Abstract
Recent efforts have attempted to describe the population structure of common chimpanzee, focusing on four subspecies: Pan troglodytes verus, P. t. ellioti, P. t. troglodytes, and P. t. schweinfurthii. However, few studies have pursued the effects of natural selection in shaping their response to pathogens and reproduction. Whey acidic protein (WAP) four-disulfide core domain (WFDC) genes and neighboring semenogelin (SEMG) genes encode proteins with combined roles in immunity and fertility. They display a strikingly high rate of amino acid replacement (dN/dS), indicative of adaptive pressures during primate evolution. In human populations, three signals of selection at the WFDC locus were described, possibly influencing the proteolytic profile and antimicrobial activities of the male reproductive tract. To evaluate the patterns of genomic variation and selection at the WFDC locus in chimpanzees, we sequenced 17 WFDC genes and 47 autosomal pseudogenes in 68 chimpanzees (15 P. t. troglodytes, 22 P. t. verus, and 31 P. t. ellioti). We found a clear differentiation of P. t. verus and estimated the divergence of P. t. troglodytes and P. t. ellioti subspecies in 0.173 Myr; further, at the WFDC locus we identified a signature of strong selective constraints common to the three subspecies in WFDC6—a recent paralog of the epididymal protease inhibitor EPPIN. Overall, chimpanzees and humans do not display similar footprints of selection across the WFDC locus, possibly due to different selective pressures between the two species related to immune response and reproductive biology.
Keywords: WFDC, natural selection, chimpanzees, serine protease inhibitor, reproduction, innate immunity
Introduction
Common chimpanzees and bonobos are different species of the Pan genus (Pan troglodytes and P. paniscus, respectively), separated by the geographical barrier of the Congo River. Common chimpanzees are further divided into subspecies across tropical Africa (Gonder et al. 1997, 2011). The issue of genomic diversity and substructure among the different chimpanzee subspecies is controversial and of great interest. Briefly, P. troglodytes was traditionally divided in three subspecies: P. t. verus, located in western Africa occupying the Upper Guinea region; P. t. troglodytes extending throughout central Africa; and P. t. schweinfurthii living in eastern Africa. Later, the analysis of mitochondrial DNA (mtDNA) variation led to the proposal of a fourth chimpanzee subspecies, P. t. ellioti (also known as P. t. vellerosus), occurring in the Gulf of Guinea (Nigeria and Cameroon) in a region limited by the Niger and Sanaga rivers (supplementary fig. S1, Supplementary Material online) (Patten and Unitt 2002; Becquet et al. 2007; Gonder et al. 2006, 2011). Recent studies support the differentiation of P. t. ellioti from P. t. troglodytes using ancestry-informative markers, enabling the identification of the four subspecies (Gonder et al. 2011, 2012). P. t. verus branched from the last common chimpanzee ancestor ∼0.46 Ma and P. t. ellioti diverged from P. t. troglodytes and P. t. schweinfurthii ∼0.32 Ma. Even though occasional hybridization occurs between P. t. ellioti and P. t. troglodytes in the wild, these subspecies remain as major genetic isolates (Gonder et al. 2011). Studies regarding chimpanzee simian immunodeficiency virus (SIVcpz) also support these findings, given that only P. t. troglodytes and P. t. schweinfurthii are infected in the wild (>30% prevalence) and P. t. ellioti only get infected when kept in captivity with P. t. troglodytes (Keele et al. 2006; Van Heuverswyn et al. 2007). SIVcpz is one of many infectious agents transferred to humans from chimpanzees, and this zoonotic infection likely provided the ancestor to the human immunodeficiency virus (HIV) (Jones et al. 2008). Therefore, a better characterization of episodes of natural selection in chimpanzees may provide ways to further understand susceptibility to pathogens in hominoids and to improve the conservation of wild chimpanzees.
Ecological changes in the natural habitat of P. troglodytes have served to shape evolutionary immune responses to pathogens and adaptive responses in reproduction-related phenotypes (Chimpanzee Sequencing and Analysis Consortium 2005). A genomic locus that is involved in both immune response and reproduction is the whey acidic protein (WAP) four-disulfide core domain (WFDC) locus (fig. 1). WFDC genes (17 in total) encode small serine protease inhibitors with functions of regulating endogenous proteases (Clauss et al. 2005; Lundwall 2007). Neighboring genes semenogelin 1 and 2 (SEMG1 and SEMG2) encode the main proteins of the seminal coagulum (Peter et al. 1998; de Lamirande 2007; Lundwall 2007). WFDC and SEMG genes evolved from the same common ancestor and maintain some similar functions involving antimicrobial, immune, and male reproduction activities (Yenugu et al. 2004; Bingle and Vyakarnam 2008; Clauss et al. 2011). Well-characterized genes at the WFDC locus include peptidase inhibitor 3 (PI3; also known as elafin) and secretory leucocyte proteinase inhibitor (SLPI), both pleiotropic molecules synthesized at mucosal surfaces that play a role in the surveillance against microbial and viral infections, including HIV-1 (Williams et al. 2006). This locus also includes the epididymal protease inhibitor EPPIN (also known as SPINLW1), which coats the surface of human spermatozoa, binds to SEMG1, and modulates the activity of prostate-specific antigen (PSA) altogether providing antimicrobial protection for spermatozoa (Wang et al. 2005; Edstrom et al. 2008; Zhao et al. 2008). SEMG1 and SEMG2 play critical roles in semen clotting and in antimicrobial and antiviral protection for spermatozoa in the female reproductive tract (Edstrom et al. 2008; Martellini et al. 2009). WFDC and SEMG genes have been shown to be targets of adaptive evolution in primates, where SEMGs dN/dS values were positively correlated with female promiscuity. Specifically, in monoandrous primates, in which females mate with a single male (e.g., humans, gorillas, and gibbons), the ejaculate is gelatinous, whereas in polyandrous primates, in which females mate with multiple partners per ovulatory period (e.g., chimpanzees and macaques), the ejaculate forms a rigid copulatory plug that prevents the insemination of females by competing males (Dixson and Anderson 2002; Dixson and Anderson 2004). In chimpanzees, the copulatory plug formation is associated with a SEMG1 modular-based length expansion causing an increase in protein crosslinking (Jensen-Seaman and Li 2003).
Fig. 1.—
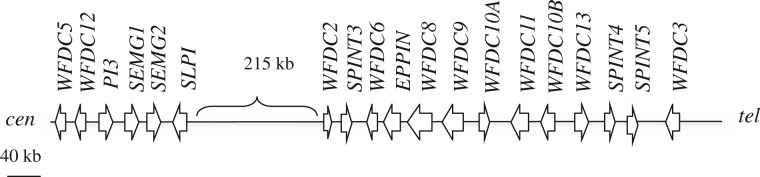
Schematic representation of the 20q13 WFDC locus, showing the relative positions of the WFDC genes. As depicted, the WFDC locus spans 700 kb and its genes are organized into two subloci (centromeric and telomeric; WFDC-CEN and WFDC-TEL, respectively), separated by 215 kb of unrelated sequence.
In human populations, the WFDC locus presents complex selective signals, including recent balancing selection on WFDC8 in Europeans and positive selection in SEMG1 in Asians (Ferreira et al. 2011; Ferreira et al. 2013). In order to evaluate the patterns of genomic variation and selection at the WFDC locus in chimpanzees, we sequenced 18 WFDC and SEMG genes and 47 control regions in 68 common chimpanzees from the subspecies P. t. troglodytes, P. t. verus, and P. t. ellioti. Overall, we generated a total of ∼13 Mb of high-quality sequence data, describing 1,268 single-nucleotide polymorphisms (SNPs), and we calculated summary statistics of population variation for 71 loci. We reconstructed the demographic history of chimpanzees and found a clear differentiation of P. t. verus from P. t. troglodytes and P. t. ellioti subspecies and in general, for WFDC genes we did not find departures from diversity levels observed in neutral evolving regions. Nevertheless, we identified a signature of strong selective constraints common to the three studied subspecies and centered in the EPPIN-like gene, WFDC6. In several primate species, WFDC6 has lost the ability to inhibit PSA and in others it appears to have accumulated different deleterious mutations. Conversely, in chimpanzees and humans, the seven disulfide bridges, known to confer antimicrobial properties to WFDC genes, are preserved in WFDC6. The fact that chimpanzees have a polyandrous mating system, and as a promiscuous species they are particularly likely to be exposed to sexually transmitted pathogens, leads us to propose that strong conservation of WFDC6 function has been necessary in chimpanzees due to its crucial role in innate immunity of the reproductive tract.
Materials and Methods
DNA Samples and Sequence Generation
The DNA samples include 68 wild-caught unrelated chimpanzees (Becquet et al. 2007; Gonder et al. 2006, 2011), including Central African subspecies, P. t. troglodytes (15 individuals), Western African subspecies, P. t. verus (22 individuals), and the Gulf of Guinea subspecies, P. t. ellioti (31 individuals) (supplementary table S1, Supplementary Material online). We studied the genetic variation in the WFDC locus by Sanger sequencing the coding regions of 18 WFDC and SEMG genes (comprising a total of 66 exons) and a number of intervening noncoding regions (spaced every ∼10 kb). Additionally, we sequenced 47 pseudogenes located in unrelated, neutrally evolving regions across the chimpanzee genome, used as control regions, as previously described (Andrés et al. 2010; Ferreira et al. 2013) (supplementary table S2, Supplementary Material online).
Primers for amplification and sequencing were designed based on the Human Genome Reference Sequence (March 2006 assembly - v36.1), available at the UCSC Genome Browser (genome.ucsc.edu, last accessed December 24, 2013). All samples were polymerase chain reaction (PCR)-amplified and analyzed by bidirectional Sanger sequencing. Further details about PCR and DNA sequencing are available from the authors upon request. The sequences were aligned to the Human Genome v26.1 and polymorphic sites and fixed differences were detected with phred-phrap-consed package (Nickerson et al. 1997). To ensure sequence quality, we discarded variant sites in the first and last 75 bp of each amplicon segment. We manually curated sites found to have discordant genotypes in different amplicons. The ancestral state of each SNP was inferred by comparison with the human, orangutan, and macaque genome sequences (Gibbs et al. 2007; Andrés et al. 2010; genome.ucsc.edu).
Statistical Analysis
We assessed the subspecies differentiation levels by calculating the population differentiation (FST statistic) and by performing principal component analysis (PCA) on the SNP data (Excoffier 2002; Patterson et al. 2006). We used a locus-by-locus analysis of molecular variance, using 20,000 simulations, which was performed by Arlequin using its default values (constant model; Excoffier et al. 2005). The EIGENSOFT software package was used for PCA (Patterson et al. 2006). We performed cluster analysis using STRUCTURE version 2.3 (Pritchard et al. 2000), assuming admixture and correlated allele frequencies. Fifty iterations of the data at each K = 1–5 with 500,000 Markov chain Monte Carlo (MCMC) burn-in steps and 500,000 MCMC iterations. STRUCTURE output was processed with CLUMPP and plotted with DISTRUCT (Conrad et al. 2006). We used STRUCTURE harvester to determine the best K estimate. Population structure analyses were performed blinded to a priori population labels.
A model of chimpanzee demography was inferred using BP&P program which implements Bayesian inference with a MCMC and accommodates a species phylogeny as well as lineage sorting due to ancestral polymorphism (Rannala and Yang 2003; Yang and Rannala 2010). We used a proposed phylogenetic tree of the three chimpanzee subspecies (Gonder et al. 2011) and included human as the outgroup. A gamma prior G(2, 1,000), with mean 2/2,000 = 0.001, was used on the population size parameters (θs), and the age of the root in the species tree (τ0) was assigned to the gamma prior G(25, 5,000). This was based on the assumption of divergence time between chimpanzee and humans of 5 Myr and a mutation rate of 10−9 per site/per year (2 × 10−8 per site per 20-year generation). The other divergence time parameters were assigned to the Dirichlet prior (Yang and Rannala, 2010). The analyses were run twice to confirm consistency, and parameters of historical demographic events are expressed as the mean and 95% confidence intervals of the posterior distributions.
Summary statistics of population genetic variation were calculated using SLIDER (http://genapps.uchicago.edu/slider/index.html, last assessed December 24, 2013). We assessed statistical significance of summary statistics using an empirical comparison to the control regions, by calculating the upper and lower 2.5 percentiles of each distribution. Specifically, we used the sequenced control regions to perform an empirical comparison of nucleotide diversity (π) and Tajima’s D values for each WFDC gene in each population. For the WFDC genes, we ran 105 coalescent simulations using “ms” (Hudson 2002) and mutation rate parameters estimated from the sequenced data with SLIDER. For the population recombination parameter, we used the PANMAP estimates of chimpanzee recombination. We assumed demographic models that included constant population size, and historic events as previously described (Hey 2010; Wegmann and Excoffier 2010), and as inferred by us for each subspecies. For every model, we calculated a null distribution of summary statistics values and calculated the 2.5th and 97.5th percentiles.
We performed HKA tests considering all subspecies in DNAsp 5.1 and using a maximum-likelihood method that incorporates values for multiple neutrally evolving regions (Hudson et al. 1987). McDonald–Kreitman test (MKT) was calculated in DNAsp v5.1 using humans as the outgroup and assuming two types of sites: putatively neutral sites (Syn) and functional sites (NSyn) (Rozas and Rozas 1995; Rozas 2009).
Haplotype phasing for all samples was inferred separately for the WFDC centromeric and telomeric subloci (WFDC-CEN and WFDC-TEL, respectively; see fig. 1 for WFDC locus substructure) using PHASE2.1 (Stephens et al. 2001; Stephens and Donnelly 2003). Haplotypes were independently inputted in Haploview 4.2 (Barrett 2009) to calculate linkage disequilibrium (LD) statistics, r2 and D′, and to identify LD and haplotype blocks (Gabriel et al. 2002). The potential functional effects at the protein level of non-synonymous (NSyn) SNPs and fixed differences were inferred using PolyPhen v2 (Adzhubei et al. 2010) and SIFT (Kumar et al. 2009).
Maximum-likelihood estimates of dN/dS (ω; dS – synonymous substitution rate and dN – non-synonymous substitution rate) were carried out using the codeml program from the software package Phylogenetic Analysis by Maximum Likelihood – PAML version 4.2 (Yang 2007). To run PAML, we first reconstructed a phylogenetic tree (DNAml from Phylogeny Inference Package [PHYLIP]; http://evolution.genetics.washington.edu/phylip.html, last accessed December 24, 2013). We used the genomic sequences from human (Homo sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), gibbon (Nomascus leucogenys), rhesus monkey (Macaca mulatta), baboon (Papio anubis), and marmoset (Callithrix jacchus). They were retrieved from public databases using EPPIN isoform 1 (Uniprot: O95925) and WFDC6 isoform 1 (Uniprot: Q9BQY6) as BLAT templates. Pan paniscus was not included in the analysis as the cDNA sequence is equal to P. troglodytes. The phylogenetic tree diverged from the known primate phylogeny in the position of orangutan and gibbon branches. To test for variable selection pressures among branches, we performed the branch model using either the null model (one ω ratio for the entire tree) or nested models (two-ratios, three-ratios, four-ratios for the tree) (Yang 1997; Bielawski and Yang 2003). The values of ω > 1 were considered as evidences of positive selection, and the values ω < 1 were considered as an indication of purifying selection. The test statistic was constructed as twice the difference in the log of the likelihoods (−2Δl), and significance was assessed by comparing this to the χ2 statistic.
Results
We sequenced with Sanger technology 68 chimpanzees (supplementary table S1, Supplementary Material online) for all WFDC and SEMG exons distributed across 54 amplicons and 47 neutrally evolving control regions, for a total of 13 Mb (supplementary table S2, Supplementary Material online). This resulted in the identification of 419 SNPs in the control regions and 849 SNPs in the WFDC locus.
Chimpanzee Genetic Structure and Demography
We first characterized the levels of differentiation between subspecies at the control regions using FST (Excoffier 2002) and PCA (Patterson et al. 2006). The pairwise FST values show lower differentiation between P. t. troglodytes and P. t. ellioti (0.104) than each of them compared with P. t. verus (0.392 and 0.400, respectively). The first two principal components (PC1 and PC2) separate P. t. verus from the other two subspecies, and the third principal component appears to not separate completely P. t. troglodytes from P. t. ellioti (supplementary fig. S2, Supplementary Material online). Contrary to previous studies, we could not separate the three subspecies using PCA (Gonder et al. 2011; Bowden et al. 2012). We further examined the shared ancestry levels of the individuals by performing a Bayesian model-based clustering approach available in the STRUCTURE software (Pritchard et al. 2000; Falush et al. 2003). The analysis was performed blinded to a population label and two groups (K = 2) were recovered (supplementary fig. S3, Supplementary Material online). The subspecies P. t. troglodytes and P. t. ellioti could not be confidently distinguished even after the inclusion of the 849 SNPs from the WFDC locus (results not shown).
We inferred the demographic history of the three chimpanzee subspecies from control regions data using a Bayesian MCMC approach, based on the phylogenetic tree proposed by previous studies (Gonder et al. 2011; Bowden et al. 2012). The estimated effective population sizes were ∼52,000 and ∼21,000 for P. t. troglodytes and P. t. verus, respectively (table 1). These values are within the range of previous models of chimpanzee demography as indicated by the overlaps between confidence intervals (table 1), and the 0.31 Myr estimate for divergence from P. t. verus is in the same time frame from previous studies (table 1; Becquet et al. 2007; Hey 2010; Wegmann and Excoffier 2010; Gonder et al. 2011). For P. t. ellioti, we estimated a recent divergence from P. t. troglodytes around 0.173 Ma and an effective size of ∼43,000 individuals, demonstrating values that were of similar order of magnitude to P. t. troglodytes. These two subspecies appear to have preserved a similar effective population size to their ancestral population (fig. 2 and table 1). Conversely, the origin of P. t. verus is associated with a bottleneck and effective population size reduction (fig. 2), providing a good fit to previous models of chimpanzee demography (Becquet et al. 2007; Hey 2010; Wegmann and Excoffier 2010; Gonder et al. 2011).
Table 1.
Estimated Parameters Using the 47 Control Regions
| Current Studya | Gonder et al. (2011) | Wegmann and Excoffier (2010)b | Hey (2010)a | Bequet (2007)a | |
|---|---|---|---|---|---|
| NPtt | 51,975 (19,737–69,162) | — | 134,900 (75,900–251,200) | 26,900 (16,100–43,900) | 23,100 (8,600–59,700) |
| NPte | 43,512 (19,925–56,175) | — | — | — | — |
| NPtv | 21,062 (16,500–27,637) | — | 9,800 (5,000–72,400) | 7,400 (5,400–10,000) | 10,100 (7,700–21,100) |
| NAPtt-Pte | 57,412 (18,762–126,287) | — | — | — | — |
| NAPtt-Pte-Ptv | 70,525 (58,850–91,900) | — | — | — | — |
| NAPan | 22,787 (1,025–47,175) | — | 89,100 (36,300–245,500) | 7,100 (3,500–12,500) | 32,900 (22,200–48,700) |
| TDIVHomoPan (Myr) | 6.58 (4.34–8.54) | — | — | — | — |
| TDIV Ptt-Pte-Ptv (MY) | 0.31 (0.236–0.405) | 0.46 (0.37–0.53) | 0.55 (0.34–0.91) | 0.46 (0.35–0.65) | 0.44 (0.32–1.10) |
| TDIV Ptt-Pte (Myr) | 0.173 (0.034–0.237) | 0.11 (0.09–0.13) | — | — | — |
NPtt, P. t. troglodytes effective population size; NPte, P. t. ellioti effective population size; NPtv, P. t. verus effective population size; NAPtt-Pte, ancestral effective population size of P. t. troglodytes and P. t. ellioti; NAPtt-Pte-Ptv, ancestral effective population size of the three subspecies; NAPan, ancestral effective population size of common chimpanzee; TDIVHomoPan, human–chimpanzee divergence time in million years; TDIV Ptt-Pte-Ptv, P. t. verus divergence time from P. t. troglodytes and P. t. ellioti; TDIV Ptt-Pte, P. t. troglodytes and P. t. ellioti divergence time in million years.
aConfidence intervals are 95% highest posterior density intervals.
bConfidence intervals are 90% highest posterior density intervals.
Fig. 2.—
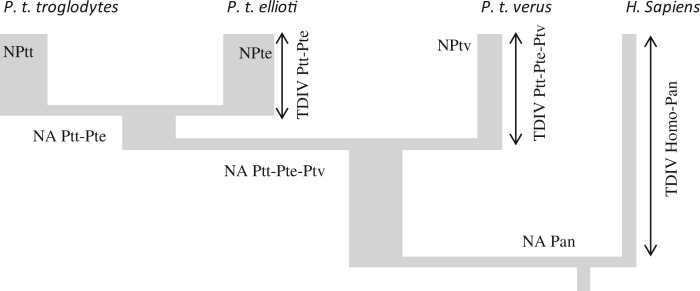
Schematic representation of the inferred demographic history of the three subspecies: Pan troglodytes troglodytes (Ptt); P. t. verus (Ptv); and P. t. ellioti (Pte). NA, ancestral effective population size; N, effective population size; TDIV, divergence time.
WFDC Locus Sequence Diversity
The patterns of variation in SEMG1 among individuals include a polymorphic highly repetitive region (9–13 modules encoded by SEMG1 exon 2) (Jensen-Seaman and Li 2003). Because the modular nature of SEMG1 precluded a consistent and unambiguous sequence alignment, the SNPs located in this repetitive region were removed from the analysis for quality purposes. A total of 766 fixed differences were identified in comparison with the human genome reference. Twenty-five indels (insertions/deletions) were found, 24 of which were located in introns, UTRs, and intergenic regions. One remaining indel located in WFDC6 was identified in a single chromosome (f = 0.02). Indels were excluded from all analyses, due to their distinct mutation rate and the low overlap with functional regions, which are unlikely to affect protein function or expression. Additionally, from the 456 human–chimpanzee fixed differences located in the WFDC locus, only 19 were within coding regions and chimpanzee-specific. Of these, 17 were non-synonymous (NSyn) and most of them were classified as benign by Polyphen v2 and SIFT (supplementary table S3, Supplementary Material online).
To characterize the within-subspecies variation, we analyzed the folded site frequency spectrum (SFS) for all SNPs and Syn and NSyn sites among WFDC genes (fig. 3). Additionally, we analyzed the deleterious effects of each coding substitution using SIFT and Polyphen (Kumar et al. 2009; Adzhubei et al. 2010; supplementary table S3, Supplementary Material online). Despite the higher number of NSyn sites in the WFDC genes, these are maintained at low frequencies in the overall species, consistent with the predicted mildly deleterious effects (supplementary table S3, Supplementary Material online). For each WFDC gene, we calculated summary statistics such as nucleotide diversity (π), Tajima’s D (Tajima 1989), Fu and Li’s D (Fu and Li 1993), Fay and Wu’s H (Fay and Wu 2005; Zeng et al. 2006), and HKA (Hudson et al. 1987) (table 2).
Fig. 3.—
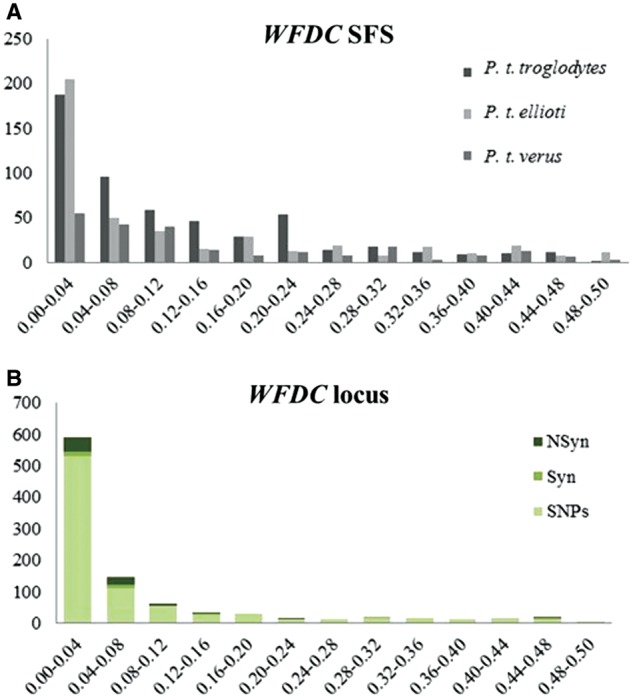
Folded SFS for the species that were resequenced. The x axis depicts the frequency of the allele frequency bin in the generated data set, whereas the y axis represents the number of alleles found within each frequency bin. Syn, synonymous changes; NSyn, nonsynonymous changes. (A) Folded SFS in WFDC locus; (B) folded SFS of WFDC locus highlighting coding mutations.
Table 2.
Summary Statistics for All the WFDC Genes
| Gene | Subspecies | L | S | π (10−4) | θw | D | D* | H | P(HKA) |
|---|---|---|---|---|---|---|---|---|---|
| WFDC5 | P. t. troglodytes | 5,536 | 32 | 11.87 | 8.077 | −0.4202 | −0.3432 | −1.6644 | 0.3858 |
| WFDC12 | P. t. troglodytes | 1,323 | 16 | 3.026 | 4.039 | −0.6318 | 0.4407 | −0.6437 | 0.1866 |
| PI3 | P. t. troglodytes | 3,377 | 25 | 7.748 | 6.310 | −0.3775 | 0.1365 | 1.5448 | 0.9625 |
| SEMG1 | P. t. troglodytes | 3,305 | 23 | 4.015 | 5.806 | −1.172 | −0.8307 | −1.2782 | 0.7463 |
| SEMG2 | P. t. troglodytes | 4,324 | 31 | 8.471 | 7.825 | −0.8778 | 0.2350 | −2.4092 | 0.9241 |
| SLPI | P. t. troglodytes | 4,709 | 30 | 16.84 | 7.573 | 0.5078 | 0.6260 | 2.5195 | 0.9095 |
| WFDC2 | P. t. troglodytes | 3,984 | 24 | 5.850 | 6.058 | −0.7272 | 0.3391 | −3.6322 | 0.4648 |
| SPINT3 | P. t. troglodytes | 3,727 | 35 | 10.72 | 8.835 | −0.8613 | −0.9377 | 14.278 | 0.0604 |
| WFDC6 | P. t. troglodytes | 2,807 | 19 | 1.269 | 4.796 | −2.073** | −2.290 | −1.7839 | 0.1907 |
| EPPIN | P. t. troglodytes | 3,233 | 23 | 2.356 | 5.806 | −1.811* | −2.218 | 5.2828 | 0.1362 |
| WFDC8 | P. t. troglodytes | 7,179 | 36 | 9.139 | 9.087 | −1.169 | −1.054 | 0.5977 | 0.5583 |
| WFDC9/10A | P. t. troglodytes | 6,863 | 44 | 16.64 | 11.11 | −0.8419 | −0.8662 | 2.0736 | 0.2643 |
| WFDC11 | P. t. troglodytes | 5,037 | 58 | 37.48 | 14.64 | −0.3621 | −0.1044 | 9.0184 | 0.4493 |
| WFDC10B/13 | P. t. troglodytes | 7,365 | 41 | 13.99 | 10.35 | −0.9045 | −0.8869 | −5.0023 | 0.882 |
| SPINT4 | P. t. troglodytes | 3,527 | 14 | 2.296 | 3.534 | −0.7060 | −0.1432 | −1.9862 | 0.9805 |
| WFDC3 | P. t. troglodytes | 7,572 | 51 | 20.48 | 12.87 | −0.9501 | −0.6436 | −0.5163 | 0.4458 |
| WFDC5 | P. t. ellioti | 5,536 | 24 | 9.751 | 5.110 | 0.8670 | 0.4099 | −1.9799 | 0.1653 |
| WFDC12 | P. t. ellioti | 1,323 | 7 | 1.277 | 1.491 | 0.8178 | 0.4173 | −0.0063 | 0.9754 |
| PI3 | P. t. ellioti | 3,377 | 14 | 3.947 | 2.981 | 0.9111 | −0.4956 | 1.1021 | 0.3997 |
| SEMG1 | P. t. ellioti | 3,305 | 28 | 3.491 | 5.962 | −1.250 | −0.5904 | −0.9815 | 0.321 |
| SEMG2 | P. t. ellioti | 4,324 | 22 | 2.378 | 4.685 | −1.193 | −0.4544 | −0.1861 | 0.2107 |
| SLPI | P. t. ellioti | 4,709 | 30 | 5.464 | 6.388 | −0.8478 | −1.875 | 4.6113 | 0.9302 |
| WFDC2 | P. t. ellioti | 3,984 | 25 | 7.905 | 5.323 | 0.2832 | −0.1960 | 5.2226 | 0.5012 |
| SPINT3 | P. t. ellioti | 3,727 | 14 | 3.866 | 2.981 | 0.8654 | 0.5262 | 0.7953 | 0.2557 |
| WFDC6 | P. t. ellioti | 2,807 | 13 | 1.364 | 2.768 | −0.7455 | −1.177 | −1.6838 | 0.3477 |
| EPPIN | P. t. ellioti | 3,233 | 23 | 3.936 | 4.898 | −0.6386 | −0.0079 | 12.0571 | 0.054 |
| WFDC8 | P. t. ellioti | 7,179 | 29 | 5.759 | 6.175 | −0.6888 | −1.990 | 4.7842 | 0.3548 |
| WFDC9 10A | P. t. ellioti | 6,863 | 28 | 10.18 | 5.962 | 0.3822 | 0.9311 | 3.1793 | 0.0392 |
| WFDC11 | P. t. ellioti | 5,037 | 42 | 15.62 | 8.943 | −0.1915 | −0.1990 | 4.7848 | 0.2656 |
| WFDC10B | P. t. ellioti | 7,365 | 29 | 6.956 | 6.175 | −0.4046 | −0.2142 | 0.055 | 0.2968 |
| SPINT4 | P. t. ellioti | 3,527 | 14 | 0.8043 | 2.981 | −1.498*** | −1.007 | 4.3977 | 0.8262 |
| WFDC3 | P. t. ellioti | 7,572 | 59 | 14.79 | 12.56 | −1.182 | −0.8554 | 15.3739 | 0.7554 |
| WFDC5 | P. t. verus | 5,536 | 14 | 1.743 | 3.218 | −0.8086 | 0.6041 | 9.148 | 0.4733 |
| WFDC12 | P. t. verus | 1,323 | 7 | 1.396 | 1.609 | 0.7787 | −1.027 | −0.8393 | 0.1127 |
| PI3 | P. t. verus | 3,377 | 7 | 1.979 | 1.609 | 1.6256 | 0.4908 | 0.8076 | 0.6147 |
| SEMG1 | P. t. verus | 3,305 | 20 | 2.323 | 4.598 | −1.2464 | −0.8163 | −1.4884 | 0.0296 |
| SEMG2 | P. t. verus | 4,324 | 9 | 0.883 | 2.069 | −0.7344 | −1.207 | −0.5666 | 0.2473 |
| SLPI | P. t. verus | 4,709 | 14 | 2.797 | 3.218 | −0.0454 | 1.071 | 0.8203 | 0.8173 |
| WFDC2 | P. t. verus | 3,984 | 3 | 0.212 | 0.690 | −0.4387 | −0.3775 | 0.7653 | 0.3611 |
| SPINT3 | P. t. verus | 3,727 | 9 | 1.865 | 2.069 | 0.5711 | 0.0750 | 3.3425 | 0.6688 |
| WFDC6 | P. t. verus | 2,807 | 9 | 0.629 | 2.069 | −1.1725 | −0.5657 | −1.0973 | 0.0616 |
| EPPIN | P. t. verus | 3,233 | 7 | 1.523 | 1.609 | 0.9748 | −0.2682 | −0.4207 | 0.2532 |
| WFDC8 | P. t. verus | 7,179 | 7 | 0.815 | 1.609 | −0.2537 | 0.4908 | −1.1501 | 0.0954 |
| WFDC9/10A | P. t. verus | 6,863 | 22 | 3.333 | 5.057 | −1.002 | −0.2720 | 3.2896 | 0.4979 |
| WFDC11 | P. t. verus | 5,037 | 25 | 7.986 | 5.747 | 0.0283 | −0.3329 | 2.408 | 0.0605 |
| WFDC10B/13 | P. t. verus | 7,365 | 14 | 3.356 | 3.218 | 0.3020 | 1.071 | 0.4249 | 0.4759 |
| SPINT4 | P. t. verus | 3,527 | 6 | 1.062 | 1.379 | 0.6795 | −0.4940 | −0.4989 | 0.856 |
| WFDC3 | P. t. verus | 7,572 | 32 | 17.27 | 7.356 | 0.6888 | 0.3704 | 12.222 | 0.8309 |
L, length sequenced (bp); S, number of segregating sites; π, nucleotide diversity per base pair (× 10−4); θw, Watterson’s estimator of Ө (4Neµ) (Watterson 1975) per base pair (× 10−4); D, Tajima’s D statistic (Tajima 1989); D*, Fu and Li’s D* test (Fu and Li 1993); H, Fay and Wu H test (Fay et al. 2002; Zeng et al. 2006); P(HKA), HKA test P value (Hudson et al. 1987).
*P value ≤0.025 using three different demographic models (constant size, our best-fit model, and Hey 2010).
**P value <0.01 using three different demographic models (constant size, our best-fit model, and Hey 2010).
***P value ≤0.025 using our best-fitting model.
Analysis of the SFS and summary statistics show P. t. troglodytes as the subspecies with the highest nucleotide diversity levels and P. t. verus as the most homogeneous subspecies (supplementary fig. S4, Supplementary Material online, and table 2). Both P. t. troglodytes and P. t. ellioti Tajima’s D values are skewed toward negative values, mostly due to their large effective population size and a population expansion that is estimated to have occurred around 50,000 years ago (Wegmann and Excoffier 2010). Nonetheless, P. t. verus Tajima’s D values are less negative than the other two subspecies, which is likely to result from an extreme decrease in population size and genetic drift (Caswell et al. 2008; Hey 2010; Wegmann and Excoffier 2010). Overall, the analysis of the summary statistics of WFDC genes shows no widespread significant departure from neutrality but instead reveals only mildly negative or positive values (table 2).
Selection Tests
To determine whether specific WFDC genes have been under selective pressures in one or all chimpanzee subspecies, we started by comparing the summary statistics for each WFDC gene with the empirical distribution of Tajima’s D in the control regions (fig. 4). Only WFDC6 and EPPIN show unusual patterns in P. t. troglodytes (fig. 4). Although in this subspecies the allele frequency spectrum is generally skewed toward rare alleles, the Tajima’s D values of WFDC6 (−2.073; P value = 1−4) and EPPIN (−1.811; P value = 0.025) present the lowest values of control and WFDC regions. WFDC6 also presented a low Tajima’s D value (−2.1039) and significant HKA (P = 0.013) when combining all individuals sequenced (supplementary table S4, Supplementary Material online). The other WFDC genes did not show strong significant P values pointing to a neutral evolution based on subspecies genetic diversity (table 2). To confirm WFDC departures from neutrality, we performed 105 coalescent simulations for each subspecies under different demographic scenarios: constant model (P. t. troglodytes, P. t. ellioti, P. t. verus), our best-fit model (P. t. troglodytes, P. t. ellioti, P. t. verus), Hey 2010 model (P. t. troglodytes), and Wegmann and Excoffier 2010 model (P. t. verus). WFDC6 and EPPIN present significantly negative Tajima’s D value compared with all models, but while WFDC6 shows always values below 1st percentile, EPPIN values lie between the 1st and 2.5th percentile (table 2).
Fig. 4.—
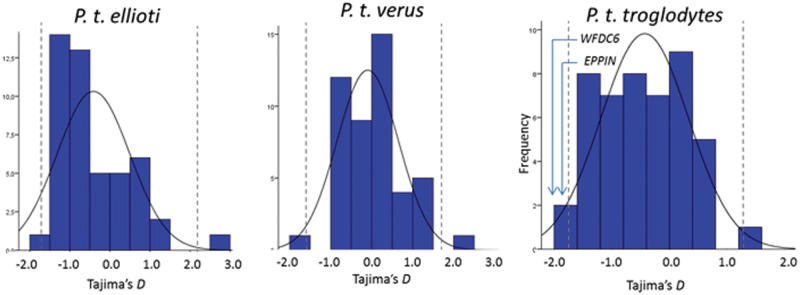
Empirical comparisons generated from the 47 control regions. Tajima’s D (Tajima 1989) was calculated for each region using SLIDER and plotted with the 2.5 and 97.5 percentiles represented as dashed lines.
The hypothesis of a recent positive selection was excluded due to the absence of LD blocks and homogeneous haplotypes in P. t. troglodytes (supplementary fig. S5, Supplementary Material online), which prevents long-range haplotype tests from being calculated. To address the hypothesis of an older selective sweep, we performed then MKT, which did not show departures from neutrality in either WFDC6 or EPPIN (results not shown). Notwithstanding, FST statistic in the WFDC6–EPPIN region is the lowest in the WFDC locus (supplementary fig. S6, Supplementary Material online), and the networks built to assess the WFDC6 and EPPIN haplotype structure show that WFDC6 has a star-shaped genealogy shared among all subspecies (fig. 5 and supplementary fig. S7, Supplementary Material online). The findings show that in WFDC6 all NSyn variants are maintained a very low frequencies and that the fixed difference K79E, predicted to alter protein function in EPPIN, is also present in bonobos (>1 Ma). This suggests strong purifying selection as the likeliest cause for WFDC6 patterns of diversity.
Fig. 5.—
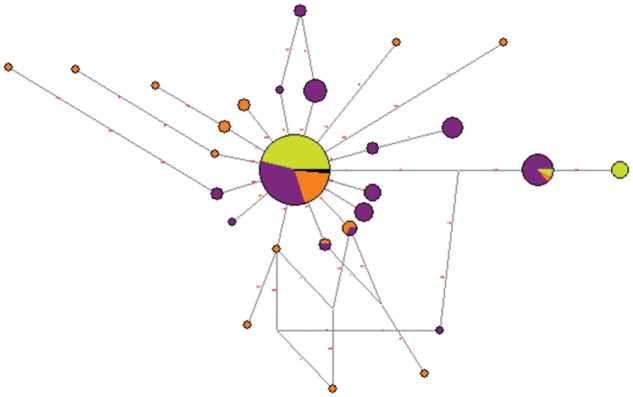
Inferred haplotype network at the WFDC6. Each circle represents a unique haplotype, and its area is proportional to its frequency. Within each circle, Pan troglodytes verus, P. t. ellioti, and P. t. troglodytes are labeled in green, purple, and orange, respectively. The mutations that differentiate each haplotype are shown along each branch.
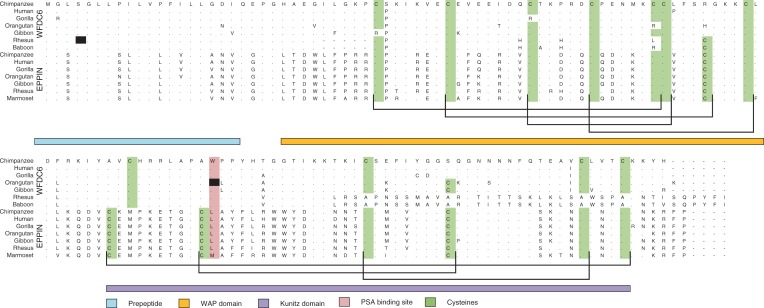
To determine the levels of selective constraints operating at WFDC6 and EPPIN, we aligned the publicly available sequences of both genes for eight primate species (chimpanzee, human, gorilla, orangutan, gibbon, rhesus monkey, baboon, and marmoset). The alignment shows that EPPIN has been conserved in all the species. WFDC6 has released constraints with signals of pseudogenization in orangutan, rhesus, and baboon and appears to be absent in marmoset (fig. 6). Evidence for WFDC6 pseudogenization includes a premature stop codon (W86X) in orangutan, a very early stop codon (S4X) and a five amino acid deletion (28 to 32) in rhesus monkey, and a frameshift mutation (T99fs139X) shared between rhesus and baboon. Also striking is the loss of two disulfide bridges in the Kunitz domain for all the primates, the species-specific loss of one disulfide bridge in the WAP domain from gorilla and gibbon, and the loss of the SEMG1 binding residue in gibbon (C102A). Furthermore, the active site conferring PSA inhibitory activity to EPPIN in WFDC6 was modified from a leucine to a tryptophan (residue 87) in most primates and to a stop codon in orangutan (fig. 6).
Fig. 6.—
Amino acid alignment of WFDC6 and EPPIN. Cysteines are marked in light green; PSA binding site is marked in pink; and disulfide bridges are marked in black lines. Black squares represent stop codons.
We calculated dN/dS (ω) ratios for the paralogs WFDC6 and EPPIN, under alternative models of gene evolution, for the entire sequence data set after the exclusion of WFDC6 pseudogenes (orangutan, rhesus, and baboon sequences). In cases where no selection is operating, ω should be equal to 1, greater than 1 when purifying selection is acting to preserve protein sequence, and significantly exceed 1 when positive selection is acting to drive divergence of protein sequence. We estimated a single ω value for the entire phylogeny (one-ratio), in which we assumed no differentiation in WFDC6 and EPPIN selective pressures. The observed value (ω = 0.4739) is lower than one suggesting an overall conservation of WFDC6 and EPPIN (supplementary fig. S8 and table S5, Supplementary Material online) (Yang 2007). As these two proteins are very similar, we examined whether the two paralogs have been subject to different selective constraints and applied the two-ratio model, allowing the branches that correspond to WFDC6 and to EPPIN clades to have distinct ω values. The ω value for WFDC6 was close to 1 (ωWFDC6 = 0.8846) and almost two times higher than that for EPPIN (ωEPPIN = 0.4738), but the model fit did not differ significantly from the one-ratio model (−2Δl = 1.04; P value = 0.35). To determine whether WFDC6 was under different selective constraints in chimpanzees, we performed two more tests: a three-ratio model, where we define the human and chimpanzee and their ancestor as one clade, and another three-ratio model, where we define only the chimpanzee WFDC6 as an independent clade. Although our results suggest that WFDC6 might have experienced very different selective pressures, as shown by the human–chimpanzee ωancWFDC6 = 1.1656 and by the chimpanzee ωWFDC6 = 0.5886, none of the new tests indicate a significant departure from the two-ratio model (supplementary table S5, Supplementary Material online). Note that the lack of significance is likely due to the limited statistical power of this small data set (only 131 codons).
Discussion
Here, we studied the sequence diversity at the WFDC locus and 47 neutrally evolving regions chosen to control for demographic effects in three P. troglodytes subspecies, P. t. troglodytes, P. t. ellioti, and P. t. verus. In our data set, we inferred the strength of the selective pressures acting in WFDC locus after retrieving the structural and geographical differentiation of the three chimpanzee subspecies. This analysis shows that P. t. verus is the least diverse subspecies and a clear defined genetic entity, while the recently separated subspecies P. t. ellioti and P. t. troglodytes are more diverse and hardly discriminated even with a set of 1,268 autosomal SNPs. In the WFDC locus, we pinpointed a single selective signal, which has a high degree of interpopulation homogeneity and identifies WFDC6 as a gene under purifying selection in chimpanzees. We hypothesize that these selective constraints were driven by a response to sexually transmitted pathogens, as P. troglodytes is a promiscuous species and gets infected by a plethora of infectious diseases in the wild.
The contribution of our data to the complex question of chimpanzee demographic history is significant when considering the first model of P. t. ellioti. Even though P. t. ellioti and P. t. troglodytes were hardly discriminated as two distinct populations, we were capable to reconstruct the demographic history of the three chimpanzee subspecies. We detected a consistent differentiation of P. t. verus and confirmed that the nucleotide diversity of P. t. verus is more similar to humans. The low differentiation of P. t. troglodytes and P. t. ellioti is consistent with their recent divergence 0.173 Ma and their larger effective population sizes (>40,000 diploids). A history of population expansion in P. t. troglodytes and P. t. ellioti is plausible if a subdivision of ancestral population (Ne = 57,412) is considered, and this could explain the negative Tajima’s D trend in both subspecies (Fischer et al. 2004; Won and Hey 2005; Fischer et al. 2006; Caswell et al. 2008; Wegmann and Excoffier 2010; Gonder et al. 2011; Bowden et al. 2012).
The signatures of selection identified in the human WFDC locus are mainly associated with homogeneous long-range haplotypes and variants located at SEMG1 (Asians), WFDC8 (Europeans), and SPINT4 (Africans), indicating a recent increase in the frequency by selection and not by demographic events (Ferreira et al. 2011; Ferreira et al. 2013). In chimpanzees, we assessed the signatures of positive selection by comparing the WFDC genes with the empirical distribution built from 47 neutrally evolving regions and with simulated null distributions of chimpanzee demography. Even though we could not detect LD blocks or extended haplotypes in our sequenced data, we found lower levels of nucleotide diversity in WFDC6 and EPPIN genes while compared with other chimpanzee loci. The significantly negative HKA P value obtained for the total sample set together with the low subspecies differentiation indicated by FST and the low frequencies of NSyn variants is suggestive of a signature of an old event of purifying selection in WFDC6 and EPPIN in P. troglodytes.
To our knowledge, no experimental studies were performed to determine WFDC6 biological functions. However, WFDC6 is considered a recent paralog of EPPIN with 71% sequence similarity and sharing of the same protein functional domains (WAP and Kunitz). EPPIN is known to protect SEMG1 from premature cleavage by its natural protease, PSA, a protease inhibitor activity conferred by L87 residue (P1 reactive site) located in the Kunitz domain. Other recognized roles of EPPIN are its antimicrobial and antiviral activities, providing protection of the spermatozoa (Yenugu et al. 2004). Due to its important functions in reproduction in primates, it is not unexpected that some level of purifying selection is acting on EPPIN to prevent NSyn mutations from altering its important biological functions. Even in primates experiencing lower levels of postcopulatory selection and lower semen coagulum thickness like gorilla (Dorus et al. 2004), it seems that the role of EPPIN in modulating the cleavage of SEMG1 is not affected. However, WFDC6, which shows the strongest signature of purifying selection in chimpanzees, does not share the same leucine residue at the reactive site, instead it has in position 87 a tryptophan (W). It is also noticeable that the majority of the replacements seen in WFDC6 include cysteines from the Kunitz domain, which in EPPIN are engaged in disulfide bonds. Therefore, we hypothesize that the serine-protease activity of WFDC6 would be impaired or targeted to a different protease other than PSA. On the other hand, the WAP domain has a highly conserved amino acidic composition between both genes, and the maintenance of the disulfide bridges suggests that the antimicrobial properties of this domain will be maintained (Wilkinson et al. 2011).
Disease transmission during mating provides a connection between reproduction and immunity, where sexually transmitted diseases (STDs) can affect fitness of individuals by imposing different selective pressures on their hosts. Previous studies found a positive correlation between levels of leukocytes (indicator of immunocompetence) and several proxies of female sexual promiscuity among species of primates with different mating systems (Nunn et al. 2000; Nunn 2003, 2002). The lack of associations with several other social, ecological, and life history variables led to the hypothesis that increased levels of transmission of STDs in promiscuous species have resulted in the evolution of a greater investment in immune response (Holmes 2004; Wlasiuk and Nachman 2010). Chimpanzees are classified as one of the most promiscuous primate species, where previous signals of rapid evolution of sperm proteins (SEMG1 and SEMG2) were found (Jensen-Seaman and Li 2003; Dorus et al. 2004; Hurle et al. 2007). Instead, humans, gorillas and gibbons are not promiscuous, maintaining a monoandrous mating system being less subject to STDs.
We hypothesize that chimpanzees, as a promiscuous species, are likely to be more exposed to STD (Nunn et al. 2000; Wlasiuk and Nachman 2010; Garamszegi and Nunn 2011). After the duplication event that originated WFDC6, an episode of rapid evolution may have occurred allowing for the accumulation of amino acid replacements. Later in chimpanzee evolution, the newly originated WFDC6 appears to have been preserved by strong selective constraints, perhaps representing an adaptive response to a higher load of pathogens. Conversely, in less promiscuous species like orangutan, rhesus, and baboon, the signals of pseudogenization present on WFDC6 seem to be associated with more relaxed constraints (higher ω values) and lower pathogen exposure or to the exploitation of different mechanisms of immune defense (Nunn et al. 2000; Nunn 2003; Anderson et al. 2004; Holmes 2004). However, as WFDC6 biological functions and target molecules have not been explored yet, our hypothesis regarding the purifying selective pressures cannot be totally elucidated.
Overall, our data provide support for a clear genetic differentiation of P. t. verus, for a recent divergence of P. t. troglodytes and P. t. ellioti subspecies, and for a single departure of neutrality in the WFDC locus due to strong selective constrains acting on WFDC6. We hypothesize that the latter may be due to an adaptive process associated to the expanded antimicrobial spectrum of WFDCs in the male reproductive tract.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge Anh-Dao Nguyen for the help inferring the ancestral allele state of each SNP and Riverside Zoo, Sunset Zoo, Lincoln Park Zoo, the Primate Foundation of Arizona, New Iberia Research Center, and Texas Biomed for sample donation. The authors would also like to thank John Kiyang and Felix Lanekster of the Limbe Wildlife Centre for sample collection. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute, by the Portuguese Foundation for Science and Technology (FCT), financed by the European Social Funds (COMPETE-FEDER) and national funds of the Portuguese Ministry of Education and Science (POPH-QREN) fellowship SFRH/BD/45907/2008 to Z.F., grant PTDC/BEX-GMG/0242/2012 to S.S., NSF Grant 0755823 to M.K.G., NIH grants HD057974 to W.J.S. and DP1 ES022577 to S.A.T., and by the Wellcome Trust Centre for Human Genetics (WT097307) to W.W.K. IPATIMUP is an Associated Laboratory of the Portuguese Ministry of Education and Science and is partially supported by FCT.
Literature Cited
- Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Hessel JK, Dixson AF. Primate mating systems and the evolution of immune response. J Reprod Immunol. 2004;61:31–38. doi: 10.1016/j.jri.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Andrés AM, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3(4):e66. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. Maximum likelihood methods for detecting adaptive evolution after gene duplication. J Struct Funct Genomics. 2003;3:11. [PubMed] [Google Scholar]
- Bingle CD, Vyakarnam A. Novel innate immune functions of the whey acidic protein family. Trends Immunol. 2008;29:444–453. doi: 10.1016/j.it.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Bowden R, et al. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 2012;8:e1002504. doi: 10.1371/journal.pgen.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell JL, et al. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 2008;4:e1000057. doi: 10.1371/journal.pgen.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clauss A, Lilja H, Lundwall A. The evolution of a genetic locus encoding small serine proteinase inhibitors. Biochem Biophys Res Commun. 2005;333:383–389. doi: 10.1016/j.bbrc.2005.05.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss A, Persson M, Lilja H, Lundwall A. Three genes expressing Kunitz domains in the epididymis are related to genes of WFDC-type protease inhibitors and semen coagulum proteins in spite of lacking similarity between their protein products. BMC Biochem. 2011;12:55. doi: 10.1186/1471-2091-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- de Lamirande E. Semenogelin, the main protein of the human semen coagulum, regulates sperm function. Semin Thromb Hemost. 2007;33:60–68. doi: 10.1055/s-2006-958463. [DOI] [PubMed] [Google Scholar]
- Dixson AF, Anderson MJ. Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatol. 2002;73:6. doi: 10.1159/000064784. [DOI] [PubMed] [Google Scholar]
- Dixson AF, Anderson MJ. Sexual behavior, reproductive physiology and sperm competition in male mammals. Physiol Behav. 2004;83:361–371. doi: 10.1016/j.physbeh.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36:1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- Edstrom AM, et al. The major bactericidal activity of human seminal plasma is zinc-dependent and derived from fragmentation of the semenogelins. J Immunol. 2008;181:3413–3421. doi: 10.4049/jimmunol.181.5.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. Human demographic history: refining the recent African origin model. Curr Opin Genet Dev. 2002;12:8. doi: 10.1016/s0959-437x(02)00350-7. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:3. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:20. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. Detecting hitchhiking from patterns of DNA polymorphism. In: Nurminsky D, editor. Selective sweeps. Georgetown (TX): Landes Biosciences; 2005. [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C-I. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:3. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Ferreira Z, Hurle B, Rocha J, Seixas S. Differing evolutionary histories of WFDC8 (short-term balancing) in Europeans and SPINT4 (incomplete selective sweep) in Africans. Mol Biol Evol. 2011;28:2811–2822. doi: 10.1093/molbev/msr106. [DOI] [PubMed] [Google Scholar]
- Ferreira Z, et al. Reproduction and immunity-driven natural selection in the human WFDC locus. Mol Biol Evol. 2013;30:938–950. doi: 10.1093/molbev/mss329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M. Evidence for a complex demographic history of chimpanzees. Mol Biol Evol. 2004;21:799–808. doi: 10.1093/molbev/msh083. [DOI] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:15. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garamszegi LZ, Nunn CL. Parasite-mediated evolution of the functional part of the MHC in primates. J Evol Biol. 2011;24:184–195. doi: 10.1111/j.1420-9101.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gonder MK, Disotell TR, Oates JF. New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. Int J Primatol. 2006;27:1103–1127. [Google Scholar]
- Gonder MK, et al. A new west African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- Gonder MK, et al. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc Natl Acad Sci U S A. 2011;108:12. doi: 10.1073/pnas.1015422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol Biol Evol. 2010;27(4):921–933. doi: 10.1093/molbev/msp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. Adaptation and immunity. PLoS Biol. 2004;2:E307. doi: 10.1371/journal.pbio.0020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:6. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurle B, Swanson W, Program NCS, Green ED. Comparative sequence analyses reveal rapid and divergent evolutionary changes of the WFDC locus in the primate lineage. Genome Res. 2007;17:276–286. doi: 10.1101/gr.6004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Li WH. Evolution of the hominoid semenogelin genes, the major proteins of ejaculated semen. J Mol Evol. 2003;57:261–270. doi: 10.1007/s00239-003-2474-x. [DOI] [PubMed] [Google Scholar]
- Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:3. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lundwall A. A locus on chromosome 20 encompassing genes that are highly expressed in the epididymis. Asian J Androl. 2007;9:540–544. doi: 10.1111/j.1745-7262.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Martellini JA, et al. Cationic polypeptides contribute to the anti-HIV-1 activity of human seminal plasma. FASEB J. 2009;23:3609–3618. doi: 10.1096/fj.09-131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn CL. A comparative study of leukocyte counts and disease risk in primates. Evolution. 2002;56:177–190. doi: 10.1111/j.0014-3820.2002.tb00859.x. [DOI] [PubMed] [Google Scholar]
- Nunn CL. Behavioural defences against sexually transmitted diseases in primates. Anim Behav. 2003;66:37–48. [Google Scholar]
- Nunn CL, Gittleman JL, Antonovics J. Promiscuity and the primate immune system. Science. 2000;290:1168–1170. doi: 10.1126/science.290.5494.1168. [DOI] [PubMed] [Google Scholar]
- Patten MA, Unitt P. Diagnosability versus mean differences of sage sparrow subspecies. Auk. 2002;119: 9. [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A, Lilja H, Lundwall A, Malm J. Semenogelin I and semenogelin II, the major gel-forming proteins in human semen, are substrates for transglutaminase. Eur J Biochem. 1998;252:5. doi: 10.1046/j.1432-1327.1998.2520216.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:14. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11:621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Wang Z, Widgren EE, Sivashanmugam P, O'Rand MG, Richardson RT. Association of eppin with semenogelin on human spermatozoa. Biol Reprod. 2005;72:1064–1070. doi: 10.1095/biolreprod.104.036483. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Excoffier L. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 2010;27:1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- Wilkinson TS, Roghanian A, Simpson AJ, Sallenave JM. WAP domain proteins as modulators of mucosal immunity. Biochem Soc Trans. 2011;39:1409–1415. doi: 10.1042/BST0391409. [DOI] [PubMed] [Google Scholar]
- Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci. 2006;110:21–35. doi: 10.1042/CS20050115. [DOI] [PubMed] [Google Scholar]
- Wlasiuk G, Nachman MW. Promiscuity and the rate of molecular evolution at primate immunity genes. Evolution. 2010;64:2204–2220. doi: 10.1111/j.1558-5646.2010.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci U S A. 2010;107:9264–9269. doi: 10.1073/pnas.0913022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenugu S, et al. Antimicrobial activity of human EPPIN, an androgen-regulated, sperm-bound protein with a whey acidic protein motif. Biol Reprod. 2004;71:1484–1490. doi: 10.1095/biolreprod.104.031567. [DOI] [PubMed] [Google Scholar]
- Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lee WH, Shen JH, Li H, Zhang Y. Identification of novel semenogelin I-derived antimicrobial peptide from liquefied human seminal plasma. Peptides. 2008;29:505–511. doi: 10.1016/j.peptides.2008.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.