Abstract
Many chemically-complex cyanobacterial polyketides and nonribosomal peptides are of great pharmaceutical interest, but the levels required for exploitation are difficult to achieve from native sources. Here we develop a framework for the expression of these multifunctional cyanobacterial assembly lines in Escherichia coli using the lyngbyatoxin biosynthetic pathway, derived from a marine microbial assemblage dominated by the cyanobacterium Moorea producens. Heterologous expression of this pathway afforded high titers of both lyngbyatoxin A (25.6 mg L-1) and its precursor indolactam-V (150 mg L-1). Production, isolation and identification of all expected chemical intermediates of lyngbyatoxin biosynthesis in E. coli also confirmed the previously proposed biosynthetic route setting a solid chemical foundation for future pathway engineering. The successful production of the nonribosomal peptide lyngbyatoxin A in E. coli also opens the possibility for future heterologous expression, characterization and exploitation of other cyanobacterial natural product pathways.
Keywords: lyngbyatoxin, cyanobacteria, heterologous expression, NRPS, terpene
Cyanobacteria are prolific producers of natural products including nonribosomal peptides (1), polyketides (2), terpenoids, alkaloids and ribosomally-synthesized peptides (3). Cyanobacterial natural products exhibit a diverse range of bioactivities, ensuring they are highly valued as lead pharmaceutical and industrial compounds (4). Large quantities of compound are required for the medicinal use, characterization or generation of new structural analogs via semisynthetic chemistry. However, the cyanobacteria that produce these compounds exhibit slow growth rates and are intractable to genetic manipulation. Consequently, there is a supply shortage of these exploitable compounds from the native producing microorganisms, which can be circumvented by the use of heterologous over-production.
Cyanobacterial complex biosynthetic pathway characterization noticeably lags behind that of other phyla, such as Streptomyces and Proteobacteria, due to an inability to heterologously express entire pathways in a readily culturable and genetically amenable host that would additionally offer opportunities for production optimization (5-14). Biosynthetically much less complex ribosomal peptide natural products from cyanobacteria, typically featuring extensive post-translational modifications, have previously been expressed in Escherichia coli (15-18). Furthermore, the expression of small stand-alone tailoring enzymes from cyanobacterial nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) pathways has also been achieved allowing characterization of their biochemistry in vitro (19-22). However, the heterologous expression of a complete cyanobacterial PKS or NRPS biosynthetic pathway, to yield the final biosynthetic product has proven very difficult to attain. The hybrid PKS/NRPS barbamide derivative, 4-O-demethylbarbamide, is to our knowledge the only successful heterologous production of a cyanobacterial compound of this type (23), although yield in a Streptomyces host was very low (< 1 μg L-1).
While not impossible, the heterologous expression of proteins in a distantly related organism can be troublesome and requires a significant amount of optimization (24, 25). Here, we selected the nonribosomal peptide-terpene hybrid lyngbyatoxin A (LTX, 1) as a model to identify suitable conditions for the heterologous expression of a cyanobacterial non-ribosomal peptide in E. coli. LTX is an indole alkaloid and a tumor promoter that elicits its bioactivity via potent activation of protein kinase C (26) and is structurally related to the actinobacterial compounds teleocidin and methylpendolmycin (Figure 1). The indole ring is highly nucleophilic and all positions are susceptible to electrophilic attack, making this an interesting precursor for semi-synthetic chemistry (27). Indolactam-V (ILV, 2), the structural core of these molecules, induces cellular differentiation of stem cells to pancreatic cells (28), with structural analogs investigated for the treatment of neurodegeneration and cancer (29, 30).
Figure 1.
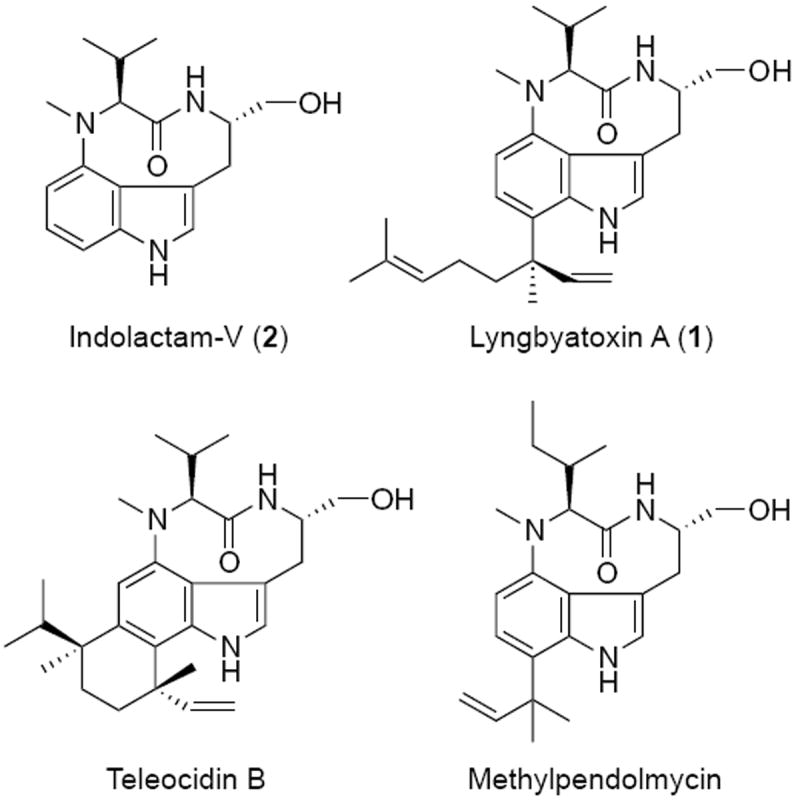
Structures of nine-membered indolactam alkaloids.
LTX was originally isolated from a cyanobacterial bloom dominated by Lyngbya majuscula (Supplementary Web-Enhanced Object) (31, 32), which has since been taxonomically reclassified as Moorea producens (33). Despite numerous attempts, we have been unable to establish a culture of an LTX-producing strain of Moorea producens. Consequently, environmental sources remain the only available routes for accessing both the compound and the genetic basis for biosynthesis. Elucidation of the lyngbyatoxin biosynthetic pathway (22) revealed four open reading frames (ltxA-D) spanning 11.3 kb, thus comprising the smallest cyanobacterial NRPS reported to date. The LtxA protein is a bimodular NRPS that condenses L-methyl-valine and L-tryptophan to form the linear dipeptide N-methyl-L-valyl-L-tryptophan which is released from the synthetase via a NADPH-dependent reductive cleavage to form the corresponding alcohol (NMVT, 3) rather than the usual thioesterase mechanism of NRPSs, which typically couple chain release and cyclization (34). A P450-dependent monooxygenase (LtxB) then catalyzes the formation of ILV through the oxidation and cyclisation of the dipeptide (21). The final step of LTX biosynthesis is performed by LtxC, a reverse prenyltransferase that transfers a geranyl pyrophosphate (GPP) to the C7 position of ILV. The isoforms lyngbyatoxin B (4) and C (5) are presumed to be formed though the action of an oxidoreductase (LtxD). Earlier endeavors to express the entire lyngbyatoxin gene cluster in Streptomyces coelicolor were not successful due to premature transcriptional termination of the ltxA gene (35). However, the tailoring enzymes LtxB and LtxC have been expressed individually in both E. coli (21, 22) and S. coelicolor (35).
We rationally adapted methodology, developed through the successful heterologous expression of NRPS and PKS gene clusters from diverse phyla (13, 14), to perform expression of the LTX biosynthetic gene cluster in E. coli. Previous accounts of the heterologous expression of cyanobacterial NRPS (35) or hybrid PKS/NRPS (23) biosynthesis pathways in Streptomyces have demonstrated limited success, which is probably – at least in part – a result of the disparity in GC content between biosynthetic genes and the host genome. Genomic GC content determines bacterial codon usage (36) and the median GC content of 38 cyanobacterial genomes is ~45%. As the lyngbyatoxin biosynthetic genes exhibit a GC content of 46%, E. coli was a logical host with a genomic GC content of 50%.
While the functionality of cyanobacterial promoters from biosynthetic pathways have been studied in E. coli using reporter genes (37), initial attempts to produce LTX from the native lyngbyatoxin biosynthesis pathway yielded no detectable levels of the compound, or any intermediates, irrespective of growth temperature or media constituents. This suggested that the native lyngbyatoxin transcriptional regulatory regions are not sufficiently functional to facilitate heterologous expression of the pathway in E. coli. However, the transcription of the pathway under the control of native promoters in E. coli was not investigated further.
The E. coli GB05-MtaA host carries a heterologous myxobacterial PPTase with broad-range substrate activity (38) and is capable of expressing a range of NRPS clusters at 30°C from the tetO tetracycline-inducible promoter (PtetO) (13, 14). We utilized a similar approach for the heterologous expression of the lyngbyatoxin biosynthetic pathway, however, under the same conditions at 30°C, production of LTX or any pathway intermediates was not observed (Supplementary Figure 1). In light of our experience and other NRPS expression studies described in the literature (9, 39), we reduced the expression temperature to 18°C yielding substantial levels of LTX in the crude methanol extract (Supplementary Figure 2). In addition to LTX, the biosynthetic intermediates NMVT and ILV were also detectable in the methanol extracts of E. coli fermentations (Figure 3, Supplementary Figure 1 and 3a-c). The total production levels of both ILV and LTX were quantified using HPLC by comparison to a standard curve derived from known quantities of analytical standards (Supplementary Figure 4 and 5). NMVT was not quantified due to lack of a standard. The average total yields, in the crude extract of cells and media, from three independent cultures of E. coli GB05-MtaA harboring tetracycline-induced pCC-Ptet-ltx were 150 ± 4.36 mg L-1 of ILV and 25.6 ± 0.77 mg L-1 of LTX. HPLC-ESI-HRMS analysis of the extract also showed very low abundance peaks corresponding to the expected masses of the lyngbyatoxin B or C isoforms (Supplementary Figure 6) in all induced cultures but were absent in the uninduced culture. To confirm the absolute identity of the compounds quantified, ILV and LTX were isolated analyzed by NMR. The 1H NMR data for LTX and ILV were in agreement with published spectra (Supplementary Tables 1 and 2) (40, 41). In comparison to the amount expected from directly scaled cultures, the yields obtained from unoptimized large-scale cultures totaled 30% of ILV and approximately 10% of LTX. This reduced yield correlated with a reduction in the culture growth rate and likely losses during purification. LTX yields from the native producer represented 0.02% of the extracted environmental material (31). As the native producer has yet to be established in culture, production in E. coli is a necessary measure to enable exploitation of these compounds.
Figure 3.

Production of LTX in E. coli. HPLC-ESI-HRMS chromatograms of the tetracycline-induced and the uninduced cultures. EIC mass ranges m/z 302-305 and m/z 435-440. Compounds and experimentally observed masses ([M+H+]) are indicated with their respective peaks. Retention times (min) NMVT 3.3, ILV 7.2, LTX 13.5. Theoretical exact masses (amu) are NMVT 303.1947, ILV 301.1790, LTX 437.3042.
The soluble protein fraction was visualized by SDS-PAGE (Supplementary Figure 7) to shed light as to whether or not the high production levels were due to an over-expression of the biosynthesis enzymes. The 276 kDa LtxA NRPS was faintly visible as a minor constituent of the total soluble protein on day four of the cultivation. The tailoring enzymes LtxB (52 kDa) and LtxC (43 kDa) were difficult to differentiate from endogenous host proteins in the soluble fraction, presumably due to their low relative abundance. Therefore, we speculate that the high compound levels may be influenced by the relatively low burden that production of the small product places on the cell following the initial synthesis of the biosynthetic machinery, and the very high culture density achieved during cultivation.
Despite the absence of ILV from M. producens extracts, it has been isolated from teleocidin-producing strains of Streptomyces (42). Due to the structural relatedness of the lyngbyatoxins and teleocidin, ILV was predicted to be a biosynthetic intermediate and was confirmed via in vitro prenylation by LtxC (22). The levels of ILV present in the heterologous host suggest an inefficient prenylation reaction with only 10% of ILV being converted to LTX. Due to the hybrid NRPS-terpenoid nature of LTX, yields would most likely be improved by augmentation of the isopentyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) availability. This can be achieved via an enhancement of flux though the deoxyxylulose 5-phosphate (DXP) pathway (43) or heterologous expression of a mevalonate pathway (44) to increase the levels of available geranyl pyrophosphate for prenylation. However, our present focus was to create a foundation for cyanobacterial secondary metabolite heterologous biosynthesis and was beyond the scope of this study. In future, LTX yield optimization using a metabolically-engineered E. coli strain may be possible, however, we also observe that ILV is a useful candidate for semi-synthetic chemistry and was produced in abundance by this system.
Extracellular export of intermediates prior to conversion is an alternate reasoning for their accumulation, hence the levels of NMVT, ILV, and LTX in the cell-free spent culture media were examined and compared to the total production levels. Extractions of spent cell-free culture media showed that approximately 50% of the total yield of NMVT is extracellular, while approximately 20% of both ILV and LTX were extracellular (Figure 4). The large proportion of extracellular NMVT relative to ILV/LTX indicates that this dipeptide may be actively exported from the cell and may negatively influence the conversion of NMVT to ILV. However, this does not seem to be the case with ILV and LTX, with the poor conversion rate most likely due to geranyl pyrophosphate limitation.
Figure 4.
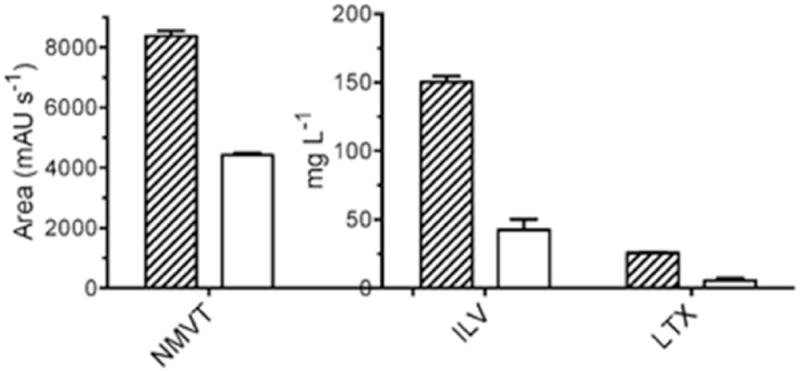
Production levels of N-methyl-L-valyl-L-tryptophanol (NMVT), indolactam-V (ILV) and lyngbyatoxin A (LTX) in E. coli. Striped bars, total culture production levels; unfilled bars, extracellular levels. ILV and LTX were quantified by HPLC using analytical standards, NMVT levels are depicted as the area under the respective HPLC trace peak. Values represent the average levels from three independent cultures (N=3) depicted with SEM.
The replacement of the native promoter and regulatory regions of the lyngbyatoxin biosynthetic pathway with an inducible promoter and new ribosome-binding site eventuated in the successful heterologous expression of LTX in the genetically unrelated host E. coli. Following successful production of LTX from the modified pathway, LTX production from the native pathway was reassessed in parallel with identical conditions to confirm the necessity for these modifications (Supplementary Figure 4). The inability to detect LTX production under the control of the native promoter, supports the notion that these elements do not function in E. coli, however, the possibility remains that suitable induction conditions were simply not achieved. Additional features, which we speculate may have been vital for our success, include the use of a host with a similar GC-content to the biosynthetic genes. The moderate production levels rather than overexpression of biosynthetic enzymes, in particular the NRPS LtxA (24), was additionally influenced by the intentional use of a very low-copy-number and, by our experience, a stable plasmid backbone. As well as the combination of using a low growth temperature, to allow time for the correct folding of the large NRPS, with a rich buffered growth media providing an abundance of essential amino acids for biosynthesis. Whilst none of these factors were shown to be unequivocally essential, the combination of these factors in addition to the presence of the heterologous phosphopantetheinyl transferse MtaA (38), ensuring efficient posttranslational activation of the NRPS (13, 14, 45), proved sufficient for the heterologous expression of the cyanobacterial NRP LTX.
Due to fast growth rates, extensive characterization, the availability of a suite of genetic tools and a relatively robust tolerance to exogenous proteins, E. coli is considered the preferred option for heterologous expression. The relative simplicity of the lyngbyatoxin biosynthetic pathway compared to most NRPSs makes it an ideal choice for the production of new compounds targeting protein kinase C. For the generation of new compound analogs it is essential that all downstream enzymatic reactions tolerate any upstream changes to the substrate (46, 47). The tailoring enzymes, LtxB and LtxC, exhibit relaxed specificity for NMVT analogs where the valyl group is substituted by other aliphatic groups (21). Therefore, our E. coli heterologous system provides an extremely feasible route for the generation of new ILV-based protein kinase C modulators.
Cyanobacteria are prolific producers of natural products exhibiting a wide range of biological activities. We have demonstrated the biological synthesis, via a multienzyme thiotemplate mechanism, and overproduction of such molecules in a heterologous expression system. This technological development will allow the exploitation of molecules with commercial significance from sources whereby previously the only feasible route was synthetic chemistry. The next step for cyanobacterial natural products will utilize the amenability of heterologous hosts to genetically engineer pathways for the generation of structural analogs with modified pharmacological properties.
METHODS
Bacterial strains and culture conditions
E. coli strain GB05-red (13) was used for propagation and recombineering, cultured in Luria Bertani (LB) medium 37°C supplemented with the appropriate antibiotic (chloramphenicol 15 μg mL-1, gentamicin 2 μg mL-1). E. coli strain GB05-MtaA, with the promiscuous PPTase MtaA from the myxobacterium Stigmatella aurantiaca chromosomally integrated via transposition (13), was used for expression and cultured in 50 mL Terrific Broth (TB) supplemented with antibiotics in 250 mL Erlenmeyer flasks stoppered with a cotton bung.
Expression vector construction
The fosmid fos-DE3-86 (22) carries ~ 35 kb of genomic DNA from Moorea producens, including the 11.3 kb lyngbyatoxin biosynthesis pathway, within a pCC1FOS (Epicentre) backbone. The tetracycline-inducible promoter cassette, consisting of gentamicin resistance (aacC1), tetracycline repressor (tetR) and PtetO regions (gentaR-tetR-PtetO) from plasmid pR6K-Tps-Genta (14) was inserted immediately upstream of the ltxA ATG start codon of fos-DE3-86 (Supplementary Figure 8) using the primers ltx-genta5 (5’-TATTCCTTATACCATCAAGATCCTTTGATTTTGAGGGCTAGAAGGCACGAACCCAGTTGAC-3’) and ltx-tetR3 (5’-TATCGCCCGCCTACTTCCACTCCAAGGTTGATTCATAATCATGTCGATCCTC TTCTCTATCACTGATAGG-3’). Sequences homologous to fos-DE3-86, to facilitate recombineering, are underlined. This new construct was named pCC-Ptet-ltx. In detail, PCR amplification was performed using Phusion DNA polymerase (New England Biolabs). The product was purified and concentrated by ethanol precipitation. E. coli GB-red harboring plasmid fos-DE3-86 were inoculated (1:40 dilution) from an overnight culture and grown in a thermomixer (Eppendorf) at 37°C shaking 950 rpm. At OD600 0.2, 0.2% (w/v) L-arabinose was added to induce expression of recombinases. Cells were cultured to an OD600 of 0.4, harvested, washed three times with ice-cold dH2O, and resuspended in 40 μl dH2O. The cells were mixed with 0.2-0.3 μg of PCR product and transferred to a sterile 1 mm electroporation cuvette. Electroporation was performed at 1300 V using an Electroporator 2510 (Eppendorf), followed by the immediate addition of 1 mL LB media. The cells were transferred to 37°C shaking 950 rpm for 75 min, concentrated, and plated on LB agar plates containing 15 μg mL-1 chloramphenicol and 1 μg mL-1 gentamicin. The plates were incubated overnight at 37°C. Correct recombinants were determined by restriction digestion.
Expression and analysis of lyngbyatoxin A production
Overnight cultures of four different clones of GB05-MtaA harboring the pCC-Ptet-ltx construct were inoculated into 50 mL TB media and grown at 30°C, shaking at 200 rpm to an OD600 of 0.17-0.18. Cultures were transferred to 18°C, shaking 200 rpm, for 30 min prior to induction of three of the cultures at an OD600 of ~0.2 by the addition of 0.5 μg mL-1 tetracycline (final concentration). Cultures were transferred to 18°C, shaking at 200 rpm, for 5 days and 2% (v/v) Amberlite XAD-16 polymeric adsorbent resin (Sigma) was added to adsorb extracellular compounds. Cultures were returned to 18°C for an additional 48 h prior to harvesting the cells and resin by centrifugation. Cells and resin were extracted with methanol and evaporated to dryness. Extracts were redissolved in 1 mL methanol and analyzed by HPLC-ESI-HRMS/MS. Separation was performed on a Dionex UltiMate 3000 RSLC system using a Waters BEHC18 (100 × 2.1 mm, 1.7 μm dp) column by injection of 2 μl of concentrated sample. Separation was achieved using a linear gradient with (A1) water + 0.1% formic acid to (B1) acetonitrile + 0.1% formic acid at a flow rate of 550 μL min-1 and 45°C. The gradient was initiated by a 0.39 min isocratic step with 5% B1 followed by an increase to 95% B1 within 18 min. HPLC was coupled to a Thermo Fisher Orbitrap via an Advion Triversa Nanomate nano-ESI system. Mass spectra were acquired in centroid mode with a range of 200-2000 m/z at a resolution of R=30000. Fragmentation by collision-induced dissociation was performed on ions corresponding to 302.20, 304.20, 438.31 m/z. Extracts from induced cultures were compared to an uninduced negative control. The yields of ILV and LTX were quantified in duplicate by comparing to a standard curve (Supplementary Figure 5) derived from known amounts of each compound on a HP Agilent series 1100 HPLC using an Agilent Zorbax SB-C18 (4.6 × 250 mm) column. Separation was achieved using a linear gradient with (A2) water + 0.1% trifluoroacetic acid to (B2) acetonitrile at a flow rate of 1.1 mL min-1 and 27.5°C. The gradient was initiated with 5% B2 and increased to 95% B2 within 30 min, followed by 95% B2 for 10 min. Results were analyzed and production levels with SEM were determined using Prism 5.0 for Mac (GraphPad Software).
Supplementary Material
Figure 2.
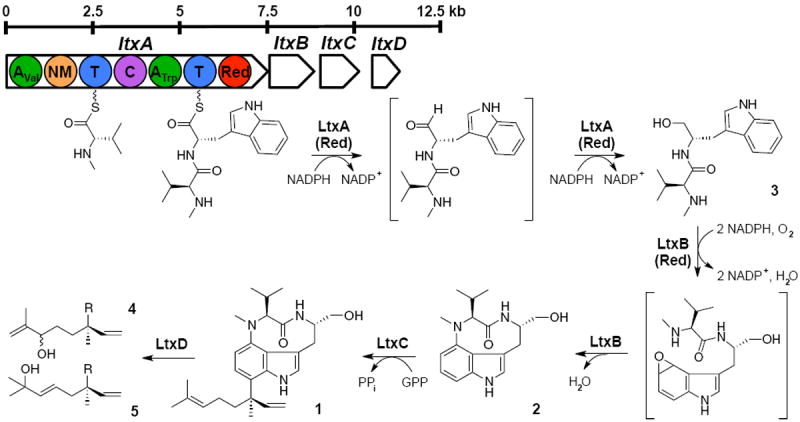
Lyngbyatoxin biosynthetic genes and proposed pathway for lyngbyatoxin A (1), B (4) and C (5). LtxB. P450-dependent monooxygenase; LtxC, prenyltransferase; LtxD, oxidoreductase; GPP, geranyl pyrophosphate; PPi, pyrophosphate.
Acknowledgments
We would like to kindly thank A. Jones and D. Edwards for providing valuable assistance towards the preparation of this manuscript, L. Gerwick for provision of strains and analytical standards, and R. C. Coates for M. producens photographic material. T. Hoffmann’s contributions to this work regarding HPLC-ESI-HRMS Orbitrap measurements in Saarbrücken are highly appreciated. This work was funded by the Australian Research Council (grants FF0883440 and LP110201096) and Diagnostic Technology P/L (BAN), as well as, NIH CA108874 (WHG). Work in RM’s laboratory was supported by the Deutsche Forschungsgemeinschaft and the Bundesministerium für Bildung und Forschung. SEO was supported by an Australian Postgraduate Award and an Adrian Lee Travel Scholarship. XB is grateful to the China Scholarship Council (CSC) for a PhD scholarship.
Footnotes
SUPPORTING INFORMATION.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sieber S, Marahiel M. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 2.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 3.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burja AM, Banaigs B, Abou-Mansour E, Burgess JG, Wright PC. Marine cyanobacteria - a prolific source of natural products. Tetrahedron. 2001;57:9347–9377. [Google Scholar]
- 5.Gomez-Escribano JP, Bibb MJ. Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol. 2012;517:279–300. doi: 10.1016/B978-0-12-404634-4.00014-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang P, Kim W, Pickens LB, Gao X, Tang Y. Heterologous expression and manipulation of three tetracycline biosynthetic pathways. Angew Chem Int Ed. 2012;51:11136–11140. doi: 10.1002/anie.201205426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Boghigian BA, Armando J, Pfeifer BA. Methods and options for the heterologous production of complex natural products. Nat Prod Rep. 2010;28:125–151. doi: 10.1039/c0np00037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzel S, Müller R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotechnol. 2005;16:594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Wenzel SC, Gross F, Zhang Y, Fu J, Stewart AF, Müller R. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via Red/ET recombineering. Chem Biol. 2005;12:349–356. doi: 10.1016/j.chembiol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Chai Y, Shan S, Weissman KJ, Hu S, Zhang Y, Müller R. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using Red/ET recombineering and inactivation mutagenesis. Chem Biol. 2012;19:361–371. doi: 10.1016/j.chembiol.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Fu J, Wenzel SC, Perlova O, Wang J, Gross F, Tang Z, Yin Y, Stewart AF, Müller R, Zhang Y. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res. 2008;36:e113. doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binz TM, Wenzel SC, Schnell H-J, Bechthold A, Müller R. Heterologous Expression And Genetic Engineering of the Phenalinolactone Biosynthetic Gene Cluster by Using Red/ET Recombineering. ChemBioChem. 2008;9:447–454. doi: 10.1002/cbic.200700549. [DOI] [PubMed] [Google Scholar]
- 13.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF, Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 14.Bian X, Huang F, Stewart AF, Xia L, Zhang Y, Müller R. Direct cloning, genetic engineering and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. ChemBioChem. 2012;13:1946–1952. doi: 10.1002/cbic.201200310. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt E, Nelson J, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long P, Dunlap W, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 17.Ziemert N, Ishida K, Quillardet P, Bouchier C, Hertweck C, de Marsac NT, Dittmann E. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl Environ Microbiol. 2008;74:1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl Environ Microbiol. 2010;76:3568–3574. doi: 10.1128/AEM.02858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muenchhoff J, Siddiqui KS, Neilan BA. Identification of two residues essential for the stringent substrate specificity and active site stability of the prokaryotic L-arginine:glycine amidinotransferase CyrA. FEBS J. 2012;279:805–815. doi: 10.1111/j.1742-4658.2012.08472.x. [DOI] [PubMed] [Google Scholar]
- 20.Muenchhoff J, Siddiqui KS, Poljak A, Raftery MJ, Barrow KD, Neilan BA. A novel prokaryotic L-arginine:glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 2010;277:3844–3860. doi: 10.1111/j.1742-4658.2010.07788.x. [DOI] [PubMed] [Google Scholar]
- 21.Huynh M, Elston M, Hernandez NM, Ball DB, Kajiyama S-i, Irie K, Gerwick WH, Edwards DJ. Enzymatic production of (−)- indolactam V by LtxB, a cytochrome P450 monooxygenase. J Nat Prod. 2010;73:71–74. doi: 10.1021/np900481a. [DOI] [PubMed] [Google Scholar]
- 22.Edwards D, Gerwick W. Lyngbyatoxin biosynthesis: sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 23.Kim EJ, Lee JH, Choi H, Pereira AR, Ban Y-H, Yoo YJ, Kim E, Park JW, Sherman DH, Gerwick WH, Yoon YJ. Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Org Lett. 2012;14:5824–5827. doi: 10.1021/ol302575h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- 25.Menzella HG, Reisinger SJ, Welch M, Kealey JT, Kennedy J, Reid R, Tran CQ, Santi DV. Redesign, synthesis and functional expression of the 6-deoxyerythronolide B polyketide synthase gene cluster. J Ind Microbiol Biotechnol. 2005;33:22–28. doi: 10.1007/s10295-005-0038-3. [DOI] [PubMed] [Google Scholar]
- 26.Basu A, Kozikowski AP, Lazo JS. Structural requirements of lyngbyatoxin A for activation and downregulation of protein kinase C. Biochemistry. 1992;31:3824–3830. doi: 10.1021/bi00130a013. [DOI] [PubMed] [Google Scholar]
- 27.Williams RM, Stocking EM, Sanz-Cervera JF. Biosynthesis of prenylated alkaloids derived from tryptophan. Top Curr Chem. 2000;209:97–173. [Google Scholar]
- 28.Chen S, Borowiak M, Fox JL, Maehr R, Osafune K, Davidow L, Lam K, Peng LF, Schreiber SL, Rubin LL, Melton D. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa Y. Artificial analogs of naturally occurring tumor promoters as biochemical tools and therapeutic leads. Biosci Biotechnol Biochem. 2012;76:1262–1274. doi: 10.1271/bbb.120162. [DOI] [PubMed] [Google Scholar]
- 30.Nelson TJ, Alkon DL. Neuroprotective versus tumorigenic protein kinase C activators. Trends Biochem Sci. 2009;34:136–145. doi: 10.1016/j.tibs.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Cardellina JH, Marner FJ, Moore RE. Seaweed dermatitis: structure of lyngbyatoxin A. Science. 1979;204:193–195. doi: 10.1126/science.107586. [DOI] [PubMed] [Google Scholar]
- 32.Aimi N, Odaka H, Sakai S, Fujiki H, Suganuma M, Moore RE, Patterson GM. Lyngbyatoxins B and C, two new irritants from Lyngbya majuscula. J Nat Prod. 1990;53:1593–1596. doi: 10.1021/np50072a035. [DOI] [PubMed] [Google Scholar]
- 33.Engene N, Rottacker EC, Kaštovský J, Byrum T, Choi H, Ellisman MH, Komárek J, Gerwick WH. Moorea producens gen. nov., sp. nov. and Moorea bouillonii comb. nov., tropical marine cyanobacteria rich in bioactive secondary metabolites. Int J Syst Evol Microbiol. 2012;62:1171–1178. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read JA, Walsh CT. The lyngbyatoxin biosynthetic assembly line: chain release by four-electron reduction of a dipeptidyl thioester to the corresponding alcohol. J Am Chem Soc. 2007;129:15762–15763. doi: 10.1021/ja077374d. [DOI] [PubMed] [Google Scholar]
- 35.Jones AC, Ottilie S, Eustáquio AS, Edwards DJ, Gerwick L, Moore BS, Gerwick WH. Evaluation of Streptomyces coelicolor A3(2) as a heterologous expression host for the cyanobacterial protein kinase C activator lyngbyatoxin A. FEBS J. 2012;279:1243–1251. doi: 10.1111/j.1742-4658.2012.08517.x. [DOI] [PubMed] [Google Scholar]
- 36.Lightfield J, Fram NR, Ely B. Across bacterial phyla, distantly-related genomes with similar genomic GC content have similar patterns of amino acid usage. PLoS ONE. 2011;6:e17677. doi: 10.1371/journal.pone.0017677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones AC, Gerwick L, Gonzalez D, Dorrestein PC, Gerwick WH. Transcriptional analysis of the jamaicamide gene cluster from the marine cyanobacterium Lyngbya majuscula and identification of possible regulatory proteins. BMC Microbiol. 2009;9:247. doi: 10.1186/1471-2180-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaitatzis N, Hans A, Müller R, Beyer S. The mtaA gene of the myxothiazol biosynthetic gene cluster from Stigmatella aurantiaca DW4/3-1 encodes a phosphopantetheinyl transferase that activates polyketide synthases and polypeptide synthetases. J Biochem. 2001;129:119–124. doi: 10.1093/oxfordjournals.jbchem.a002821. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe K, Hotta K, Praseuth AP, Koketsu K, Boddy CN, Wang CCC, Oguri H, Oikawa H. Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nat Chem Biol. 2006;2:423–428. doi: 10.1038/nchembio803. [DOI] [PubMed] [Google Scholar]
- 40.Endo Y, Shudo K, Itai A, Hasegawa M, Sakai S-i. Synthesis and stereochemistry of indolactam-V, an active ragnent of teleocidins. Structural requirements for tumor-promoting activity. Tetrahedron. 1986;42:5905–5924. [Google Scholar]
- 41.Izumikawa M, Khan ST, Komaki H, Takagi M, Shinya K. JBIR-31, a new teleocidin analog, produced by salt-requiring Streptomyces sp. NBRC 105896 isolated from a marine sponge. J Antibiot. 2010;63:33–36. doi: 10.1038/ja.2009.113. [DOI] [PubMed] [Google Scholar]
- 42.Irie K, Hirota M, Hagiwara N, Koshimizu K, Hayashi H, Murao S, Tokuda H, Ito Y. The Epstein-Barr virus early antigen inducing indole alkaloids, (-)-indolactam V and its related compounds produced by Actinomyces. Agric Biol Chem. 1984;48:1269–1274. [Google Scholar]
- 43.Reiling KK, Yoshikuni Y, Martin VJJ, Newman J, Bohlmann J, Keasling JD. Mono and diterpene production in Escherichia coli. Biotechnol Bioeng. 2004;87:200–212. doi: 10.1002/bit.20128. [DOI] [PubMed] [Google Scholar]
- 44.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 45.Lai J, Koglin A, Walsh C. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 46.Han JW, Kim EY, Lee JM, Kim YS, Bang E, Kim BS. Site-directed modification of the adenylation domain of the fusaricidin nonribosomal peptide synthetase for enhanced production of fusaricidin analogs. Biotechnol Lett. 2012;34:1327–1334. doi: 10.1007/s10529-012-0913-8. [DOI] [PubMed] [Google Scholar]
- 47.Villiers B, Hollfelder F. Directed evolution of a gatekeeper domain in nonribosomal peptide synthesis. Chem Biol. 2011;18:1290–1299. doi: 10.1016/j.chembiol.2011.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


