer than 5 amino acids and all PTMs observed in conotoxins except glycosylation were allowed, 21.8% of all nonredundant masses (1879 of 8604) identified in C. flavidus venom could be matched to Fla15.7 within 10 ppm mass deviation (supplemental Table S2). However, this matching is most likely unreliable. Thus, in the current study, only masses with corresponding MS/MS spectra were taken as identified conotoxin components.
In the study of conotoxins, one particular difficulty is the prediction of the mature toxin boundary from the precursor sequence, as multiple cleavages are often involved in the toxin maturation process, and the cleavage sites are, as noted above, currently not predictable. This leads to a situation in which matching a partial peptide fragment to the precursor sequence is not sufficient to determine the primary sequence of the conotoxin, which is distinct from the general situation of bottom-up proteomic studies (36). In an effort to overcome this difficulty, two considerations were taken into account in this study. First, in order to obtain the full-length sequences of conotoxins present in the C. flavidus venom duct, the peptides in the venom extract were not digested with proteases prior to MS/MS analysis. Given the generally short length of conotoxins, the full-length peptides were well ionized and fragmented. Second, nonspecific enzyme was applied for the theoretical digestion of conotoxin precursor sequences in the database for the searching of MS/MS spectra. This unbiased processing resulted in the identification of several unexpected cleavage sites employed for conotoxin maturation.
In high-throughput proteomic analysis of conotoxins, some additional difficulties arise from the neutral loss of certain labile PTMs during fragmentation and the lack of a comprehensive database. Consequently, special attention was paid in interpreting the MS/MS spectra, and two sequences were manually corrected after automatic assignment (Fig. 2): conantokin-fla and fla6e. With regard to the MS/MS spectrum of conantokin-fla, the precursor ion with an MH+ value of 1998.8379 matched three possible conantokin mature peptides derived from the C. flavidus cDNA library, with a deviation of <2 ppm: GYEDHREIAETIREL, containing one sulfation modification and two Gla residues; GYEEDWEIWENIRK, containing three Gla residues; and GYEEDREIAETIREL, containing four Gla residues. Because the loss of the sulfation on Tyr may happen in MS mode (37), whereas no corresponding desulfation mass was found in our MS spectra, the first sequence seems unlikely. During HCD fragmentation, the sulfation of Tyr and the carboxylation of Glu are labile and thus are not maintained in the fragment ions (21, 34). The MS/MS spectrum was manually examined against these three sequences with no PTM (Fig. 2A). Matching with the third versus the first or second sequence gave a much better interpretation of the MS/MS spectra, especially of doubly charged fragment ions; therefore, this sequence was taken as the most confident result, although the automatic annotation was assigned to the first sequence.
Regarding the MS/MS spectrum of O-conotoxin fla6e, the sequence was automatically assigned as SCGHSGAGCYTRPCCPGLHCSGGHAGGLCV with hydroxylation on Pro13 (Fig. 2B, panel i). However, manual examination revealed that the two most intense ions were either not assigned or incorrectly assigned on the second isotopic peak. By manually analyzing the spectrum with pLabel, we found that an L28V point mutation gave a beautiful fragment ion series from b27++++ to b23++++, and a T11S point mutation further led to the assignment of b12++, b15++, b17++, b18++, b19++, and b20+++ ions, as well as y11+ and y12++ ions. To match the precursor ion mass, Gly23 was mutated to Thr, which was supported by the b22+++ ion (Fig. 2B, panel ii). Therefore, the sequence of fla6e was finally corrected to SCGHSGAGCYSRPCCPGLHCSGTHAGGVCV without PTM, the cDNA of which has not yet been reported. These two examples highlight the fact that manual examination of conotoxin MS/MS spectra is required in order to achieve confident results for further analysis.
Diversification of Conotoxin mRNA Sequences
The analysis of the C. flavidus cDNA library identified 69 nonredundant conotoxin precursors that belonged to 11 superfamilies, including the newly identified Q-superfamily. Within each superfamily, the mature peptide region was generally more variable than the other regions (Figs. 1B, 1C, 5A, and 6A), so that diversified similar conotoxins could be produced, as observed in previous studies (38, 39). Apart from local variations due to missense mutations, fragment insertion/deletion also caused pronounced changes in conotoxin sequences, often resulting in a change of cysteine frameworks (Fig. 1C). Another interesting way is the mutation-induced premature termination of the coding region, which was observed for the first time in this study in the newly identified Q-superfamily (Fig. 5B). This diversity strategy seems quite effective relative to the others, taking into consideration the comparatively small change in nucleotide sequence coupled with the resulting large change in protein sequence.
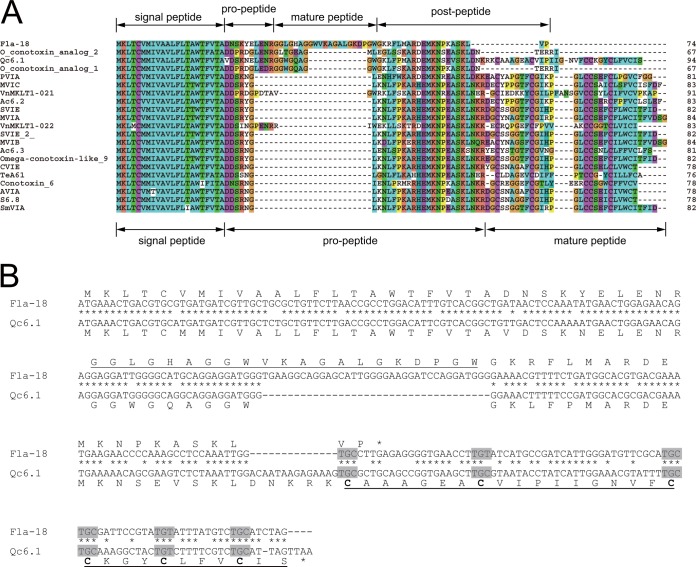
Fig. 6.
Comparison of the cDNA-encoded precursor sequence of Fla-18, a cysteine-free O1-superfamily conotoxin identified in this work, with the 20 most similar sequences of known O1-conotoxins. A, precursor sequence alignment shows that the organization of the Fla-18 precursor is distinct from that of the precursors of the canonical O1-conotoxins. The mature peptide region of Fla-18 is inserted into the propeptide region, and the C-terminal cysteine-rich region is missing in Fla-18. B, comparison of the cDNA and protein sequences of Fla18 and Qc6.1 shows that the absence of the cysteine-rich region in Fla-18 is caused by a 14-bp deletion in its cDNA sequence, which leads to the premature termination of the coding region.
Most strikingly, multiple diversifications were simultaneously observed upon O1-superfamily sequence alignment (Fig. 6). The sequences with cysteine framework VI/VII are similar to each other, owing to local missense mutations. However, the precursor organization of Fla-18 is clearly different from that of the other O1-superfamily conotoxins, because of the insertion of the mature peptide region of Fla-18 into the propeptide region of canonical O1-conotoxins and the deletion of the Cys-rich mature peptide sequence in the Fla-18 precursor. Comparing the nucleotide sequences of these O1-superfamily cDNAs showed that the shortening of the coding region of Fla-18 at the C terminus was actually caused by a 14-bp fragment deletion and the resulting premature termination of the coding region.
It is also interesting that, according to our transcriptome analysis, most conotoxin superfamilies have variable cysteine frameworks. In addition, the same cysteine framework may exist in different superfamilies. One notable example is that of cysteine framework VI/VII, which is present in V-, A-, Q-, and O1-superfamilies (Figs. 1B, 1C, 5A, and 6A). Although these conotoxins belonging to different superfamilies share the same cysteine framework, they have different lengths, cysteine codon usage, and sequences (supplemental Fig. S28). This suggests that these conotoxins evolved from different ancestors, supporting the argument that cysteine configuration is not phylogenetically relevant for conotoxins (11).
Conotoxin Diversification at the Protein Level
Our MS/MS analysis of C. flavidus venom not only identified 31 conotoxins, but also revealed two major strategies of conotoxin diversity at the protein level: alternative PTM and alternative cleavage. Alternative PTM was observed for proline hydroxylation (in the case of fla14a, fla14b, and fla14c and in the case of fla6a, fla6b, and fla6c), glutamate carboxylation (in fla01 versus fla02 and in fla3c versus fla3d), and even C-terminal amidation (in fla04 versus fla05). These alternative PTMs multiply the number of peptide products from the same precursor, thereby contributing to conotoxin diversity.
Our results also demonstrated that alternative cleavage, another efficient method for conotoxin diversity at the protein level, may occur more ubiquitously than previously thought. Alternative cleavage was observed in conomarphin, a cysteine-free conotoxin belonging to the M-superfamily (40). Here, we show that alternative cleavage occurs in several conotoxins belonging to the A-, O1-, T-, and Q-superfamilies (Table I). More interestingly, the alternative cleavage at the N terminus shows some sequence preference, with the cleavage site generally at the carboxyl side of Gly, or occasionally at the carboxyl side of Ala. However, the cleavage at the C terminus seems rather position-dependent. It is therefore highly likely that alternative cleavages at the two different termini of conotoxins are mediated by different enzymatic machineries. The proteolysis at the N terminus may be mediated by a signal peptidase-like enzyme, thus showing sequence preference for small side-chain residues (41), whereas the cleavage at the C terminus is possibly carried out by a carboxypeptidase (42). Further study would be required in order to identify these enzymes and their substrate selectivity.
In addition to the cleavages at the termini of conotoxins, the most surprising cleavage was observed in fla15a (Fig. 3 and Table I). This cleavage occurred in the middle of the predicted mature toxin region, rather than at the N or C terminus, producing a new cysteine framework relative to the framework in the precursor sequence. This result indicates that the cysteine framework of conotoxins can be changed not only at the gene level, but also at the protein level. This result further complicates the maturation process of conotoxins, as well as the mechanisms behind conotoxin diversity.
CONCLUSIONS
Our venomic study of C. flavidus, combining transcriptomic and proteomic analyses, identified 69 nonredundant conotoxin cDNA sequences, 31 conotoxin peptide sequences, and a new Q-superfamily. More importantly, we found that conotoxin-encoding transcripts can be diversified via hypermutation, fragment insertion/deletion, and mutation-induced premature termination, and that a single mRNA species can generate multiple toxin products through alternative PTMs and alternative cleavages of the translated precursor. The combination of these strategies at different levels is probably responsible for the structural diversity of conotoxins.
Supplementary Material
Acknowledgments
We thank Dr. Yong Zhang, Dr. Liang Lin and Dr. Jin Zi (Beijing Genomic Institute, Shenzhen, China) for the help in MS data acquisition. We are grateful to Dr. Si-Min He, Dr. Meng-Qiu Dong, Dr. Ping Xu and the pFind team members for their insightful discussions and advice. The cDNA sequences obtained via the RACE method in this work have been deposited in GenBank with the accession numbers KC582357–KC582377.
Footnotes
* This work was supported by the Chinese Ministry of Science and Technology (grants 2010CB529802 and 2012AA092201), a Specialized Research Fund for the Doctoral Program of Higher Education of China (SRFDP) (grant 20120072120031), and the Fundamental Research Funds for the Central Universities (grant 2011KJ039).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- bp
- base pair
- GO
- Gene Ontology
- HCD
- higher-energy C-trap dissociation
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1. Terlau H., Olivera B. M. (2004) Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 84, 41–68 [DOI] [PubMed] [Google Scholar]
- 2. Miljanich G. P. (2004) Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 11, 3029–3040 [DOI] [PubMed] [Google Scholar]
- 3. Lynch S. S., Cheng C. M., Yee J. L. (2006) Intrathecal ziconotide for refractory chronic pain. Ann. Pharmacother. 40, 1293–1300 [DOI] [PubMed] [Google Scholar]
- 4. Davis J., Jones A., Lewis R. J. (2009) Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 30, 1222–1227 [DOI] [PubMed] [Google Scholar]
- 5. Biass D., Dutertre S., Gerbault A., Menou J. L., Offord R., Favreau P., Stocklin R. (2009) Comparative proteomic study of the venom of the piscivorous cone snail Conus consors. J. Proteomics 72, 210–218 [DOI] [PubMed] [Google Scholar]
- 6. Laht S., Koua D., Kaplinski L., Lisacek F., Stocklin R., Remm M. (2012) Identification and classification of conopeptides using profile Hidden Markov Models. Biochim. Biophys. Acta 1824, 488–492 [DOI] [PubMed] [Google Scholar]
- 7. Kaas Q., Yu R., Jin A. H., Dutertre S., Craik D. J. (2012) ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40, D325–D330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duda T. F., Jr., Palumbi S. R. (1999) Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. U.S.A. 96, 6820–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conticello S. G., Gilad Y., Avidan N., Ben-Asher E., Levy Z., Fainzilber M. (2001) Mechanisms for evolving hypervariability: the case of conopeptides. Mol. Biol. Evol. 18, 120–131 [DOI] [PubMed] [Google Scholar]
- 10. Ye M., Khoo K. K., Xu S., Zhou M., Boonyalai N., Perugini M. A., Shao X., Chi C., Galea C. A., Wang C., Norton R. S. (2012) A helical conotoxin from Conus imperialis has a novel cysteine framework and defines a new superfamily. J. Biol. Chem. 287, 14973–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puillandre N., Koua D., Favreau P., Olivera B. M., Stocklin R. (2012) Molecular phylogeny, classification and evolution of conopeptides. J. Mol. Evol. 74, 297–309 [DOI] [PubMed] [Google Scholar]
- 12. Buczek O., Bulaj G., Olivera B. M. (2005) Conotoxins and the posttranslational modification of secreted gene products. Cell. Mol. Life Sci. 62, 3067–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pi C., Liu J., Peng C., Liu Y., Jiang X., Zhao Y., Tang S., Wang L., Dong M., Chen S., Xu A. (2006) Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics 88, 809–819 [DOI] [PubMed] [Google Scholar]
- 14. Hu H., Bandyopadhyay P. K., Olivera B. M., Yandell M. (2011) Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 12, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu H., Bandyopadhyay P. K., Olivera B. M., Yandell M. (2012) Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics 13, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pi C., Liu Y., Peng C., Jiang X., Liu J., Xu B., Yu X., Yu Y., Jiang X., Wang L., Dong M., Chen S., Xu A. L. (2006) Analysis of expressed sequence tags from the venom ducts of Conus striatus: focusing on the expression profile of conotoxins. Biochimie 88, 131–140 [DOI] [PubMed] [Google Scholar]
- 17. Haack J. A., Rivier J., Parks T. N., Mena E. E., Cruz L. J., Olivera B. M. (1990) Conantokin-T. A γ-carboxyglutamate containing peptide with N-methyl-D-aspartate antagonist activity. J. Biol. Chem. 265, 6025–6029 [PubMed] [Google Scholar]
- 18. Rigby A. C., Lucas-Meunier E., Kalume D. E., Czerwiec E., Hambe B., Dahlqvist I., Fossier P., Baux G., Roepstorff P., Baleja J. D., Furie B. C., Furie B., Stenflo J. (1999) A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx. Proc. Natl. Acad. Sci. U.S.A. 96, 5758–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakubowski J. A., Kelley W. P., Sweedler J. V. (2006) Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: an important component of conotoxin discovery. Toxicon 47, 688–699 [DOI] [PubMed] [Google Scholar]
- 20. Czerwiec E., Kalume D. E., Roepstorff P., Hambe B., Furie B., Furie B. C., Stenflo J. (2006) Novel gamma-carboxyglutamic acid-containing peptides from the venom of Conus textile. FEBS J. 273, 2779–2788 [DOI] [PubMed] [Google Scholar]
- 21. Wolfender J. L., Chu F., Ball H., Wolfender F., Fainzilber M., Baldwin M. A., Burlingame A. L. (1999) Identification of tyrosine sulfation in Conus pennaceus conotoxins alpha-PnIA and alpha-PnIB: further investigation of labile sulfo- and phosphopeptides by electrospray, matrix-assisted laser desorption/ionization (MALDI) and atmospheric pressure MALDI mass spectrometry. J. Mass Spectrom. 34, 447–454 [DOI] [PubMed] [Google Scholar]
- 22. Fainzilber M., Nakamura T., Lodder J. C., Zlotkin E., Kits K. S., Burlingame A. L. (1998) Gamma-conotoxin-PnVIIA, a gamma-carboxyglutamate-containing peptide agonist of neuronal pacemaker cation currents. Biochemistry 37, 1470–1477 [DOI] [PubMed] [Google Scholar]
- 23. Tayo L. L., Lu B., Cruz L. J., Yates J. R., 3rd. (2010) Proteomic analysis provides insights on venom processing in Conus textile. J. Proteome Res. 9, 2292–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ueberheide B. M., Fenyo D., Alewood P. F., Chait B. T. (2009) Rapid sensitive analysis of cysteine rich peptide venom components. Proc. Natl. Acad. Sci. U.S.A. 106, 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhatia S., Kil Y. J., Ueberheide B., Chait B. T., Tayo L., Cruz L., Lu B., Yates J. R., 3rd, Bern M. (2012) Constrained de novo sequencing of conotoxins. J. Proteome Res. 11, 4191–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Violette A., Biass D., Dutertre S., Koua D., Piquemal D., Pierrat F., Stocklin R., Favreau P. (2012) Large-scale discovery of conopeptides and conoproteins in the injectable venom of a fish-hunting cone snail using a combined proteomic and transcriptomic approach. J. Proteomics 75, 5215–5225 [DOI] [PubMed] [Google Scholar]
- 27. Dutertre S., Jin A. H., Kaas Q., Jones A., Alewood P. F., Lewis R. J. (2013) Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteomics 12, 312–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 29. Wang L. H., Li D. Q., Fu Y., Wang H. P., Zhang J. F., Yuan Z. F., Sun R. X., Zeng R., He S. M., Gao W. (2007) pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2985–2991 [DOI] [PubMed] [Google Scholar]
- 30. Li D., Fu Y., Sun R., Ling C. X., Wei Y., Zhou H., Zeng R., Yang Q., He S., Gao W. (2005) pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics 21, 3049–3050 [DOI] [PubMed] [Google Scholar]
- 31. Slysz G. DeconTools_Autoprocessor_v1.0.4672, released on Oct 16, 2012, from Biological MS Data and Software Distribution Center in Pacific Northwest National Laboratory, Richland, WA [Google Scholar]
- 32. Peng C., Liu L., Shao X., Chi C., Wang C. (2008) Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides 29, 985–991 [DOI] [PubMed] [Google Scholar]
- 33. Santos A. D., McIntosh J. M., Hillyard D. R., Cruz L. J., Olivera B. M. (2004) The A-superfamily of conotoxins: structural and functional divergence. J. Biol. Chem. 279, 17596–17606 [DOI] [PubMed] [Google Scholar]
- 34. Nakamura T., Yu Z., Fainzilber M., Burlingame A. L. (1996) Mass spectrometric-based revision of the structure of a cysteine-rich peptide toxin with gamma-carboxyglutamic acid, TxVIIA, from the sea snail, Conus textile. Protein Sci. 5, 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye M., Hong J., Zhou M., Huang L., Shao X., Yang Y., Sigworth F. J., Chi C., Lin D., Wang C. (2011) A novel conotoxin, qc16a, with a unique cysteine framework and folding. Peptides 32, 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 37. Medzihradszky K. F., Darula Z., Perlson E., Fainzilber M., Chalkley R. J., Ball H., Greenbaum D., Bogyo M., Tyson D. R., Bradshaw R. A., Burlingame A. L. (2004) O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol. Cell. Proteomics 3, 429–440 [DOI] [PubMed] [Google Scholar]
- 38. Yuan D. D., Han Y. H., Wang C. G., Chi C. W. (2007) From the identification of gene organization of alpha conotoxins to the cloning of novel toxins. Toxicon 49, 1135–1149 [DOI] [PubMed] [Google Scholar]
- 39. Kaas Q., Westermann J. C., Craik D. J. (2010) Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon 55, 1491–1509 [DOI] [PubMed] [Google Scholar]
- 40. Zhang L., Shao X., Chi C., Wang C. (2010) Two short D-Phe-containing cysteine-free conopeptides from Conus marmoreus. Peptides 31, 177–179 [DOI] [PubMed] [Google Scholar]
- 41. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 42. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.