Abstract
Aims/hypothesis
We sought to determine the mRNA transcriptome of all major human pancreatic endocrine and exocrine cell subtypes, including human alpha, beta, duct and acinar cells. In addition, we identified the cell type-specific distribution of transcription factors, signalling ligands and their receptors.
Methods
Islet samples from healthy human donors were enzymatically dispersed to single cells and labelled with cell type-specific surface-reactive antibodies. Live endocrine and exocrine cell subpopulations were isolated by FACS and gene expression analyses were performed using microarray analysis and quantitative RT-PCR. Computational tools were used to evaluate receptor–ligand representation in these populations.
Results
Analysis of the transcriptomes of alpha, beta, large duct, small duct and acinar cells revealed previously unrecognised gene expression patterns in these cell types, including transcriptional regulators HOPX and HDAC9 in the human beta cell population. The abundance of some regulatory proteins was different from that reported in mouse tissue. For example, v-maf musculoaponeurotic fibrosarcoma oncogene homologue B (avian) (MAFB) was detected at equal levels in adult human alpha and beta cells, but is absent from adult mouse beta cells. Analysis of ligand–receptor interactions suggested that EPH receptor–ephrin communication between exocrine and endocrine cells contributes to pancreatic function.
Conclusions/interpretation
This is the first comprehensive analysis of the transcriptomes of human exocrine and endocrine pancreatic cell types—including beta cells—and provides a useful resource for diabetes research. In addition, paracrine signalling pathways within the pancreas are shown. These results will help guide efforts to specify human beta cell fate by embryonic stem cell or induced pluripotent stem cell differentiation or genetic reprogramming.
Keywords: Alpha cell, Beta cell, Paracrine signalling, Transcription factor
Introduction
The human beta cell transcriptome has been examined in studies of whole pancreatic islets [1–4], beta cells laser-captured from frozen tissue sections [5] or transduced beta cells [6], but these observations may not reflect the nature of normal beta cells. Moreover, the transcriptomes of human exocrine pancreatic cell subtypes have not yet been reported, despite the importance of these populations for pancreatic function and their interrelationship with the endocrine pancreas. In mice, pancreatic cell subtype expression profiling has been performed using lineage marked transgenics [7, 8], but significant differences between the mouse and human pancreatic transcriptional programmes exist. Programming of pluripotent stem cells or somatic progenitors by induced transcription factor production is a promising approach for beta cell generation [9, 10], which would benefit substantially from a more complete list of factors differentially produced in human beta cells.
The importance of transcription factor fate specification is well recognised. The v-maf musculoaponeurotic fibrosarcoma oncogene homologue (avian) (MAF) family of basic leucine zipper transcription factors has been strongly implicated in the determination of islet cell fate. Mafa, a regulator of insulin expression, has been shown to be expressed only in beta cells in mice [11]. Conversely, although Mafb is expressed in both alpha and beta cells during murine embryonic development, in adult mice it is found only in the alpha cells [12]. Among the NK-related homeobox gene family, Nkx6-1 shows adult expression restricted to beta cells [13] like Mafa, whereas Nkx2-2 is detected in alpha and beta cells [14]. The paired box (PAX) group also contains members involved in islet cell fate specification. Pax4 is required for the development of mouse beta cells, but is absent from mature islets [15], whereas Pax6 is found in mature alpha and beta cells [16]. Other important endocrine transcriptional regulators include Neurog3 and Pdx1. Both are expressed at intervals during endocrine cell development, but only the latter is retained in adult rodent islets (specifically beta cells) [17]. The expression patterns of these murine factors are frequently used to guide efforts to reprogramme non-beta cells to a beta cell identity, as reviewed by Baeyens et al. [18], or to differentiate embryonic stem cells or induced pluripotent stem cells into beta cells, as reviewed by van Hoof et al. [19]. It is therefore important to identify the differences between human and mouse endocrine cell transcriptional regulation.
Pancreatic exocrine and endocrine cell subtypes are also distinguishable by their differential participation in paracrine signalling. Islet hormones are best known for activating cells in remote tissues to maintain glucose homeostasis, but also locally affect secretion by acinar and duct cells. The significance of pancreatic paracrine signalling is highlighted by the observation that duct-secreted cytokines can inhibit insulin secretion and contribute to immune responses in islets [20, 21]. Duct– islet association is also critical during pancreatic development, when epithelial duct cells differentiate into endocrine precursors. Understanding these relationships may hold the key to transdifferentiating duct cells to beta cells.
Here we used novel monoclonal antibodies to purify populations of live human alpha, beta, duct and acinar cells for a comprehensive study of their gene expression by microarray and quantitative RT-PCR. Transcriptional regulators known to be important for cell type specification in mouse pancreas were surveyed and compared with those in human cells. An important divergence was observed in MAFB, which was expressed in adult human beta cells. Several transcriptional regulators without previously reported expression restricted to pancreatic and/or exocrine cell subtype were observed, including HOPX (pan-islet), HDAC9 (beta cell), CDX2 (duct) and BATF2 (acinar). We also evaluated potential heterotypic cell×cell interactions between endocrine and exocrine populations and identified 121 ligand–receptor pairs: 27 ligands of seven transmembrane domain (7TM) receptors, 26 ephrins (EFNs) and 14 TGF-β family ligands. Ephrins and their receptors were over-represented, suggesting an important role for these molecules.
Methods
Tissue sources and cell isolation
Human islet samples from normal human donors were provided by the National Institutes of Health Integrated Islet Distribution Program. The use of human tissue was approved by our local Institutional Ethics Committee. Clinical information regarding these samples is listed in electronic supplementary material (ESM) Table 1. Islets were collected after 100 to 700 min of cold ischaemia and cultured in CRML 1066 for 6 to 48 h prior to overnight shipment. Viable (trypan blue-excluding) cell frequency was 95% to 99%. A single cell suspension was prepared by incubation for 10 min in 0.05% (vol./vol.) HyQ Trypsin (Hyclone, Logan, UT, USA) at 37° C, with gentle dispersal by a p1000 micropipettor (Gilson, Middleton, MI, USA) every 3 min. Undispersed material was removed with a 40 μm strainer (BD Falcon, Bedford, MA, USA). Cells were then washed and resuspended in CMRL+2% (vol./vol.) FBS (Hyclone)+0.1 mg/ml trypsin/chymotrypsin inhibitor (Sigma-Aldrich, St Louis, MO, USA). For tissue section labelling, an intact human donor pancreas was provided by S. Orloff (Oregon Health and Science University); portions of the organ were embedded in Tissue-tek cryomatrix (Sakura, Tokyo, Japan) and stored at −86°C.
Indirect immunofluorescent staining
Adult human pancreas cryosections (5 μm) were cut with a CM1950 cryostat (Leica Biosystems, Nussloch, Germany) and dehydrated in acetone for 10 min at −20°C. Non-specific labelling was blocked with 2% (vol./vol.) goat serum (Hyclone). To produce primary antibody, the hybridoma lines listed in ESM Table 2 were grown to super-confluence in 50 ml DMEM+10% (vol./vol.) FBS. Antibody-containing supernatant fractions were collected, tested for reactivity and stored at 4°C. Primary labelling used hybridoma supernatant fractions diluted 1:50 in DPBS for 30 min; secondary labelling was with 1:200 dilution of DyLight488-conjugated anti-mouse IgM (μ chain) and Cy3-conjugated anti-mouse IgG (1+2a+3) (Jackson ImmunoResearch, West Grove, PA, USA) for 20 min. Anti-EPH receptor (EPH) B2 (R&D Systems, Minneapolis, MN, USA) and EFNB3 (LifeSpan BioSciences, Seattle, WA, USA) were diluted 1:100 and detected with the same secondary antibodies. Polyclonal rabbit anti-keratin 19 (KRT19), a gift from X. Wang (Shanghai Institutes for Biological Sciences), was used at 1:400 and detected with Cy3-conjugated anti-rabbit IgG (Jackson ImmunoResearch). Nuclei were labelled with Hoechst 33342 (Molecular Probes, Eugene, OR, USA). An Axioskop 2 plus (Carl Zeiss, Jena, Germany) was used for imaging.
Flow cytometry
Dissociated islet cells were incubated for 30 min at 4°C in hybridoma supernatant fraction diluted 1:50 in CMRL+2% (vol./vol.) FBS+0.1 mg/ml trypsin/chymotrypsin inhibitor. This dilution permitted consistent and specific cell type labelling over a broad range of cell concentrations (0.1×106−5×106 cells/ml). Cells were then washed with cold CMRL and resuspended in a 1:200 dilution of secondary antibodies (PE-conjugated anti-mouse IgM [μ chain] and Dylight488-conjugated anti-mouse IgG [1+2a+3]; Jackson ImmunoResearch). Purified human pancreas islet (-specific antibody) (HPi2) and human pancreas alpha (-specific antibody) (HPa2) antibodies (Novus Biologicals, Littleton, CO, USA) diluted at 1:200 were tested and found to be equally effective as hybridoma supernatant fraction. Dead cells were marked with propidium iodide (10 μg/ml). Cells were analysed with a FACScalibur or sorted by an inFlux V-GS (BD Biosciences, San Jose, CA, USA for both) at 15 psi using a 100 μm nozzle. The forward scatter (FSC): pulse width gating excluded cell doublets from sorts. Data were analysed using FlowJo (Treestar, Ashland, OR, USA).
RNA isolation and quantitative RT-PCR
Cells were sorted into Trizol Liquid Sample (Invitrogen, Carlsbad, CA, USA). First strand cDNA synthesis and quantitative RT-PCR reactions were performed as previously described [22]. Primer sequences are listed in ESM Table 3. Gene expression levels are reported as the difference between baseline-corrected, curve-fitted cycle thresholds for the gene of interest minus the average cycle thresholds of the housekeeping genes Lamin A/C and 18S rRNA. Statistical mean and SD were obtained with Microsoft Excel.
Western blotting
Cell lysates were generated by resuspension of flash-frozen pellets of FACS-sorted populations in SDS-PAGE loading buffer and separated by 12% to 20% gradient Bis-Tris SDS-PAGE (Bio-Rad, Hercules, CA, USA). After semi-dry electrophoretic transfer, membrane-bound proteins were labelled with antibodies recognising histone deacetylase 9 (HDAC9), HOP homeobox (HOPX) (Santa Cruz Biotechnology, Santa Cruz, CA, USA for both) or β-actin (Cell Signaling Technology, Danvers, MA, USA), and visualised by electrochemiluminescence (Perkin Elmer, Waltham, MA, USA). Two patient samples were used, in which >105 alpha and beta cells were available (one man, 30 years old, type O, BMI 32.3 kg/m2; one woman, 50 years old, type unknown, BMI 35.6 kg/m2).
Microarray analyses
cDNA was synthesized from RNA (isolated as described above) and amplified (WT-Ovation Pico; NuGEN Technologies, San Carlos, CA, USA). Amplified cDNA (2 μg) was labelled using BioPrime (Invitrogen) with Cy3/Cy5-labelled nucleotides (GE Amersham Biosciences, Piscataway, NJ, USA). Of the four biological replicates, two were labelled with Cy3 and the others with Cy5; reciprocal labelling controlled for dye bias. We amplified 12 whole-islet cDNA preparations, then pooled and divided them into portions to create four reference samples. Labelled samples were hybridised overnight to the 4×44 Whole Human Genome Array (Agilent, Santa Clara, CA, USA) and scanned with a DNA microarray scanner (G2565B; Agilent). Data were normalised by the print-tip loess method using linear models for microarray data in R [23]. Subsequently, 1000 consistent probes were selected with a maximum of 1.2-fold change across all comparisons. Final expression ratios were generated using loess normalisation. Differentially expressed genes were called using the Significance Analysis of Microarrays one class response package [24] with a false discovery rate (FDR) of 20%. Log2 expression ratios for probes were quantile-normalised and converted to a ratio for each gene (the median of the median of all the values of each probe for that gene). Differentially expressed genes (5038) were identified using PaGE [25] (95% CI). Replicate results were hierarchically clustered using TIGR (www.tm4.org/mev.html, accessed 1 February 2011) [26, 27]. K-means clustering using Pearson correlation was performed to identify patterns for Ingenuity analysis (Ingenuity Systems, Redwood City, CA, USA).
Results
Isolation of human pancreatic endocrine and exocrine subpopulations
To determine the gene expression profiles of the major human pancreatic cell types, we used the collection of cell surface-labelling monoclonal antibodies [22] listed in ESM Table 2. As shown in Fig. 1a–c, dual labelling with the pan-islet marker HPi2 and the alpha cell marker HPa2 allowed separation of alpha (HPi2+HPa2+) and beta (HPi2+HPa2−) cells. To mark exocrine, rather than endocrine cell subpopulations, a three-antibody combination containing the acinar marker human pancreas exocrine (-specific antibody) (HPx1), pan-duct marker HPd3 and the large duct-specific antibody HPd1 was employed. Figure 1d–g shows cells from the same human islet sample sorted with this exocrine marking combination; acinar (HPx1+), large duct (HPd1+HPd3+) and small duct (HPd1−HPd3+) populations are indicated. These example plots show flow cytometric analysis of a sample of relatively low purity (<50% islets), but viable acinar and duct cells were recoverable from all samples. Antibody-labelled endocrine and exocrine subpopulations are illustrated in intact pancreatic tissue in Fig. 1h–j. As expected, HPa2+ alpha cells comprised a subset of HPi2+ islet cells (Fig. 1h), HPx1+ acinar cells were distinct from HPd3+ ducts (Fig. 1i) and large ducts (HPd1+) were seen to be a subset of cells labelled by pan-duct marker HPd3 (Fig. 1j). The pan-duct labelling specificity of HPd3 was also confirmed by co-labelling for keratin 19 (KRT19) [28]; a complete overlap was observed (Fig. 1k).
Fig. 1.
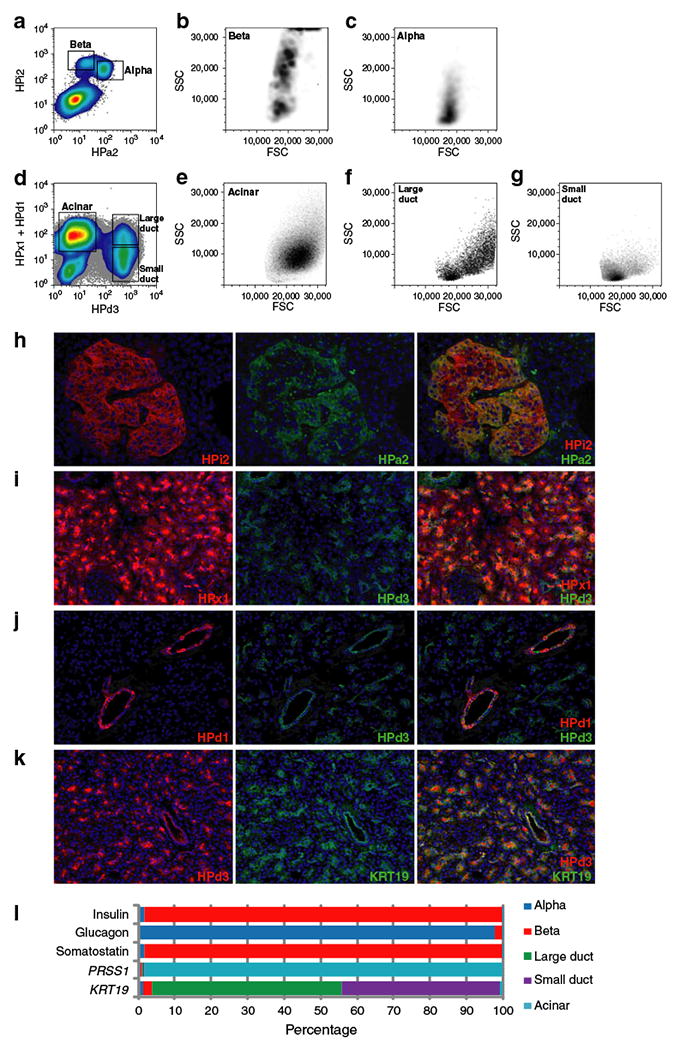
The isolation of subsets of live human pancreatic endocrine and exocrine cells. a The FACS-detected fluorescence of pancreatic islet cells co-labelled with endocrine cell markers HPi2 and HPa2, with back-scattering visualisation of (b) beta and (c) alpha cells to reveal their size/granularity (FSC/side scatter [SSC]) characteristics. d As above (a), but co-labelled with exocrine markers HPx1, HPd1 and HPd3, and with back-scattering visualisation of the populations defined as acinar (e), large duct (f) and small duct (g). h Dual immunofluorescent labelling of adult human pancreatic cryosections simultaneously labelled with the same (a, b) groups of endocrine or (i, j) exocrine cell type-specific antibodies, respectively. k The comprehensive duct labelling of HPd3 is demonstrated by co-labelling with KRT19. l The relative levels of markers of beta (insulin), alpha (glucagon), delta (somatostatin), acinar (PRSS1) or duct (KRT19) cell identity. Quantitative RT-PCR results obtained from FACS-isolated populations were calculated as ΔCt values relative to the mean of Lamin A/C and 18S rRNA. The total signal detected for each marker gene is indicated as a percentage
Pancreatic cell type purity assessment
Figure 1l shows the relative levels of mRNA encoding the prototypical cell type markers insulin, glucagon, somatostatin, trypsin and KRT19, as measured in the seven samples listed in ESM Table 1. The HPi2+HPa2− beta cell fraction contained 115-fold more insulin transcripts than any of the other populations, indicating that any beta cell contamination of the other populations was low. However, the presence of the majority of the somatostatin mRNA in this fraction indicated that delta cells were also HPi2+HPa2−. For comparison, HPi2+HPa2+ alpha cells contained 45-fold more glucagon, the HPx1+ acinar population had 396-fold more trypsin and the duct fractions (HPd3+HPd1+/−) had 25-fold and 21.2-fold more KRT19 mRNA than their counterparts. Thus, our sorting scheme permitted separation and molecular analysis of these populations.
Microarray analyses of pancreatic cell subpopulations
Having verified the purity of these populations, we assessed their global mRNA expression profile by microarray analysis using four biological replicates per population. These data are available through the ArrayExpress database (www.ebi.ac.uk/arrayexpress/, accessed 1 July 2011; accession numbers E-MTAB-463 and E-MTAB-465), and gene lists can be found at http://137.53.250.24/grompelab/ (accessed 1 July 2011). Cluster analysis of the global gene expression profiles was performed using five sorted cell populations and focusing on 5,038 genes that were differentially expressed between any pair of cell types. As shown in Fig. 2, the two endocrine populations are closely related, but a group of genes was expressed differently in alpha and beta cells. The duct and acinar populations appear on a separate branch of the tree. Large and small ducts are nearly identical, but unique blocks are found in duct and acinar populations. K-means clustering revealed five major expression patterns: endocrine (1,086 genes), beta cell-specific (528), non-endocrine (845), acinar-specific (925) and duct-specific (457). Genes expressed specifically in endocrine cells (or beta cells only) were significantly enriched for diabetes (p=5× 10−11) and neurological disorder (p=1× 10−20) categories according to Ingenuity analysis, indicating an association of neurological secretion pathways with endocrine cells. The top pathway in the acinar-specific genes was protein synthesis (p=6× 10−17). Several pathways were strongly enriched among genes specific to ducts, namely: cell migration (p=3 × 10−15), cell-to-cell signalling, especially adhesion (p=6× −14), cell death (p=7× 10−14) and tumorigenesis (p=9× 10−11).
Fig. 2.
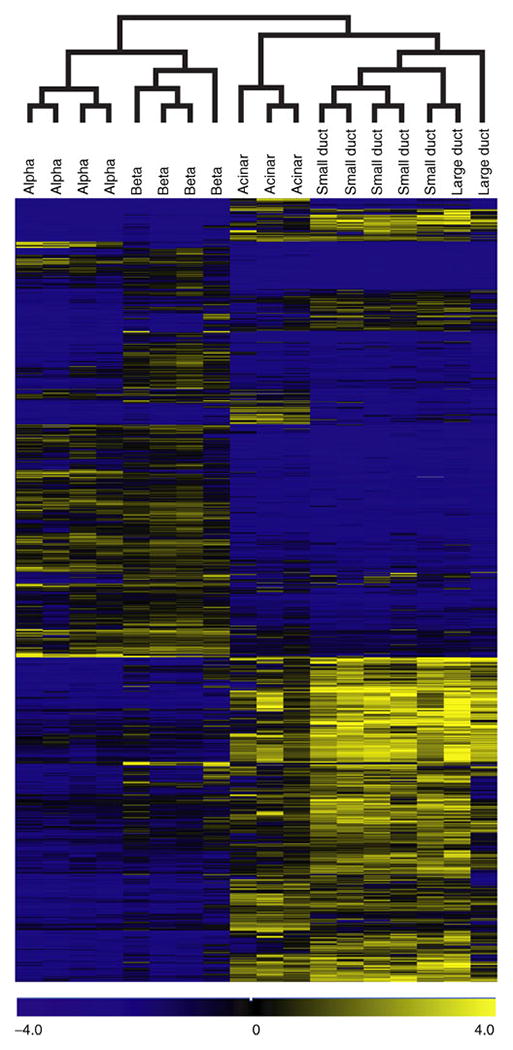
Hierarchical clustering of microarray-assessed gene expression in human pancreatic endocrine and exocrine cell subpopulations. We used PaGE algorithm to identify 5,038 differentially expressed genes (in any pair-wise comparison; 95% CI), and clustered them hierarchically using TIGR Multiexperiment Viewer
Comparative gene expression in beta vs alpha cells
To assess the major differences in gene expression between alpha and beta cells, we performed two-colour microarray analyses. These experiments permitted a direct comparison of these highly related endocrine populations using cells subjected to identical handling (islet isolation from the same individual, followed by simultaneous enzymatic dispersal, antibody labelling and FACS). The most strongly differentially expressed genes in human beta and alpha cells are listed in Tables 1 and 2. Genes with strong beta cell-specific expression included known factors, e.g. HADH [29], IL1B [30], IAPP [31] and PTGS2 [32], as well as several novel genes. Alpha cell-selective genes have been less well studied, but transcription factors shown to contribute to alpha cell fate specification (IRX2, ARX, see below) were strongly upregulated in this population. To measure whether delta cell-selective genes might have been misidentified as beta cell-selective, ten of the top candidates were examined in cells enriched by Newport Green dye positivity rather than antibody selection. As illustrated in ESM Table 4, this method provided weaker beta cell enrichment, but substantially reduced delta cell contamination (NG+ cells were only twofold enriched for somatostatin). Other than DCX, each of the genes examined was also enriched in the NG+ population and therefore beta cell-selective. Certain known beta cell-selective genes showed differential expression, but did not meet statistical significance (e.g. PDX1, which was ninefold enriched in beta cells, but with >10% FDR) or were not detected by the array (e.g. MAFA), suggesting that low abundance transcripts such as transcription factors were being missed. Because these include important regulators of pancreatic cell fate, we specifically examined transcriptional regulators by quantitative RT-PCR to augment the microarray results.
Table 1.
Genes with >20-fold elevated expression in sorted human beta cells relative to alpha cells
| Gene | Transcript | Description | FC | FDR | AdjP |
|---|---|---|---|---|---|
| SSTa | NM_001048 | Somatostatin | 107.8 | 0 | 4.4× 10−3 |
| CLIC6 | NM_053277 | Chloride intracellular channel 6 | 76.6 | 0 | 5.2 ×10−3 |
| BM678403 | BM678403 | cDNA clone UI-E-EO0-ahw-c-09-0-UI 3′ | 70.1 | 0 | 4.4× 10−3 |
| CB305813 | CB305813 | cDNA clone UI-CF-EN1-aeg-d-07-0-UI 3′ | 65.1 | 0 | 3.4 × 10−3 |
| TFCP2L1 | NM_014553 | Transcription factor CP2-like 1 | 56.3 | 0 | 3.4 × 10−3 |
| ITIH5 | NM_030569 | Inter-alpha (globulin) inhibitor H5 | 56.1 | 0 | 9.1× 10−3 |
| DGKB | NM_004080 | Diacylglycerol kinase, beta 90 kDa | 54.9 | 0 | 1.0 × 10−2 |
| INS | NM_000207 | Insulin | 45.9 | 0 | 1.8× 10−2 |
| HADH | NM_005327 | Hydroxyacyl-coenzyme A dehydrogenase | 43.6 | 0 | 8.0× 10−3 |
| SOD3 | NM_003102 | Superoxide dismutase 3, extracellular | 42.2 | 0 | 3.4 × 10−3 |
| HLA-DQB1 | NM_002123 | MHC class II HLA-DQ-beta | 40.9 | 0 | 2.1 × 10−2 |
| C9orf135 | NM_001010940 | Chromosome 9 open reading frame 135 | 37.9 | 0 | 4.4× 10−3 |
| DLK1 | NM_003836 | Delta-like 1 H. log (Drosophila) | 37.9 | 0 | 1.8× 10−2 |
| BX438895 | BX438895 | cDNA clone IMAGE:785925 3′ | 34.5 | 0 | 1.8× 10−2 |
| SSPN | NM_005086 | Sarcospan (Kras oncogene-associated gene) | 34.1 | 0 | 7.1 × 10−3 |
| PTPRK | NM_002844 | Protein tyrosine phosphatase, receptor type, K | 33.8 | 0 | 4.7× 10−3 |
| ASB9 | NM_001031739 | Ankyrin repeat and SOCS box-containing 9 | 32.2 | 0 | 5.2 × 10−3 |
| DCXa | NM_000555 | Doublecortex; lissencephaly, X-linked | 30.8 | 0 | 4.9× 10−3 |
| IAPP | NM_000415 | Islet amyloid polypeptide | 30.6 | 0 | 1.0× 10−2 |
| RBP4 | NM_006744 | Retinol binding protein 4, plasma | 30.4 | 0 | 5.2 ×10−3 |
| PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 | 29.6 | 0 | 1.3 × 10−2 |
| SCD5 | NM_001037582 | Stearoyl-CoA desaturase 5 | 29.6 | 0 | 5.2 × 10−3 |
| DAPL1 | NM_001017920 | Similar to death-associated protein | 29.3 | 0 | 3.4 × 10−3 |
| RGS16 | NM_002928 | Regulator of G-protein signalling 16 | 28.9 | 0 | 4.3× 10−3 |
| PLCH2 | NM_014638 | Phospholipase C, eta 2 | 28.8 | 0 | 7.8× 10−3 |
| BG818013 | BG818013 | cDNA clone IMAGE:4915128 5' | 28.5 | 0 | 6.6× 10−3 |
| LOC284033b | AK095052 | cDNA FLJ37733 fis, clone BRHIP2020827 | 28.5 | 0 | 9.3× 10−3 |
| TNS1 | NM_022648 | Tensin 1 | 27.2 | 0 | 4.7× 10−3 |
| GLT25D2 | NM_015101 | Glycosyltransferase 25 domain containing 2 | 27.2 | 0 | 6.4× 10−3 |
| CALD1 | NM_033138 | Caldesmon 1, transcript variant 1 | 27.0 | 0 | 4.7× 10−3 |
| PRPH | NM_006262 | Peripherin | 26.3 | 0 | 6.0× 10−3 |
| IL1B | NM_000576 | Interleukin 1, beta | 26.0 | 0.15 | 3.0 × 10−2 |
| TSHZ2 | ENST00371497 | Teashirt H.log 2 (Zinc finger protein 218) | 25.0 | 0 | 1.7 × 10−2 |
| SGIP1 | NM_032291 | SH3-domain GRB2-like interacting protein 1 | 24.6 | 0 | 3.4 ×10−3 |
| ESR1 | NM_000125 | Oestrogen receptor 1 | 24.4 | 0 | 4.3 × 10−3 |
| CDH22 | NM_021248 | Cadherin-like 22 | 24.2 | 0 | 8.6× 10−3 |
| IGF2 | NM_000612 | Insulin-like growth factor 2 (somatomedin A) | 23.7 | 0 | 3.4 ×10−3 |
| ANTXR1 | NM_032208 | Anthrax toxin receptor 1 | 23.6 | 0 | 9.6× 10−3 |
| THC2656690 | THC2656690 | NA | 23.6 | 0 | 9.5× 10−3 |
| BC063022 | BC063022 | cDNA clone IMAGE:5246259, partial cds | 23.6 | 0 | 1.1 × 10−2 |
| IGSF11 | NM_152538 | Immunoglobulin superfamily, member 11 | 23.4 | 0 | 4.7× 10−3 |
| SFRP1 | NM_003012 | Secreted frizzled-related protein 1 | 23.1 | 0.25 | 4.5 × 10−2 |
| XYLT1 | NM_022166 | Xylosyltransferase I | 23.0 | 0.09 | 2.3 × 10−2 |
| ADCYAP1 | NM_001099733 | Pituitary adenylate cyclase-activating polypeptide | 22.8 | 0.09 | 2.2× 10−2 |
| EPDR1 | NM_017549 | Ependymin related protein 1 (zebrafish) | 22.8 | 0 | 4.2 ×10−3 |
| FBN2 | NM_001999 | Fibrillin 2 (congenital contractural arachnodactyly) | 22.8 | 0 | 1.8× 10−2 |
| PCDH7 | NM_002589 | BH-protocadherin, transcript variant a | 22.4 | 0 | 7.9× 10−3 |
| SLC17A6 | NM_020346 | Solute carrier family 17, member 6 | 22.2 | 0 | 1.7×10−2 |
| CNGA3 | NM_001298 | Cyclic nucleotide gated channel alpha 3 | 21.6 | 0 | 4.7× 10−3 |
| BC104421 | BC104421 | cDNA clone IMAGE:40004940 | 21.6 | 0 | 8.0× 10−3 |
| CAPN13 | NM_144575 | Calpain 13 | 20.6 | 0 | 3.4 × 10−3 |
Expression levels are the means derived using four runs of two-colour pairwise binding of FACS-sorted alpha and beta cell cDNA. Genes with an FDR of ≥5%, duplicate probe results and probes called as ‘marginal’ were all excluded
Attributed to delta cell contamination (see below);
Also known as SHISA6
AdjP, adjusted p value; FC, fold change
Table 2.
Genes with ≥20-fold elevated expression in sorted human alpha cells relative to beta cells
| Gene | Transcript | Description | FC | FDR | AdjP |
|---|---|---|---|---|---|
| GCG | NM_002054 | Glucagon | 111.1 | 0 | 1.5× 10−2 |
| POPDC3 | NM_022361 | Popeye domain containing 3 | 83.3 | 0 | 1.4 × 10−2 |
| LOXL4 | NM_032211 | Lysyl oxidase-like 4 | 83.3 | 0 | 4.3× 10−3 |
| GRIN3A | NM_133445 | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 66.7 | 0 | 3.4 ×10−3 |
| STK32B | NM_018401 | Serine/threonine kinase 32B | 58.8 | 0 | 5.4 × 10−3 |
| SPINK4 | NM_014471 | Serine peptidase inhibitor, Kazal type 4 | 55.6 | 0 | 3.4 ×10−3 |
| CRH | NM_000756 | Corticotropin releasing hormone | 43.5 | 0 | 1.2 ×10−2 |
| THC2690347 | THC2690347 | Seven TM receptor protein 224, partial | 41.7 | 0 | 1.7 ×10−2 |
| GC | NM_000583 | Group-specific component (vitamin D binding protein) | 38.5 | 0 | 5.0×10−3 |
| FA P | NM_004460 | Fibroblast activation protein, alpha | 33.3 | 0.25 | 4.1 × 10−2 |
| GJA3 | NM_021954 | Gap junction protein, alpha 3, 46 kDa (connexin 46) | 31.3 | 0 | 5.0 ×10−3 |
| MUCL1 | NM_058173 | Small breast epithelial mucin | 29.4 | 0 | 3.4×10−3 |
| AI492422 | AI492422 | cDNA clone IMAGE:2131746 3' | 29.4 | 0 | 1.3×10−2 |
| C20orf39a | NM_024893 | Chromosome 20 open reading frame 39 | 28.6 | 0.07 | 1.9×10−2 |
| VIM | NM_003380 | Vimentin | 27.0 | 0 | 8.9× 10−3 |
| STC2 | NM_003714 | Stanniocalcin 2 | 27.0 | 0 | 4.4× 10−3 |
| PTPRT | NM_133170 | Protein tyrosine phosphatase, receptor, T transcript variant 1 | 27.0 | 0 | 6.0×10−3 |
| MUC13 | NM_033049 | Mucin 13, cell surface associated | 25.6 | 0 | 4.5× 10−3 |
| BQ286187 | BQ286187 | cDNA clone IMAGE:5782164 5′ | 25.6 | 0 | 4.4× 10−3 |
| IRX2 | AY335940 | Iroquois-class homeodomain protein IRX-2 | 25.0 | 0 | 9.3 ×10−3 |
| THC2496213 | THC2496213 | cDNA clone IMAGE:2735726 3′ | 23.8 | 0 | 6.8× 10−3 |
| RP11-35N6.1b | NM_207299 | Plasticity related gene 3, transcript variant 1 | 23.8 | 0 | 1.0 ×10−2 |
| F10 | NM_000504 | Coagulation factor X | 22.7 | 0 | 5.2 ×10−3 |
| ARX | NM_139058 | Aristaless related homeobox | 22.2 | 0 | 4.7× 10−3 |
| BVES | NM_147147 | Blood vessel epicardial substance, transcript variant 5 | 21.7 | 0.22 | 3.4×10−2 |
| NPNT | NM_001033047 | Nephronectin | 20.8 | 0 | 8.9× 10−3 |
| C11orf41 | NM_012194 | G2 protein mRNA, partial cds | 20.8 | 0.32 | 4.2×10−2 |
| FXYD5 | NM_144779 | FXYD domain containing ion transport regulator 5 | 20.0 | 0 | 3.4×10−3 |
| BQ777622 | BQ777622 | cDNA clone IMAGE:6032433 3′ | 20.0 | 0 | 9.4× 10−3 |
Expression levels are the means derived using four runs of two-colour pairwise binding of FACS-sorted alpha and beta cell cDNA. Genes with an FDR of ≥5%, duplicate probe results and probes called as ‘marginal’ were all excluded
C20orf39, also known as SYNDIG1;
RP11-35N6.1, also known as LPPR1
AdjP, adjusted p value; FC, fold change
Cell type-restricted expression of transcriptional regulators
Of the islet samples listed in ESM Table 1, five were used to examine specific transcription factor expression by quantitative RT-PCR. Several known regulators of mouse endocrine pancreatic cell fate were surveyed (Fig. 3a) in human alpha and beta cell mRNA. Among these we found that the expression of PDX1 and MAFA was restricted to beta cells and that of IRX2 and ARX to alpha cells, as expected from prior data obtained in rodents. MAFB, NKX2-2, NKX6-1, PAX6 and HNF4A expression was detected in both cell types, whereas PAX4 and NEUROG3 mRNA was not detected in either population (data not shown). Of these results, the most notable was that MAFB is expressed in human beta cells at a level comparable to that in alpha cells (whereas adult mouse beta cells lack Mafb) [12]. We also report the first detection of IRX2 in adult alpha cells; prior observations were confined to fetal mouse tissue [33].
Fig. 3.
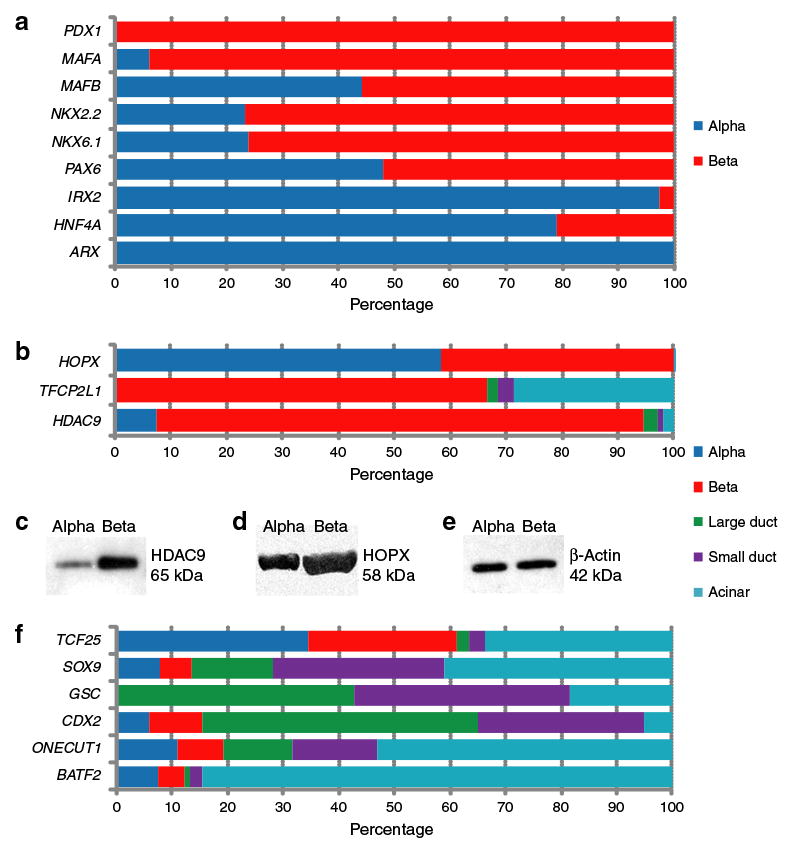
Gene expression distribution of known and novel transcriptional regulators in pancreatic cell subpopulations. a, b, f Quantitative RT-PCR results obtained from RNA isolated from FACS-isolated human pancreatic cell populations were calculated as ΔCt values relative to the mean of housekeeping genes Lamin A/C and 18S rRNA. The total signal detected for each gene in each pancreatic cell subpopulation is indicated in per cent. c–e Protein levels of HDAC9, HOPX and β-actin were determined by SDS-PAGE of lysates obtained from sorted populations and by western blotting
Microarray analysis also revealed differential expression of regulators not previously described in pancreatic islets. Figure 3b shows quantitative RT-PCR confirmation that expression of HOPX, TFCP2L1 and HDAC9 in the human pancreas is restricted to cell types. HOPX mRNA was detected in alpha and beta cells, but was absent from any exocrine population. TFCP2L1 was expressed only by beta cells within the islet, but its mRNA was also detectable (at a lower level) in acinar cells. HDAC9 was detected at much higher levels in beta cells than in any other pancreatic cell population. As illustrated in Fig. 3c–e, western blotting indicated that the mRNA beta cell-selective and pan-islet expression patterns were also reflected at the protein level for HDAC9 and HOPX, respectively.
We also identified a few transcription factors that were differentially expressed in exocrine pancreatic populations (Fig. 3f). These included genes with an alpha and acinar cell-specific transcript distribution (TCF25), a pan-exocrine expression pattern (SOX9 and GSC), duct-specific gene activation (CDX2) and acinar-specific expression (ONECUT1 and BATF2). Of these, only SOX9 and ONECUT1 had previously been reported to mark pancreatic cell lineages; the proteins encoded by these, SRY-box containing gene 9 and one cut domain, family member 1, are markers of developing ducts and islets in mice [34, 35].
Heterotypic cell×cell interaction in the human pancreas
To evaluate signalling interactions between different human pancreatic cell subpopulations, we conducted differential expression analysis using PaGE with a 20% FDR for ligand- and receptor-encoding genes for each pair of cell types (each cell type vs each of the remaining four cell types). Table 3 lists the matches compared with a curated human receptor–ligand database (Human Plasma Membrane Receptome [HPMR], www.receptome.org/HPMR/, accessed 1 February 2011). To specifically evaluate interactions between islet hormones and duct-resident hormone receptors, we compared our RNA-based observations for these populations with those of Bertelli and Bendayan [36], who recently published a protein-based survey of secondary islet hormones reported to affect duct and/or acinar secretion (Table 4). Substantial agreement was observed.
Table 3.
Ligand–receptor pairs supported by HPMR and by HPRD protein × protein interaction prediction
| Population with ligand | Population with receptor | HPMR (n)a | HPRD (n)a |
|---|---|---|---|
| Acinar | Alpha | 5 | 5 |
| Acinar | Beta | 1 | |
| Acinar | Small duct | 3 | |
| Alpha | Acinar | 8 | 16 |
| Alpha | Beta | 1 | 2 |
| Alpha | Large duct | 9 | 15 |
| Alpha | Small duct | 12 | 18 |
| Beta | Acinar | 18 | 28 |
| Beta | Alpha | 2 | 2 |
| Beta | Large duct | 22 | 29 |
| Beta | Small duct | 20 | 32 |
| Large duct | Acinar | 1 | 1 |
| Large duct | Alpha | 8 | 8 |
| Large duct | Beta | 3 | 7 |
| Small duct | Acinar | 4 | 5 |
| Small duct | Alpha | 5 | 15 |
| Small duct | Beta | 3 | 12 |
Total number of pairs: HPMR 121, HPRD 199
Table 4.
Expression pattern of known islet hormone and ductal receptor genes
| Hormone precursor | Expression pattern | Hormonelocalisationa | Receptor | Expression pattern |
|---|---|---|---|---|
| IAPP | ↑ in beta, alpha | Beta+delta | ||
| ADM | ↑ in small duct, beta | PP | RAMP2 | ↑ in alpha |
| CALCA | ↑ in acinar, alpha | Subpopulation of delta | ||
| INS | Beta | |||
| CHGA | CHGB ↑ in alpha, beta | Alpha+beta | ||
| SCG2 | ↑ in alpha, beta | Alpha+beta | ||
| GHRL | n.d.e. | Epsilon | ||
| CORT | ↑ in alpha, beta | Single islet cells | SSTR3 | ↑ in large duct, acinar, small duct |
| HCRT | Alpha+beta | HCRTR1 | ↑ in acinar | |
| RETN | n.d.e. | Beta | ||
| UCN | ↑ in large duct, alpha | Beta | ||
| CRH | ↑ in alpha, beta | Alpha | CRHRb | ↑ in large duct |
| PYY | ↑ in large duct | Alpha, some PP | PPYR1 | ↑ in acinar, large/small duct |
| NPYR5c | ↑ in large/small duct | |||
| INS | ↑ in beta, alpha | INSR | ↑ in acinar, large duct | |
| INSL3/5 | ↑ in large/small duct | |||
| GCG | ↑ in alpha, beta | GCGR | ↑ in large duct | |
| SST | ↑ in beta | SSTR1 | ↑ in alpha and beta | |
| SSTR2 | ↑ in alpha | |||
| SSTR3 | ↑ in large duct, acinar, small duct | |||
| SSTR4 | ↑ in large/small duct | |||
| GLP1R | ↑ in beta |
Vertical arrow symbols (↑) denote increased expression.
As reported by Bertelli and Bendayan [36];
CRHR, also known as CRHR1;
NPYR5, also known as NPY5R
IAPP, islet amyloid polypeptide; n.d., not determined; n.d.e., no differential expression; PP, polypeptide
Predicted heterotypic ligand–receptor pairs
For ten pancreatic cell-pairs and two directions (ligand–receptor), 121 ligand–receptor pairs were identified (Table 3, ESM Table 5). These included 62 non-redundant relationships previously reported in non-pancreatic tissues or tumours. Others have been described previously in the pancreas, such as EPHA4+ EFNA5, vascular endothelial growth factor+neuropilin, angiotensinogen+AGT receptor and betacellulin+EGF receptor. We identified 89 pairs of endocrine ligands (60 beta, 29 alpha) and exocrine receptors, and 24 pairs of endocrine receptors and exocrine ligands. For the remaining eight pairs, ligand and receptor were in the same compartment. ESM Fig. 1 illustrates these interactions; the detailed relationships are listed in ESM Table 6.
Ligand–receptor family composition
To obtain an overview of 121 ligand–receptor pairs, the distribution of ligands among the families was categorised in HPMR. The ligand distribution among families was uneven, with ligands of 7TM receptors, EFN and TGF-beta family ligands being the most frequent (ESM Table 6). This was mirrored in the corresponding receptor subfamilies (ESM Table 6). After accounting for ligand family representation, EFNs and EPH receptors were substantially over-represented. This is consistent with a recent publication that examined intra-islet paracrine signalling and identified EFN family gene expression [37].
From in vivo and/or in vitro protein×protein interaction, 199 ligand–receptor pairs from the Human Protein Reference Database (HPRD) were inferred (Table 3, ESM Table 5). Table 5, ESM Table 7 and ESM Fig. 1 show comparisons of these results with the HPMR observations described above. Of the additional protein×protein interactions identified only in the HPRD database, most belong to the TGF-β and fibroblast growth factor families.
Table 5.
EPH receptor–EFN pairs predicted in inter-tissue (exocrine–endocrine) and intra-tissue (alpha–beta) cell type pairs
| Population with ligand | Population with receptor | Ligand | Recepto |
|---|---|---|---|
| Acinar | Alpha | EFNA1 | EPHA4 |
| Small duct | Alpha | EFNA1 | EPHA4 |
| Acinar | Alpha | EFNA2 | EPHA4 |
| Alpha | Small duct | EFNA5 | EPHA2 |
| Alpha | Small duct | EFNA5 | EPHB2 |
| Beta | Large duct | EFNA5 | EPHA2 |
| Beta | Small duct | EFNA5 | EPHA2 |
| Beta | Alpha | EFNA5 | EPHA4 |
| Beta | Large duct | EFNA5 | EPHA4 |
| Beta | Large duct | EFNA5 | EPHA5 |
| Beta | Acinar | EFNA5 | EPHA8 |
| Beta | Large duct | EFNA5 | EPHA8 |
| Beta | Small duct | EFNA5 | EPHA8 |
| Beta | Acinar | EFNA5 | EPHB2 |
| Beta | Large duct | EFNA5 | EPHB2 |
| Beta | Small duct | EFNA5 | EPHB2 |
| Small duct | Alpha | EFNB2 | EPHA4 |
| Alpha | Large duct | EFNB3 | EPHB2 |
| Alpha | Small duct | EFNB3 | EPHB2 |
| Alpha | Large duct | EFNB3 | EPHB3 |
| Beta | Large duct | EFNB3 | EPHA4 |
| Beta | Acinar | EFNB3 | EPHB2 |
| Beta | Large duct | EFNB3 | EPHB2 |
| Beta | Small duct | EFNB3 | EPHB2 |
| Beta | Large duct | EFNB3 | EPHB3 |
| Beta | Small duct | EFNB3 | EPHB3 |
The expression of EPH receptor–EFN gene families revealed distinct usage of protein isoforms by cell types. EFNB3 and EFNA5 were upregulated in endocrine (alpha and beta) cells, whereas EFNB2 and EFNA1 were produced at higher levels in the exocrine compartment (large, small duct and acinar). For the EPH receptors, each exocrine cell type had at least two genes upregulated, but none were differentially activated in alpha or beta cells (Table 5). This pattern suggests that communication between exocrine and endocrine cells occurs and can be mediated by EPH receptor–EFN signalling. ESM Fig. 2 illustrates the distribution of receptor–ligand pair EFNB3 and EPHB2 protein in pancreas tissue. Consistent with the RNA-based observations, these proteins were observed primarily in islets or ducts, respectively.
Discussion
In this study, we generated the first gene expression atlas of adult human pancreatic alpha, beta, duct and acinar cells. The expression of known transcriptional regulators was similar—but not identical—to that in mouse islets, and novel transcriptional regulators were discovered in islet and exocrine cells. In addition, a computational ligand–receptor survey revealed potential paracrine regulatory circuits, including several EFN family members. These observations extend the understanding of human pancreatic cell regulation and should aid future studies of cell fate determination and function.
Transcriptional regulator expression in endocrine and exocrine cells
Although MAFB is often described as an alpha cell marker in adult pancreatic tissue [12], several exceptions have been observed. In the islets of adult Meriones jirds (gerbils), MAFB is found in alpha and beta cells [38]; even in mice, MAFB is detectable in the beta cells of adolescents and pregnant adults [39]. Coupled with our observation that MAFB is expressed in adult human beta cells, this suggests that the pattern observed in adult mice is the exception rather than the rule. MAFB has not been regarded as a useful reprogramming agent for the derivation of beta cells; our findings suggest that this should be re-evaluated. Iroquois homeobox genes like IRX2 are transcription factors with developmental roles in broad pattern specification [40]. In one report, Irx2 was shown to be restricted to alpha cells in the developing mouse pancreas [33], suggesting a potential role for the specification of this lineage. Since IRX2 is also restricted to alpha cells in adult humans, it could be a target for inhibition to suppress polyhormonal fates and promote beta cell identity. The deletion of Arx, expression of which is also restricted to alpha cells in the adult human islet, increases the ratio of beta:alpha and/or delta:alpha cells in knockout mice [41].
We observed cell type-selective expression of several transcriptional regulators not previously reported in the pancreas; three of these had particularly noteworthy cell type specificities. The transcriptional regulator HOPX contains a homeobox domain, but lacks the DNA binding motif normally associated with HOX genes [42]. In developing cardiac and pulmonary tissue, HOPX acts downstream of NK2-1 and NK2-5 (respectively) [43]. Its expression in adult human islet cells, which also express NK family genes, suggests that HOPX might contribute to the specification or maintenance of endocrine cell identity. Beta cell-specific expression of HDAC9 is of particular interest. HDAC9 is a transcriptional co-repressor characterised principally as a neuronal development factor [44]. However, it was recently implicated as a fasting-induced upstream transcription factor-1 inhibitor and an important element of a novel insulin-dependent metabolic regulatory system [45]. Beta cell-specific HDAC9 expression in the pancreas suggests that it could be a useful reprogramming agent and enforcer of beta cell identity. Information on BATF2 is extremely limited, but our observation of highly acinar cell-specific expression is potentially important. The reprogramming of acinar cells to a beta cell-like state has been demonstrated [46]; if this process could be enhanced by suppression of factors like BATF2, it could become a significant therapeutic approach.
Heterotypic cell type interactions: the roles of EPH receptor–EFN signalling in the pancreas
The ligand and receptor in EPH receptor–EFN signalling are cell surface localised; binding between these proteins requires cell–cell contact and triggers a forward signalling pathway in the receptor cell and a reverse signalling pathway in the ligand cell [47]. EPH receptor–EFN signalling has been implicated in several processes, including tissue development, cancer and glucose homeostasis. In the latter, EPH receptor–EFN signalling regulates insulin secretion, with EPH receptor signalling suppressing insulin secretion, whereas EFN signalling activates it; the balance correlates with blood sugar level [48]. Islet hormone regulation of duct cell bicarbonate fluid secretion is physiologically important [49]. However, the topographical association between islet and duct cells in the pancreas implies that cell organisation during epithelial branching morphogenesis and capillary formation is important for pancreatic development. EPH receptor–EFN signalling is important for regulating this process during neural development and may play a similar role during pancreatic tissue organisation.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIDDK grants U01 DK072477 (to M. Grompe) and R21 DK071216 (to K. H. Kaestner). We thank S. Abraham, K. Shoop, Y. Zhong and M.T. Grompe for their antibody development work, and salute E. McGonigal for his assistance with FACS sorting.
Abbreviations
- EFN
Ephrin
- EPH
Ephrin receptor
- FDR
False discovery rate
- FSC
Forward scatter
- HPa2
Human pancreas alpha (-specific antibody)
- HPi2
Human pancreas islet (-specific antibody)
- HPx1
Human pancreas exocrine (-specific antibody)
- HPd1/HPd3
Human pancreas duct (-specific antibody)
- HPMR
Human plasma membrane receptome
- HPRD
Human protein reference database
- 7TM
Seven transmembrane domain
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00125-011-2283-5) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Contribution statement CD, JS and CFL designed, analysed, interpreted data and drafted the manuscript. PSC, AJF, OS and RB analysed data and edited the manuscript. PRS, CJS, KHK and MG conceived and designed experiments, and edited the manuscript. All authors approved the final version.
Duality of interest Several of the antibodies described have been licensed to Novus Biologicals, LLC. C. Dorrell, P.R. Streeter and M. Grompe will receive royalty revenue from their sale. All other authors declare that there is no duality of interest associated with this manuscript.
Contributor Information
C. Dorrell, Papé Family Pediatric Research Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, L321, Portland, OR 97239, USA
J. Schug, Department of Genetics and Institute for Diabetes, Obesity, and Metabolism, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
C. F. Lin, Department of Genetics and Center for Bioinformatics, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
P. S. Canaday, Papé Family Pediatric Research Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, L321, Portland, OR 97239, USA
A. J. Fox, Department of Genetics and Institute for Diabetes, Obesity, and Metabolism, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
O. Smirnova, Department of Genetics and Institute for Diabetes, Obesity, and Metabolism, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
R. Bonnah, Papé Family Pediatric Research Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, L321, Portland, OR 97239, USA
P. R. Streeter, Papé Family Pediatric Research Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, L321, Portland, OR 97239, USA
C. J. Stoeckert, Jr, Department of Genetics and Center for Bioinformatics, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
K. H. Kaestner, Department of Genetics and Institute for Diabetes, Obesity, and Metabolism, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
M. Grompe, Papé Family Pediatric Research Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Rd, L321, Portland, OR 97239, USA, grompem@ohsu.edu
References
- 1.Maffei A, Liu Z, Witkowski P, et al. Identification of tissue-restricted transcripts in human islets. Endocrinology. 2004;145:4513–4521. doi: 10.1210/en.2004-0691. [DOI] [PubMed] [Google Scholar]
- 2.Gunton JE, Kulkarni RN, Yim S, et al. Loss of ARNT/HIF1β mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Kutlu B, Burdick D, Baxter D, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyttle BM, Li J, Krishnamurthy M, et al. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia. 2008;51:1169–1180. doi: 10.1007/s00125-008-1006-z. [DOI] [PubMed] [Google Scholar]
- 5.Marselli L, Thorne J, Dahiya S, et al. Gene expression profiles of beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5:e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabat M, Luciani DS, Piret JM, Johnson JD. Maturation of adult beta-cells revealed using a Pdx1/insulin dual-reporter lentivirus. Endocrinology. 2009;150:1627–1635. doi: 10.1210/en.2008-1224. [DOI] [PubMed] [Google Scholar]
- 7.Hara M, Wang X, Kawamura T, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 8.Quoix N, Cheng-Xue R, Guiot Y, Herrera PL, Henquin JC, Gilon P. The GluCre-ROSA26EYFP mouse: a new model for easy identification of living pancreatic alpha-cells. FEBS Lett. 2007;581:4235–4240. doi: 10.1016/j.febslet.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 9.Meivar-Levy I, Ferber S. Adult cell fate reprogramming: converting liver to pancreas. Methods Mol Biol. 2010;636:251–283. doi: 10.1007/978-1-60761-691-7_16. [DOI] [PubMed] [Google Scholar]
- 10.Kordowich S, Mansouri A, Collombat P. Reprogramming into pancreatic endocrine cells based on developmental cues. Mol Cell Endocrinol. 2010;323:62–69. doi: 10.1016/j.mce.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artner I, Le Lay J, Hang Y, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 13.Jensen J, Serup P, Karlsen C, Nielsen TF, Madsen OD. mRNA profiling of rat islet tumors reveals nkx 6.1 as a beta-cell-specific homeodomain transcription factor. J Biol Chem. 1996;271:18749–18758. doi: 10.1074/jbc.271.31.18749. [DOI] [PubMed] [Google Scholar]
- 14.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 15.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 16.Turque N, Plaza S, Radvanyi F, Carriere C, Saule S. Pax-QNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol Endocrinol. 1994;8:929–938. doi: 10.1210/mend.8.7.7984154. [DOI] [PubMed] [Google Scholar]
- 17.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baeyens L, Bouwens L. Can beta-cells be derived from exocrine pancreas? Diabetes Obes Metab. 2008;10(Suppl 4):170–178. doi: 10.1111/j.1463-1326.2008.00949.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Hoof D, D'Amour KA, German MS. Derivation of insulin-producing cells from human embryonic stem cells. Stem Cell Res. 2009;3:73–87. doi: 10.1016/j.scr.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Pavlovic D, Chen MC, Bouwens L, Eizirik DL, Pipeleers D. Contribution of ductal cells to cytokine responses by human pancreatic islets. Diabetes. 1999;48:29–33. doi: 10.2337/diabetes.48.1.29. [DOI] [PubMed] [Google Scholar]
- 21.Movahedi B, Van de Casteele M, Caluwe N, et al. Human pancreatic duct cells can produce tumour necrosis factor-alpha that damages neighbouring beta cells and activates dendritic cells. Diabetologia. 2004;47:998–1008. doi: 10.1007/s00125-004-1426-3. [DOI] [PubMed] [Google Scholar]
- 22.Dorrell C, Abraham SL, Lanxon-Cookson KM, Canaday PS, Streeter PR, Grompe M. Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res. 2008;1:183–194. doi: 10.1016/j.scr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Gentleman R. Bioinformatics and computational biology solutions using R and bioconductor. Springer; New York: 2005. [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant GR, Liu J, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- 26.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 27.Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 micro-array software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 28.von Burstin J, Reichert M, Wescott MP, Rustgi AK. The pancreatic and duodenal homeobox protein PDX-1 regulates the ductal specific keratin 19 through the degradation of MEIS1 and DNA binding. PLoS One. 2010;5:e12311. doi: 10.1371/journal.pone.0012311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy OT, Hohmeier HE, Becker TC, et al. Functional genomics of the beta-cell: short-chain 3-hydroxyacyl-coenzyme A dehydrogenase regulates insulin secretion independent of K+ currents. Mol Endocrinol. 2007;21:765–773. doi: 10.1210/me.2006-0411. [DOI] [PubMed] [Google Scholar]
- 30.Dinarello CA, Donath MY, Mandrup-Poulsen T. Role of IL-1β in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes. 2010;17:314–321. doi: 10.1097/MED.0b013e32833bf6dc. [DOI] [PubMed] [Google Scholar]
- 31.Cooper GJ, Willis AC, Clark A, Turner RC, Sim RB, Reid KB. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc Natl Acad Sci U S A. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson RP. Dominance of cyclooxygenase-2 in the regulation of pancreatic islet prostaglandin synthesis. Diabetes. 1998;47:1379–1383. doi: 10.2337/diabetes.47.9.1379. [DOI] [PubMed] [Google Scholar]
- 33.Petri A, Ahnfelt-Ronne J, Frederiksen KS, et al. The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J Mol Endocrinol. 2006;37:301–316. doi: 10.1677/jme.1.02096. [DOI] [PubMed] [Google Scholar]
- 34.Lioubinski O, Muller M, Wegner M, Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Ables ET, Pope CF, et al. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech Dev. 2009;126:958–973. doi: 10.1016/j.mod.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertelli E, Bendayan M. Association between endocrine pancreas and ductal system. More than an epiphenomenon of endocrine differentiation and development? J Histochem Cytochem. 2005;53:1071–1086. doi: 10.1369/jhc.5R6640.2005. [DOI] [PubMed] [Google Scholar]
- 37.Yang YH, Szabat M, Bragagnini C, et al. Paracrine signalling loops in adult human and mouse pancreatic islets: netrins modulate beta cell apoptosis signalling via dependence receptors. Diabetologia. 2011;54:828–842. doi: 10.1007/s00125-010-2012-5. [DOI] [PubMed] [Google Scholar]
- 38.Gustavsen CR, Chevret P, Krasnov B, Mowlavi G, Madsen OD, Heller RS. The morphology of islets of Langerhans is only mildly affected by the lack of Pdx-1 in the pancreas of adult Meriones jirds. Gen Comp Endocrinol. 2008;159:241–249. doi: 10.1016/j.ygcen.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Pechhold S, Stouffer M, Walker G, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat Biotechnol. 2009;27:1038–1042. doi: 10.1038/nbt.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- 41.Collombat P, Mansouri A, Hecksher-Sorensen J, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen F, Kook H, Milewski R, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 43.Yin Z, Gonzales L, Kolla V, et al. Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am J Physiol Lung Cell Mol Physiol. 2006;291:L191–L199. doi: 10.1152/ajplung.00385.2005. [DOI] [PubMed] [Google Scholar]
- 44.Sugo N, Oshiro H, Takemura M, et al. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci. 2010;31:1521–1532. doi: 10.1111/j.1460-9568.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 45.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Konstantinova I, Nikolova G, Ohara-Imaizumi M, et al. EphA-Ephrin-A-mediated beta cell communication regulates insulin secretion from pancreatic islets. Cell. 2007;129:359–370. doi: 10.1016/j.cell.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 49.Hegyi P, Rakonczay Z., Jr The inhibitory pathways of pancreatic ductal bicarbonate secretion. Int J Biochem Cell Biol. 2007;39:25–30. doi: 10.1016/j.biocel.2006.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


