Abstract
Purifying selection often results in conservation of gene sequence and function. The most functionally conserved genes are also thought to be among the most biologically essential. These observations have led to the use of sequence conservation as a proxy for functional conservation. Here we describe two genes that are exceptions to this pattern. We show that lack of sequence conservation among orthologs of CG15460 and CG15323 – herein named jean-baptiste (jb) and karr respectively – does not necessarily predict lack of functional conservation. These two Drosophila melanogaster genes are among the most rapidly evolving protein-coding genes in this species, being nearly as diverged from their D. yakuba orthologs as random sequences are. jb and karr are both expressed at an elevated level in larval males and adult testes, but they are not accessory gland proteins and their loss does not affect male fertility. Instead, knockdown of these genes in D. melanogaster via RNA interference caused male-biased viability defects. These viability effects occur prior to the third instar for jb and during late pupation for karr. We show that putative orthologs to jb and karr are also expressed strongly in the testes of other Drosophila species and have similar gene structure across species despite low levels of sequence conservation. While standard molecular evolution tests could not reject neutrality, other data hint at a role for natural selection. Together these data provide a clear case where a lack of sequence conservation does not imply a lack of conservation of expression or function.
Keywords: Orphan genes, novel genes, positive selection, testes
Introduction
A cornerstone of molecular evolution is that sequence conservation and functional conservation go hand-in-hand. This makes sense as a protein’s function is related to its amino acid sequence. Similarly, functional conservation is commonly considered an indicator of how biologically or evolutionarily essential a gene is. These principles are so universally accepted that it is common practice to use molecular evolutionary conservation to identify the most functionally important parts of proteins (Marks et al. 2011; Friedman et al. 2009; Temple, Jones, and Jones 2010). Following similar logic, “ultraconserved” elements have been identified across numerous taxa and at various evolutionary distances (Bejerano et al. 2004). These ultraconserved sequences are under strong purifying selection (Katzman et al. 2007) and as a result it is assumed that they would be required for life. Surprisingly, mice carrying knockouts for four ultraconserved elements showed no measurable defects (Ahituv et al. 2007), suggesting that ultraconserved elements may not always (or even usually) be as essential as expected. This fact hints that the relationship between sequence conservation, functional conservation and biological importance may not be as robust as commonly assumed.
At the other end of the spectrum, DNA and protein sequences can evolve rapidly for a variety of reasons—natural selection, mutational hot spots, etc. Often the most rapidly changing sequences do not have conserved function and are evolving under relaxed purifying selection. For example, pseudogenes show high rates of sequence evolution and are assumed to be nonfunctional (Li, Gojobori, and Nei 1981). Natural selection can also drive rapid sequence divergence. Van Valen (Van Valen 1973) theorized that organisms and their genes may both be forced to evolve rapidly to meet the demands of a changing environment. Empirical data support this hypothesis. Many genes vital to immunity (Obbard et al. 2009; Sackton et al. 2007) and sexual function (Turner and Hoekstra 2006) evolve at elevated rates and show molecular signatures of positive selection.
In Drosophila, male-biased genes evolve particularly rapidly, often as a result of positive selection (Meiklejohn et al 2003; Zhang, Hambuch, and Parsch 2004; Pröschel, Zhang, and Parcsh 2006; Haerty et al. 2007; Assis, Zhou, and Bachtrog 2012). Genes specific to male tissues are more likely to be orphans (have no known orthologs) and have higher rates of molecular evolution than genes expressed in other tissues or only in females. The male accessory gland proteins (Acps) in Drosophila are a classic case of sexual conflict driving rapid molecular evolution. Acps are expressed in the male, are transferred to females during sex, and perform functions that benefit males -- sometimes at the expense of females (Chapman et al. 2001; Chapman et al. 2003; McGraw et al. 2004; Adams and Wolfner 2007; Avila and Wolfner 2009). Overall, Acps are among the most rapidly evolving genes in Drosophila (Begun and Lindfors 2005), though they perform functions vital to fitness.
Some Acps are so diverged that identifying orthologs in closely related species is difficult (Wagstaff and Begun 2005a; Wagstaff and Begun 2005b; Wagstaff and Begun 2007). This finding raises the possibility that some functional genes in Drosophila are evolving even more rapidly than these Acps – perhaps so quickly that orthologs have not been identified in even the closest relatives. But what would such genes do, and can function be maintained in the face of rapid evolutionary change?
Here, we identify two genes in Drosophila melanogaster that are evolving so rapidly that they initially appeared to be lineage-specific orphans. These genes have testes-biased expression and are important to male viability. We identified putative orthologs in D. simulans, D. sechellia, D. yakuba and D. erecta and showed that their expression level and pattern was conserved despite low levels of both amino acid and nucleotide sequence conservation. Finally, while molecular evidence is inconclusive about the role of positive selection on the evolution of these genes, they are probably the two most rapidly evolving genes yet characterized in Drosophila. Because these genes are so rapidly changing but have conserved expression patterns, we propose to name CG15460 jean-baptiste (jb) CG15323 karr in homage to Jean-Baptiste Alphonse Karr, the author of the phrase “the more things change, the more they stay the same.”
Material and Methods
Screen for candidate genes
To find extremely rapidly evolving genes in D. melanogaster, we searched for genes that appeared to be lineage-specific (following Levine (2006)). Briefly, genes in D. melanogaster were compared by local BLAST to D. yakuba, D. erecta, and D. annanassae. Genes with an e- value > 0.000001 in all three species and good EST support in D. melanogaster were considered candidate D. melanogaster-subgroup specific genes (“orphans”). These candidates (a total of 15) were also used in a search for de novo protein coding genes (see Reinhardt et al 2013). We aligned candidates to all insect genomes using FlyBase’s BLAST (Tweedie et al. 2009) and removed genes that had been retained in D. melanogaster and other more diverged species. We also performed BLAST against NCBI’s nr database and removed candidates that were or contained known transposable elements, microbial genes, or other genome annotations.
We searched for the remaining candidates in other species (D. yakuba, D. simulans, D. sechellia and D. erecta) using UCSC’s whole genome chained BLASTZ alignments, which are more sensitive to highly diverged hits than BLAST or BLAT (Chiaromonte, Yap, and Miller 2002). We then used the UCSC and Flybase genome browsers to ask whether the D. yakuba, D. erecta, D. simulans, and D. sechellia chained BLASTZ alignments covered annotated genes in all four species. We retained candidate genes that matched at least one annotated gene with a similar gene structure in all four species.
Molecular evolutionary analyses
We aligned the extended gene region (5–10kb surrounding the gene) of each candidate and its putative orthologs (see Supplemental table 1) to one another using MAUVE, (Darling et al. 2004; Darling, Mau, and Perna 2010) to determine the extent of collinearity of each ortholog to the D. melanogaster gene. We performed a progressiveMAUVE multiple alignment assuming collinearity (progressiveMauve --collinear --seed-family --disable-backbone) and input the alignment into PAML’s baseml (Yang 2007). Using this alignment we estimated the per base pair rate of substitution along the gene region. We counted the number of fixed differences between D. melanogaster and D. simulans in 500 bp windows along the alignment, then aligned the 39 Drosophila melanogaster Raleigh genomes (Langley et al. 2012) to these regions and calculated polymorphism (π) in each window. We also calculated Tajima’s D (Tajima 1989) and Fu and Li’s D and F (Fu and Li 1993) for 500 base pair windows across the region using DNAsp v5 (Librado and Rozas 2009).
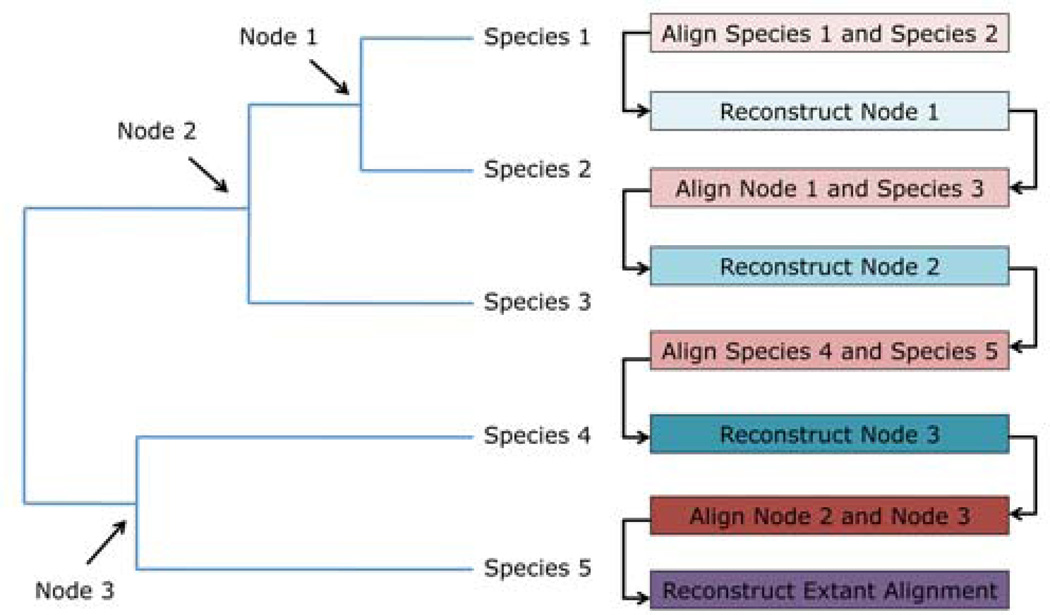
The high level of divergence between sequences made automated alignment of extant genes difficult. This is a known issue, and a common approach is to use known phylogenetic information to assist in alignment (e.g. Feng and Doolittle 1987). We reconstructed the ancestral sequences for each node using PAML’s codeml (Figure 1) and used the reconstructed nodes to facilitate alignment. The most closely related collinear extant genes were aligned pairwise by translated clustalW (Thompson, Gibson, and Higgins 2002), and then remapped to the coding sequences. We used codeml to reconstruct the most likely ancestral state from each pair of sequences. The internal nodes were aligned to one another or to related extant sequences as appropriate (Figure 1). This process was repeated until the common ancestral sequences for the D. yakuba/D. erecta orthologs were aligned to the common ancestral sequences in the D. melanogaster species subgroup. The extant sequences were then aligned to one another using these guide alignments. While ancestral sequence reconstruction is likely to improve alignment of highly diverged sequenced, as with any alignment algorithm, it is not guaranteed to reproduce the true alignment.
Figure 1. Using ancestral sequence reconstruction to guide alignment.
We aligned the amino acid sequences of the most closely related species to one another, then used PAML (codeml) to reconstruct the ancestral nucleotide sequence for each node (Methods). We continued this process until Nodes 2 and 3 could be aligned to one another. Finally, we remapped the extant sequences onto this alignment.
We used PAML’s codeml to compare several models of codon evolution (e.g. branch-selection, site-selection, neutral). We used log-ratio tests to determine if any models were significantly better than the neutral model. We used the alignment of D. melanogaster and D. simulans along with the 39 DPGP Raleigh lines (www.DPGP.org) to estimate the number of silent and non-silent fixed differences and polymorphisms within the protein coding regions. We compared these values using the McDonald-Kreitman test (McDonald and Kreitman 1991).
We assessed the potential effect of transposable elements on duplication of karr by aligning (using BLAT) one of the transposable elements near karr, INE-1, to the D. melangaster, D. simulans, and D. sechellia genomes, as well as to the flanking regions surrounding the orthologs/paralogs of karr in each species. The longest copy of INE-1 present near any paralog of karr (INE-1{5470}) was used as the query in order to hit as many partial copies of INE-1 as possible. Any match longer than 50 bp in length was counted as a hit. Overrepresentation of INE-1 in the flanking regions was assessed using a chi-square test.
Sequence similarity of D. melanogaster orthologs and rapidly evolving genes
We used EMBOSS’ water pairwise alignment program (Rice, Longden, and Bleasby 2000) to determine the sequence similarity of all D. melanogaster genes to their orthologs in D. yakuba and D. simulans. We pulled the best hit from BLAT and found the percent identity and proportion of the D. melangoaster sequence that aligned to the ortholog (proportion matching). We plotted these values using R, and compared the percent identity and proportion matching to 1) the rapidly evolving genes we identified and 2) 100 randomly generated 500 base pair sequence pairs.
Tissue collection and dissection
Male reproductive tracts were dissected on ice from whole flies (D. yakuba, D. simulans, and D. melanogaster) in PBS. Male reproductive tracts and carcasses were each pooled and then flash frozen in liquid nitrogen. Whole females and males of each species were collected and flash-frozen. D. melanogaster and D. yakuba male reproductive tracts were further dissected into accessory glands and testes in PBS and flash frozen. D. melanogaster third instar larvae were sexed by identification of genital discs following Drosophila protocols (Blair 2000), then flash-frozen. Testes were also dissected from males carrying a null mutation at the gene tombola (tombGS12862, stock generously supplied by Dr. Helen White-Cooper), and sons of females mutant for the tudor gene (Bloomington stock #1786, these flies lack a male germline).
Gene expression analyses
We mined expression information from online databases – FlyAtlas (Chintapalli, Wang, and Dow 2007), modENCODE RNAseq data (Graveley et al. 2010), Baylor RNAseq data (Daines et al. 2011), and FlyTED: testes expression database (Zhao et al. 2010). We then extracted RNA from at least two biological replicates of each dissected tissue using TRIZOL reagent (Invitrogen, Grand Island, NY #15596-026), and made cDNA using M-MLV reverse transcriptase (Invitrogen, Grand Island, NY #28025013). We performed relative qRT-PCR quantification using gene-specific primers and a control primer that worked across all species (Actin5c). All qPCR was performed using two technical replicates. 5’ and 3’ RACE were performed following manufacturer’s instructions on D. melanogaster, D. yakuba, and D. simulans testes RNA using the FirstChoice RLM-RACE kit from Ambion (Grand Island, NY #AM1700) and nested gene-specific primers.
RNAi knockdown
Virgin females from Actin-GAL4 (P{Act5C-GAL4}25FO1, Bloomington #4414) were collected and crossed to lines carrying UAS-RNAi constructs for CG15323 (karr), and CG15460 (jb) (www.VDRC.org #35689 and #43403, (Dietzl et al. 2007)). CyO (control) and straight winged (RNAi) progeny of both sexes were counted and collected. We confirmed RNAi knockdown using the same qRT-PCR methods as described above but using gpdh instead of Actin as the control gene.
Viability assays
To estimate effects on adult viability, we simply counted the number of control (CyO) and RNAi (straight-winged) progeny eclosing from each RNAi cross (described above). To determine the stage at which lethality was occurring, we crossed the same RNAi lines to a stock with the same Actin-GAL4 and CD8::UAS-GFP on the same chromosome (kindly donated by S. Chen). RNAi or control status can be ascertained at any stage (RNAi larvae/pupae/adults will express GFP). We collected larvae from the cross during the late third instar (“wandering”)/prepupal stage, and sorted by GFP expression and sex (Blair 2000). We then allowed each type to continue development and counted the number that survived, or that died prior to pupation or prior to eclosion.
Fertility assays
We used a sperm exhaustion assay to estimate the effect of RNAi knockdown of CG15460 (jb) and CG15323 (karr) on male fertility. In this assay (modified from (Sha Sun, Ting, and Wu 2004)), single males are challenged with two virgin females per day across a five-day period. Males with defects in sperm production should produce fewer offspring per female over the assay period. We used a linear model (mean_offspring = genotype + day + genotype X day + ε) to determine if there were significant effects of genotype (indicating a general fertility defect), or a genotype by day interaction effect (indicating a defect in sperm production).
Results
CG15460 (jb) and CG15323 (karr) are among the most rapidly evolving genes in Drosophila melanogaster
We identified two genes in D. melanogaster that have evolved so rapidly that orthology to collinear genes in D. yakuba and D. erecta was not readily apparent. Following Levine et al. (2006), we compared genes in D. melanogaster by local alignment (BLAST) to the D. yakuba, D. erecta, and D. annanassae genomes (Clark et al. 2007). Genes matching poorly to all three species but with EST support in D. melanogaster became candidate D. melanogaster-subgroup specific genes. We aligned these to all insect genomes and removed genes that had been retained in any other species. This eliminated genes that were selectively lost in the D. yakuba, D. erecta, and D. annanassae genomes. To distinguish rapid evolvers from de novo genes or genes that were multiply lost, we searched the BLASTZ alignments from UCSC and retained genes that matched at least one D. yakuba and D. erecta gene. This search yielded CG15460 and CG15323 hereafter referred to as jean-baptiste (jb) and karr respectively. Currently available evidence of orthologs for these genes is mixed. Although Flybase GBrowse (Marygold et al. 2012) shows only D. simulans and D. sechellia orthologs for CG15460 and no orthologs for CG15323, although the recent OrthoDB analysis (Waterhouse et al. 2012) did identify some of the same orthologs we found.
jb and karr aligned to annotated genes in all five sequenced species in the D. melanogaster subgroup, but could not be found in distantly-related species. Some rejected candidates are collinear to apparently non-coding or radically structurally diverged sequences in D. yakuba and D. erecta – these genes likely evolved de novo from the non-coding sequences (Reinhardt et al 2013, Levine et al. 2006) or may be misannotated as non-coding regions in these other species. karr (CG15323) was originally reported as a de novo gene, but the BLASTZ alignment showed weak similarity to the D. yakuba gene GE17891 and the D. erecta gene GG19692; see Supplemental table 1). The jb CDS aligned to multiple genes in D. sechellia, D. erecta and D. yakuba. One of these copies flanks the collinear jb ortholog in each species, suggesting that this gene is a tandem duplicate and one copy was lost in the D. melanogaster lineage. Additionally, D. erecta and D. yakuba also have a few distributed copies of jb (Supplemental table 1). karr has potential paralogs within D. melanogaster and matches to multiple genes in D. simulans and D. sechellia, but only matches one gene in D. yakuba and D. erecta. Though the D. yakuba and D. erecta copies are not collinear to the copies in D. melanogaster, they are collinear to one another (see Supplemental table 1).
jb and karr and their putative orthologs are among the least similar ortholog pairs in Drosophila
The CDSs of jb and karr and their D. simulans and D. yakuba orthologs have among the lowest sequence similarity of any orthologous pairs in Drosophila (Supplemental table 1, Figure 2). We also generated and aligned (EMBOSS, Rice, Longden, and Bleasby 2000) 100 pairs of randomly generated DNA sequences to determine the lowest expected similarity scores using this method. jb and karr are among the top 10% most diverged orthologous pairs in both D. simulans and D. yakuba and similarity to the D. yakuba orthologs is nearly as weak as similarity between random sequences. It is therefore unsurprising that these genes were not originally annotated as orthologs in these species. However, in contrast to some other highly diverged genes, both karr and jb align along most of their length and appear to have conserved intron/exon boundaries and splice forms (see below).
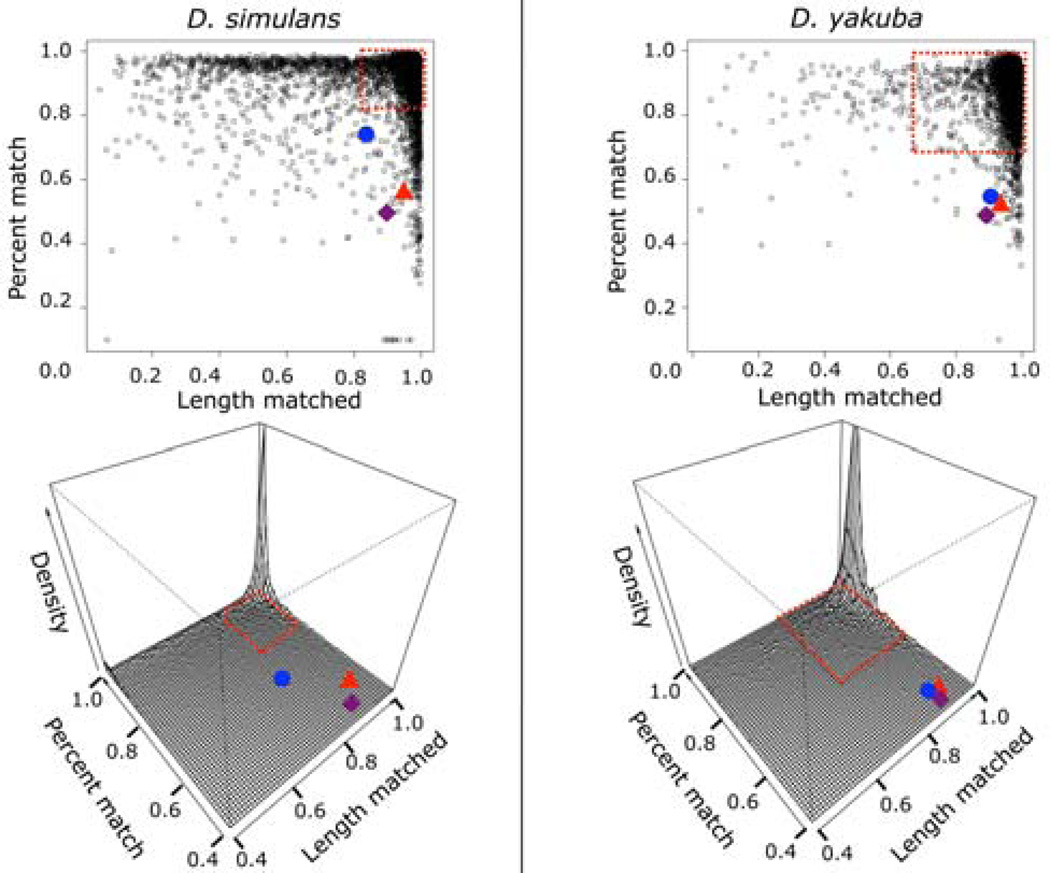
Figure 2. jb and karr are among the most diverged genes in D. melanogaster.
We aligned the nucleotide sequence from the CDS of every gene in D. melanogaster to its annotated orthologs in D. simulans and D. yakuba using EMBOSS’ water aligner (black dots). We also aligned jb (blue) and karr (red) to their putative orthologs from D. simulans and D. yakuba. The red dashed box shows where 90% of known protein-coding genes lie. Both jb and karr fall outside this box in each species. Finally, we generated 100 pairs of random 500bp nucleotide sequences and align each pair of sequences to each other to estimate the average similarity of random sequences. The average sequence conservation and length matched across the 100 replicates is in purple. Both genes are nearly as dissimilar to their D. yakuba orthologs as the average pair of randomly generated nucleotide sequences.
jb and karr are strongly expressed in male tissues
The high level of sequence divergence between these genes and their putative orthologs makes confirmation of true orthology difficult. Similar expression patterns would suggest that these divergent orthologs perform similar functions. Data from FlyAtlas (Chintapalli, Wang, and Dow 2007) and RNA-seq (Daines et al. 2011) show that expression in D. melanogaster adults is highest in male tissues, and can be detected from the third larval instar through adulthood. We confirmed these patterns by measuring expression of jb and karr in the testes, accessory glands, the remaining male carcass, and whole females. Both genes showed peak expression in the testes (Figure 3a). Expression was weak (jb) or undetectable (karr) in the accessory glands, demonstrating that karr and jb are not likely to be accessory gland proteins (ACPs). We confirmed that expression of both genes is reliant on the germline by measuring expression in testes from mutant flies lacking a male germline (sons-of-tudor, Supplemental figure 1).
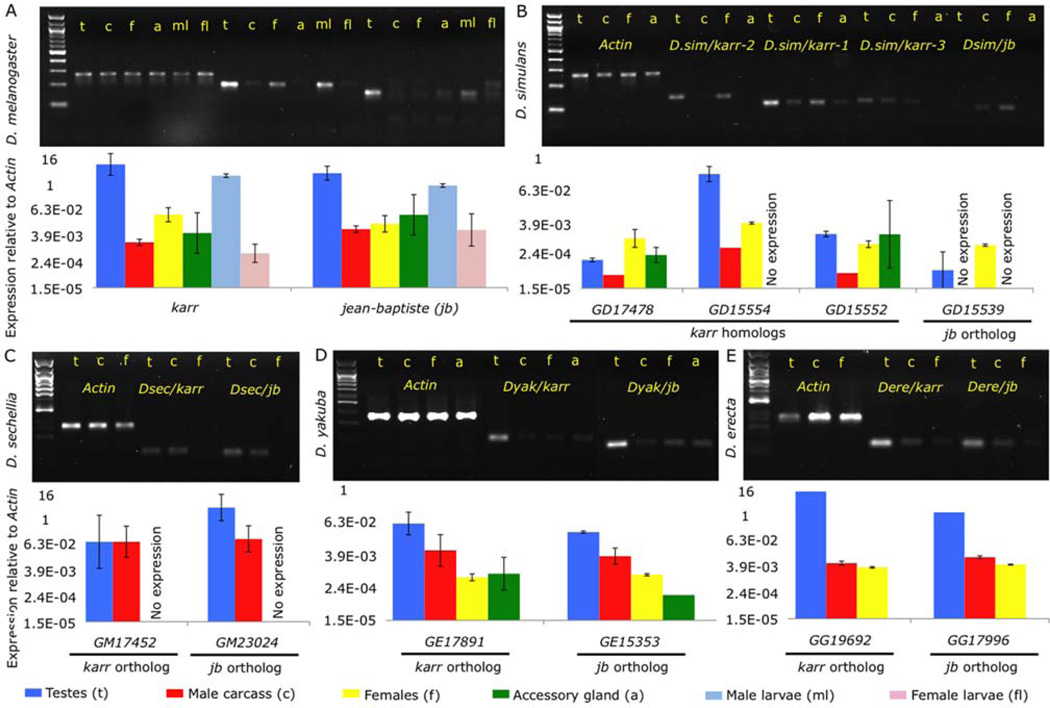
Figure 3. Expression of karr and jb are male biased and this pattern is conserved across five species.
RT-PCR (gels) and qPCR (bar graphs) measurements of gene expression are shown for D. melanogaster (A), D. simulans (B), D sechellia (C), D. yakuba (D) and D. erecta (E). In each species, expression of putative jb and karr orthologs was compared between the testes (t), the remaining male carcass (c for), and whole females (f). In D. melanogaster, D. yakuba, and D. simulans, male accessory glands were also assayed (a). Multiple orthologs of CG15323 exist in D. simulans and D. sechellia and expression was measured for the three “collinear” copies in D. simulans. Only GD15554 (Dsim/karr-1) and GM17452 (Dsec/karr-1) in D. sechellia showed the characteristic expression pattern seen in the other species. Expression of jb and karr was also measured in D. melanogaster male larvae (ml) and female larvae (fl).
Expression was greatly reduced but not absent. Many genes expressed in male meiotic cells are under the control of so-called meiotic arrest genes (e.g tombola, Jiang et al. 2007), but both karr and jb were expressed at normal levels in tombGS12862 (tombola null) testes (Supplemental figure 1). This implies both genes function in parallel to or independently of the meiotic arrest pathway.
Next, we compared expression of the presumed orthologs in adult male testes, male carcass, and female D. simulans, D.sechellia, D. yakuba, and D. erecta. We also measured expression in accessory glands from D. simulans and D. yakuba. The orthologs of both genes showed peak expression in the testes of D. sechellia, D. yakuba, and D. erecta. D. simulans was more complicated, because we measured expression of three of the duplicate copies of karr. GD15554 (Dsim/karr-1) shows a nearly identical expression pattern to D. melanogaster, but the other two copies (Dsim/karr-2 and Dsim/karr-3) have weak expression in all tissues. We next verified that expression of orthologs was not due to nonspecific “background” transcription. First, we used RT-PCR to confirm there was no expression of sequences directly up- or down-stream of the annotated mRNA in the testes (Supplemental figure 2). We eliminated the possibility that transposable elements in proximity of karr could be driving expression by confirming that flanking transposons were not expressed (Supplemental figure 2). Additionally, matching the pattern observed in the D. simulans paralogs, neither of the D. melanogaster “paralogs” of CG15323 were expressed in the testes (Supplemental figure 2A). Finally, we used 5’ and 3’ RLM-RACE to verify the expression and sequence of the mature mRNA in D. yakuba (Supplemental data). We confirmed the annotated CDS for GE17891 (Dyak/karr) using both 5’ and 3’ RACE, and found additional 5’ and 3’ sequence, presumably representing unannotated 3’ and 5’ UTRs. We only found a fragment of the 5’ RACE product for GE15353 (Dyak/jb), but this matched 55 base pairs just 5’ of the annotated CDS. The RACE results indicate that stable mRNAs are produced from the putative orthologs of jb and karr. These data imply that despite extremely rapid rates of protein divergence between species, these genes have retained the same gene structure and pattern of strong expression in the male germline.
RNAi silencing of these rapidly evolving genes is semi-lethal in male Drosophila melanogaster
We used RNA interference to knock down expression of karr and jb in D. melanogaster. We drove the expression of UAS-RNAi constructs for each gene by crossing RNAi stocks to a ubiquitous GAL4 driver (Actin-GAL4) and confirmed by qRT-PCR that expression of each gene was successfully knocked down (data not shown). We found a significant reduction in the number of RNAi male offspring compared to the other offspring classes (Two-tailed Fisher’s exact test, for karr P = 0.0045; for jb P = 0.0161, Table 1). This result was unexpected as expression appeared to be strongest in the male reproductive tract in adults. However, RNAseq data showed that both genes were expressed during larval development as well as in adults. As larvae were of mixed sex in the RNAseq experiment, we measured expression of both genes in third instar larvae after sorting by sex and found higher expression in males (Figure 3a), but some expression in females. Lethality may be occurring during development or metamorphosis phenotype. To determine the stage of lethality, we crossed RNAi stocks to an Actin-GAL4 driver stock that also contained UAS-GFP, allowing identification of RNAi offspring of any stage by GFP expression. We sorted late third instar “wandering” larvae by both sex and GFP expression, then allowed these larvae to continue development, and scored the number of each genotype surviving to pupation and eclosion. We reconfirmed that there was a significant reduction in the number of successfully eclosed male RNAi offspring when compared to controls for both genes (Table 2). In addition, in this assay we saw a small and marginally significant (χ2 = 4,08, P = 0.04342) reduction in the number of jb (but not karr) RNAi females that eclosed compared to control males, so it is possible that the viability effect extends to both sexes for this gene. The stage of lethality differed between the two genes. For jb, a comparable number of all offspring types survived to the third larval instar, but a large proportion of the RNAi male pupae failed to eclose (25% eclosion rate versus 69% for controls). jb-RNAi pupae arrested at the pharate stage, appearing fully developed inside the pupae with discernable eyes, wings, and legs. For karr, a smaller proportion of RNAi male offspring reached the third larval instar, but eclosion rates were similar across all groups. We conclude therefore that karr is important for male fly development during either embryonic or early larval stages whereas jb acts during pupation and may impact viability in both males and females.
Table 1.
RNAi of either jb or karr is semi-lethal in males
| Control | RNAi | Proportion males |
Proportion males |
Fisher's exact test |
|||
|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | RNAi | control |
P-value two-tailed |
|
| Jb (CG15460) | 584 | 633 | 423 | 565 | 0.428 | 0.480 | 0.0161 |
| Karr (CG15323) | 590 | 620 | 561 | 741 | 0.431 | 0.488 | 0.0045 |
Table 2.
karr and jb are important to larval and pupal development respectively
| UAS:RNAi /CyO |
UAS:RNAi /CyO |
UAS:RNAi /ActinGAL4; CD8:UAS- GFP |
UAS:RNAi /ActinGAL4; CD8:UAS- GFP |
Proportion RNAi males |
Proportion Control males |
Fisher′s Exact test |
||
|---|---|---|---|---|---|---|---|---|
| Surviving to | Males | Females | Males | Females | P-value | |||
| jb | 3rd instar | 106 | 105 | 122 | 134 | 0.476 | 0.502 | 0.642 |
| (CG15460) | Eclosion | 73 | 75 | 30 | 62 | 0.326 | 0.493 | 0.011 |
| karr | 3rd instar | 147 | 108 | 164 | 183 | 0.473 | 0.576 | 0.013 |
| (CG15323) | Eclosion | 114 | 88 | 122 | 148 | 0.452 | 0.564 | 0.02 |
We tested if RNAi flies had fertility defects, as would be expected given the strong expression in the testes and germline dependence of jb and karr. We set up a series of single-fly matings using RNAi and control males for both genes as well as a more intensive fertility assay – sperm exhaustion (Sun 2004). We found no difference between control and RNAi males in the number of offspring produced by either assay (Supplemental figure 3). Thus, despite being strongly testes expressed, these genes are not essential to male fertility.
jb but not karr is collinear across the five Drosophila species in which it is found
ProgressiveMAUVE (Darling, Mau, and Perna 2010) alignments of the 10kbp surrounding each putative ortholog from FlyBase in all five species showed that for jb there was a single, collinear region across all five species that included a gene with similar orientation and structure (Figure 4). The neighboring genes were present and highly conserved (although as previously mentioned, there was a tandem duplicate of jb in D. sechellia, and D. yakuba that was not present in D. melanogaster). However, the collinear orthologs to jb showed the weakest sequence similarity across the entire region. karr, on the other hand, was more complicated. A single ortholog is identifiable in D. erecta and D. yakuba, but in both D. simulans and D. sechellia multiple regions aligned suggesting recent gene duplication (Supplemental table 1, Supplemental figure 4). None of these genes are collinear to the D. melanogaster copy.
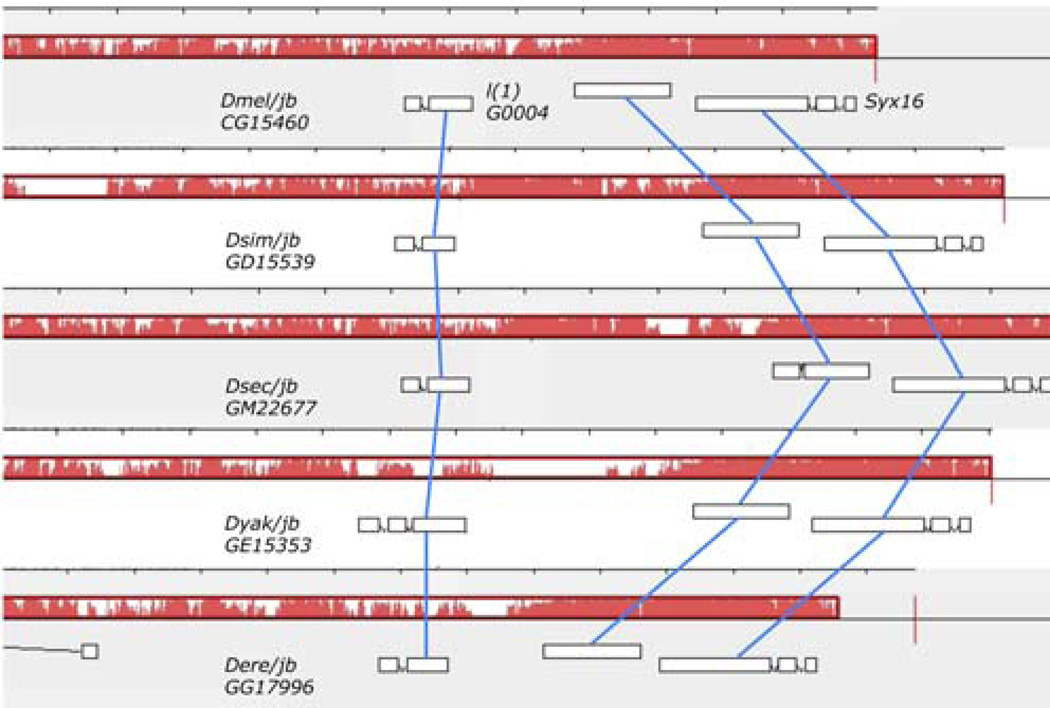
Figure 4. jb is collinear to and shares a conserved gene structure with orthologs from four other Drosophila species.
We used progressiveMAUVE to align the extended gene regions of jb and each of its four putative orthologs. We found that despite weak sequence conservation over the gene regions (red lines), the genes were collinear (blue lines), maintained their orientation relative to conserved flanking genes, and in all but one case have identical gene structure (the D. yakuba ortholog has an additional exon).
jb is evolving at an elevated rate compared to flanking sequences and other rapidly evolving genes
Because jb was collinear across all five species, we could reconstruct the evolutionary history of the gene region and the evolution of the protein. We tested the hypothesis that the high level of divergence of jb was due to positive selection rather than simple neutral drift. Genes under positive selection are predicted to show high levels of divergence (especially nonsynonymous divergence) and low levels of polymorphism compared to sequences evolving neutrally or under purifying selection. We tested this concept using baseml (Yang 2007) to estimate the number of nucleotide substitutions occurring along all branches in 500 bp windows across the MAUVE multiple alignment. We estimated polymorphism in the same windows using D. melanogaster population genomics data from DPGP (Langley et al. 2012). As a positive control, we performed the same analysis on ovulin (Acp26Aa), a male-specific protein-coding gene known to have diverged under positive selection in the D. melanogaster subgroup and is a well-studied model of rapid sequence evolution driven by positive selection in Drosophila (Aguadé 1998; Wong, Albright, and Wolfner 2006; Wong et al. 2010; Tsaur, Ting, and Wu 1998). The highest substitution rates in these gene regions (Figure 5, blue bars) were over the windows including the genes jb (Figure 5, top) and ovulin (Figure 5, bottom), suggesting that both genes are evolving more rapidly than their immediate genomic background. Conversely, polymorphism (π) was low over the windows containing jb and ovulin (Figure 5, red dots). We failed to detect recent positive selection using Tajima’s D and Fu and Li’s D and F in the windows overlapping jb. We hypothesize this is due to insufficient power because of how few polymorphic sites were present. We next tested for positive selection acting on the jb protein in the lineage leading to D. melanogaster. We used the McDonald and Kreitman test (McDonald and Kreitman 1991) with polymorphism data from DPGP (Langley, 2012) and confirmed positive selection was acting on ovulin but not jb in the North American DPGP data. The African data did not confirm strong positive selection for either gene. jb had high numbers of nonsynonymous differences between species in both populations, but few polymorphic sites (8 sites in the African sample, 3 sites in the North American sample, Table 3). Thus, the absence of a signature of positive selection (P = 0.480 in Africa and P = 0.800 in North America) may reflect weak power. In addition, the ability to accurately estimate rates of substitution relies heavily on reproducing the correct sequence alignment. Although we used an iterative approach to protein alignment (see methods), these sequences are highly diverged and it may not be possible to generate a single correct alignment. Thus, substitution rates of rapidly changing sequence may be overestimated (due to incorrectly “forcing” alignment of residues) or underestimated (due to repeated substitution at a site in a lineage).
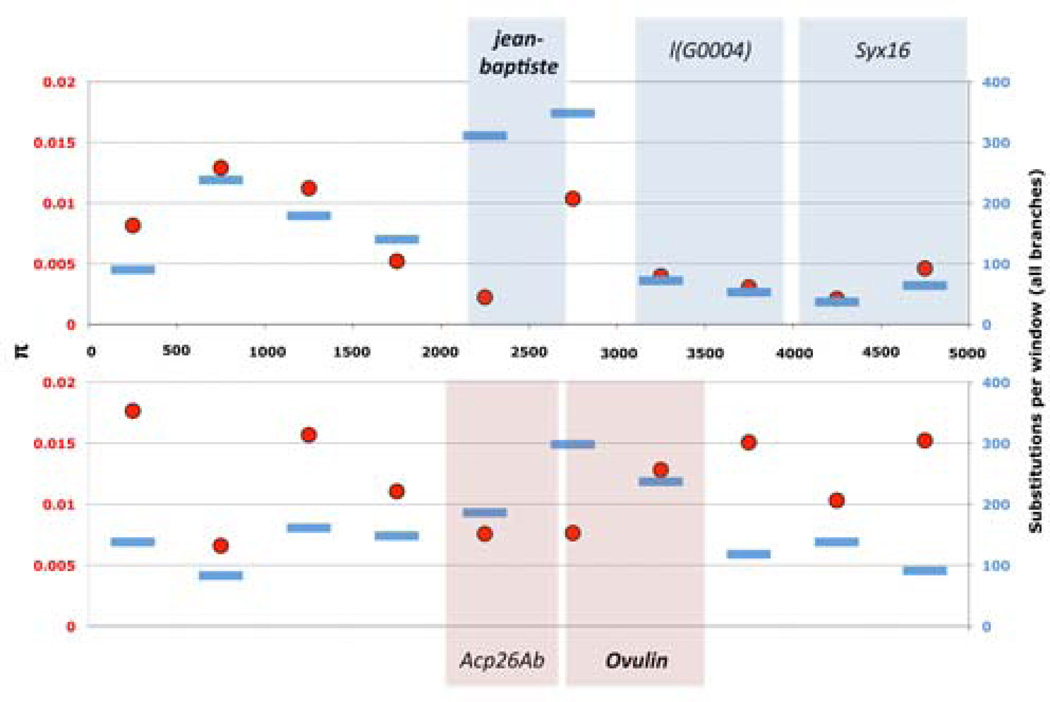
Figure 5. jb has high levels of divergence but low levels of polymorphism relative to flanking sequence.
We used PAML (baseml) to estimate the number of substitutions (blue bars) that have occurred along all branches in 500bp windows in the jb expanded gene region (top panel) and the Ovulin gene region (bottom panel), a rapidly evolving male expressed gene known to have undergone positive selection. We also measured π (red dots) in the same windows using 39 Raleigh lines from the Drosophila 50 genomes data (www.dpgp.org). Gene models are shown above and below each panel.
Table 3.
Results of tests of molecular evolution on jean-baptiste
| dN/dS | Dn | Pn | Ds | Ps | NI (Pn/Ps) /(Dn/Ds) |
α 1- (Ds*Pn)/ (Dn*Ps) |
DoS Dn/(Dn+Ds) - Pn/(Pn+Ps) |
MK test (G) |
MK test P-value |
|
|---|---|---|---|---|---|---|---|---|---|---|
| jb-NC, USA | 1.028 | 47 | 2 | 17 | 1 | 0.723 | 0.277 | 0.068 | 0.064 | 0.800 |
| jb-Malawi, Africa | 47 | 5 | 16 | 3 | 0.567 | 0.433 | 0.121 | 0.499 | 0.480 | |
| Acp26Aa-NC, USA | 0.677 | 78 | 14 | 23 | 14 | 0.295 | 0.705 | 0.272 | 7.423 | 0.006 |
| Acp26Aa-Malawi, Africa | 81 | 7 | 23 | 6 | 0.331 | 0.669 | 0.240 | 3.190 | 0.074 | |
| Acp26Aa* | 75 | 22 | 21 | 16 | 0.385 | 0.615 | 0.202 | 5.330 | 0.021 |
from Tsaur, Ting, and Wu 1998, MBE
As we were unable to distinguish whether recent evolution of jb is being driven by positive selection using polymorphism-based approaches, we next compared models of codon substitution in the jb protein across five species. If jb is evolving under positive selection, we expect to observe an elevated rate of nonsynonymous codon substitutions. Particular codons should be substituted at a level above the background of the gene (indicating positive selection acting repeatedly at these sites) or nonsynonymous substitutions should occur at an elevated rate along one specific lineage (indicating positive selection along that lineage). dN/dS for jb in a pairwise comparison with D. simulans was ~ 1 (Table 3), a value consistent with neutral molecular evolution. Given that jb is functionally important, it seems unlikely this gene is truly evolving without constraint. In order to look for signs of positive or negative selection, we contrasted site and branch models assuming selection (codeml models 2–8) to a model assuming neutrality (codeml model 1 “Nearly Neutral”, Yang 2007), but saw no statistical improvement using the selection models. Hence we were again unable reject the null hypothesis that jb is evolving under neutral drift alone, and we present results of codon evolution under the nearly neutral model (Figure 6). The rates of both synonymous and nonsynonymous protein codon substitution in jb were rapid along all lineages and, overall, almost double that of the rapidly evolving gene ovulin (Figure 6). The pattern of evolutionary change for both genes is similar, with a slower rate of evolution within the D. melanogaster subgroup than across the rest of the tree for both genes.
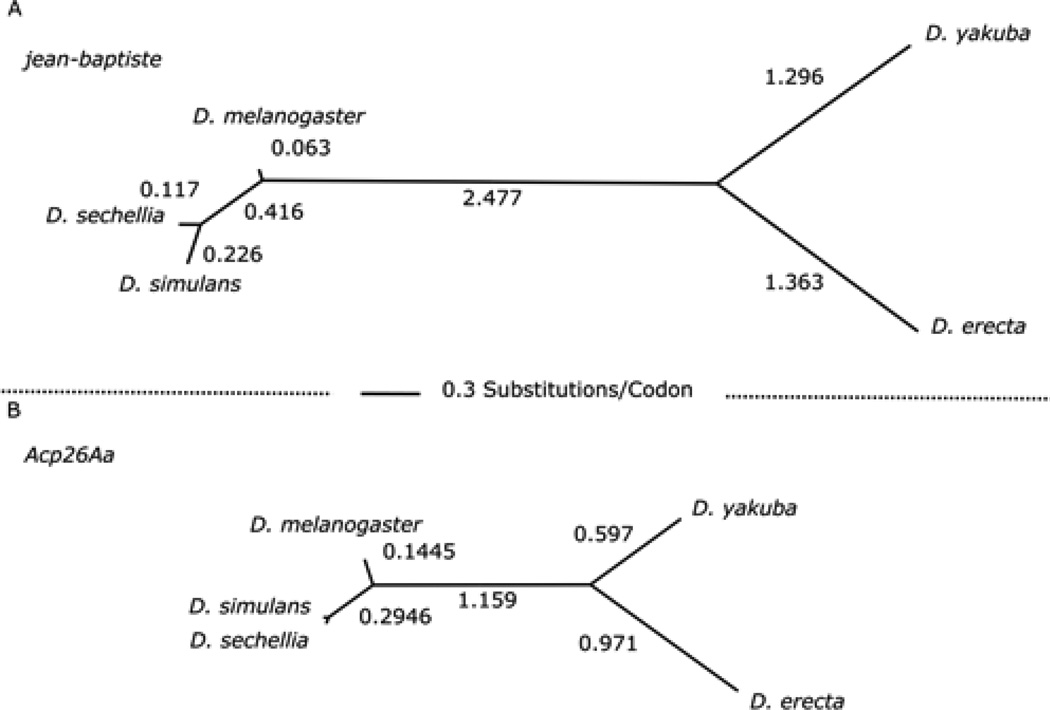
Figure 6. Jean-baptisteprotein is evolving at twice the rate of ovulin.
We used PAML (codeml) to estimate the rate of codon substitution between jb (A) and its putative orthologs, in comparison to the rapidly evolving gene ovulin (B) and found that the former had roughly double the rate of substitution along all branches. Branch lengths are to scale.
The genomic dynamics karr may be linked to the action of transposable elements
Because karr had multiple potential orthologs and paralogs in the D. melanogaster subgroup, and it was unclear which of these were “true” orthologs, we did not feel it was appropriate to use traditional tests of selection on this gene family. We instead investigated the origin of these homologs within the D. melanogaster species subgroup. We observed that karr expanded its copy number in the three species through a number of large segmental duplications and rearrangements as well as dispersed duplication (Figure 7). In contrast to jb, the location of all D. melanogaster, D. simulans, and D. sechellia copies of karr differ from that of the homologs in the D. yakuba/D. erecta clade. We noted that all three potential paralogs in D. melanogaster had annotated transposable elements nearby (diver and INE). We searched the collinear gene regions in the five species for potential TEs, and found homology to INE and diver elements near every ortholog in D. simulans and D. sechellia, but no evidence for either TE in the genomes of D. yakuba or D. erecta -- two species in which karr is single copy and collinear between these two species. Compared to their occurrence in the genome, INE is overrepresented in the regions surrounding these genes (Supplemental table 2), indicating the presence of this TE is not coincidental but instead may be connected to the duplication and dispersal of these genes in the common ancestor of D. melanogaster, D. simulans and D. sechellia, (Figure 7).
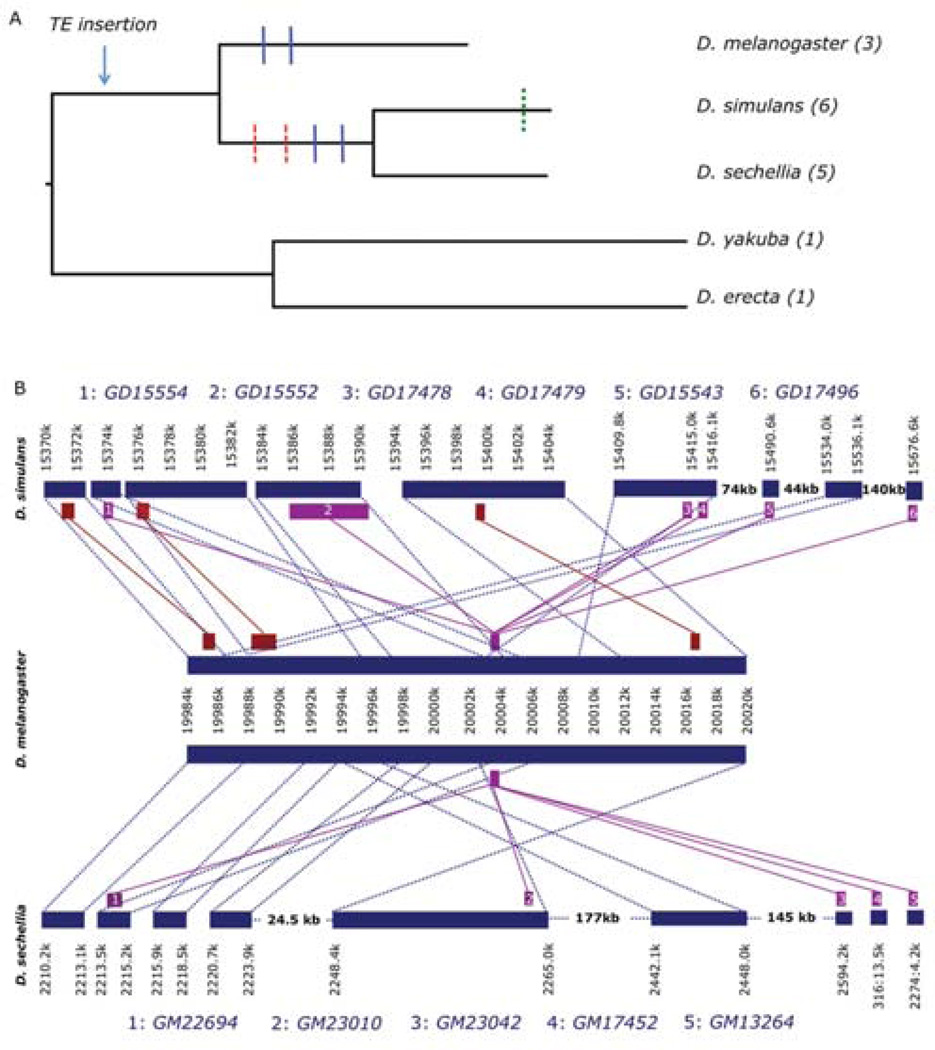
Figure 7. Multiple copies of karr exist in the D. melanogaster species subgroup and appear to be TE associated.
Panel (A) shows karr has multiple putative orthologs in each of D. melanogaster, D. simulans and D. sechellia (parenthesis show number of duplicates), and each is associated with one or more transposable elements (diver and INE). D. yakuba and D. erecta each have only a single copy and no evidence of the associated TEs. Blue bars indicate inferred large scale rearrangements, red bars gene translocations, and green bars tandem duplications. Panel (B) shows that the region of the X chromosome containing karr has been duplicated, rearranged, and transposed multiple times in D. melanogaster’s sister species D. simulans (top) and D. sechellia (bottom). The ends of collinear regions are shown in blue dotted lines, genes are brown or purple blocks, and orthologous genes are connected by solid lines. Purple genes are orthologous to karr (center). In D. simulans, all copies are found on a 300kb region of X chromosome. In D. sechellia, two of the copies are found on small, unordered scaffolds and the remainder are X-linked.
Discussion
Functionally important genes are often evolutionarily constrained because amino acid sequence must be preserved to maintain a protein’s catalytic or structural role. Here, we describe two genes that are startling exceptions to this pattern. karr and jb are among the most rapidly evolving protein-coding genes in Drosophila, yet gene structure, gene expression, and phenotypic data all suggest that the biological function of these genes is likely highly conserved. For example, these genes are expressed strongly in male larvae and adult testes, and expression is reduced in the absence of a male germline. Knockdown of these genes in D. melanogaster via RNA interference causes male-specific developmental defects leading to semi-lethality. Yet despite their functional role, the rate of sequence divergence in these genes is so great that assignment of orthology is difficult and conflicted in the current literature (Waterhouse et al. 2012; Marygold et al. 2012). Nevertheless, we found sequences syntenic to the D. melanogaster CDS out to D. yakuba and D. erecta. These orthologs showed the same intron/exon structure and expression pattern as observed in D. melanogaster. Thus, despite low sequence conservation, these genes unexpectedly appear both structurally/functionally conserved and important to fitness.
These genes are extremely rapidly evolving, and they are expressed at their highest level in the testes, yet their loss causes defects during male development. It is possible that expression in the essential tissue (not currently known) is also male biased. Alternatively, knockdown may have been more efficient in males. Regardless, our finding that these two genes are both testes biased and rapidly evolving is consistent with previous work in Drosophila (Wagstaff and Begun 2005a, 2005b, and 2007, Wong et al 2006, Haerty et al 2007, Wong et al 2010). Studies of male-specific genes and traits have focused on the evolution of sperm and seminal proteins (Aguadé 1998; Wong, Albright, and Wolfner 2006; Wong et al. 2010; Tsaur, Ting, and Wu 1998), and on male and female mating behavior (e.g. Chapman et al 2003, Demir and Dickson 2005). There is, however, little evidence from these studies that rapidly evolving male-biased genes are essential for viability. How can we explain our observation that the knockdown of testes-biased genes causes defects during development? While nearly 20% of annotated genes show male-biased expression (Graveley et al. 2010), genes expressed in male germline stem cells prior to meiosis are typically expressed in at least one other cell type (White-Cooper and Bausek 2010). Therefore, elevated expression in the testes may not always indicate a gene’s primary function is testes specific. Rather, genes may be expressed at a high level due to general transcriptional “permissiveness” in the testes (Kleene 2001; Kleene 2005). Kaessmann (Kaessmann 2010) has proposed that the testes are something of an “evolutionary playground,” where novel genes may become expressed for the first time, and later co-opted to function in other tissues. The fact that we could detect some expression in other tissues suggests this model may explain the evolution of jb and karr. Furthermore, as expression is not restricted to males, we might expect the knockdown of these genes to affect females as well. This is consistent with the weak effect of RNAi silencing of jb on viability in females.
We next must explain what forces could have led to the extremely rapid sequence evolution of genes that strongly affect male fitness. Most essential genes evolve slowly under purifying selection. The extensive protein-coding divergence of jb indicates that purifying selection was not the primary evolutionary force acting across these species. Surprisingly, we were unable to reject simple neutral sequence evolution of jb using standard tests of molecular evolution. Natural selection may still be playing a role in jb evolution – levels of polymorphism are strikingly low in spite of an overall rate of divergence far above background levels. This pattern is suggestive of recurrent selective sweeps altering the amino acid sequence and stripping polymorphism from this biologically important gene despite our failure to statistically reject the null hypothesis of neutrality. Our work compliments recent studies showing that new genes can strongly affect fitness (Chen, Zhang, and Long 2010; Ding et al. 2010). So far, however, no complete molecular explanation has been found for how or why such genes have become essential.
We found that karr was associated with transposable elements in the Drosophila genome and that TE’s may have led to expansion of this gene family in the D. melanogaster species subgroup (Supplemental table 2). Transposable elements – particularly active ones – often include regulatory machinery that can induce expression of neighboring genes, suggesting that the association with transposable elements could drive the expression of karr and its putative orthologs. Of the two putative paralogs of karr in D. melanogaster, and the three collinear D. simulans homologs, qRT-PCR shows that only one gene from each species is strongly expressed in the testes (Figure 3a and b, Supplemental figure 2, RNAseq data shows that the paralogs of karr are also expressed in males, albeit weakly). This strong testes expression pattern is apparently ancestral, as it is shared by the D. yakuba and D. erecta orthologs (Figure 3d, 3e). The diver and INE elements near to Dmel/karr were not expressed (Supplemental figure 2). We conclude that some of the putative orthologs of karr are likely to have been duplicated and carried across the genome by transposable elements, but their expression patterns are not incidental artifacts of these elements but were acquired after the genes moved.
This pair of exceptionally fast evolving genes highlights a challenge facing the study of genes that are lineage-specific in Drosophila and other species (Heinen et al 2009, Xie et al 2012, Carvunis et al 2012, Cai et al. 2008; Chen, Zhang, and Long 2010; Knowles and McLysaght 2009; Levine et al. 2006; Toll-Riera et al. 2008). It is difficult to distinguish whether lineage-specificity is due to multiple losses, rapid sequence evolution, or true de novo evolution. Genes that appear to be entirely “new” may simply be so diverged that sequence similarity is difficult to detect. In fact, karr was first identified as a de novo gene (Levine et al. 2006), based on the fact that it could not be found within the collinear region in D. yakuba or D. erecta. We found D. yakuba and D. erecta genes with weak homology to karr, that share its expression pattern but reside at another genomic locus – apparently having translocated in the D. melanogaster lineage after the split of the D. yakuba/D. melanogaster ancestor. If genes can evolve at such a rate that they cannot be identified between closely related species, we must be cautious in interpreting a simple lack of sequence similarity as true lineage specificity.
Sequence conservation is often used as a hallmark of functional conservation and an indicator of evolutionary importance. While this trend often holds genome-wide, the exceptions to this pattern – such as jb and karr – provide a window into how evolutionary novelty becomes incorporated into the essential biological processes of an organism. Our work is the converse of functional studies in mice showing that ultraconserved sequences are apparently not essential (Ahituv et al. 2007). The next critical question to answer is why these rapidly evolving essential genes exist, why they evolve so quickly, and how these genes retain their essential function in the face of this exceptional rate of molecular evolution.
Supplementary Material
Acknowledgements
The authors wish to thank Manyuan Long and three anonymous reviewers for comments and suggestions on the manuscript and study design. We thank Sidi Chen and Nicholas VanKuren from Manyuan Long’s lab for the Actin-GAL4; UAS-GFP stock, and Helen White-Cooper for the tombola null stock. We thank Teni Coker, Betty Wanjiru, Anais Monroy, Alicia Brandt, and Sophia Shih for technical assistance. This work was supported by NSF grant MCB 0920196 to CDJ and a Royster Society Fellowship to JAR.
References
- Adams Erika M, Wolfner Mariana F. Seminal Proteins but Not Sperm Induce Morphological Changes in the Drosophila Melanogaster Female Reproductive Tract During Sperm Storage. Journal of Insect Physiology. 2007 Apr;53(4):319–331. doi: 10.1016/j.jinsphys.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé M. Different Forces Drive the Evolution of the Acp26Aa and Acp26Ab Accessory Gland Genes in the Drosophila Melanogaster Species Complex. Genetics. 1998 Nov;150(3):1079–1089. doi: 10.1093/genetics/150.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahituv Nadav, Zhu Yiwen, Visel Axel, Holt Amy, Afzal Veena, Pennacchio Len A, Rubin Edward M. Deletion of Ultraconserved Elements Yields Viable Mice. PLoS Biology. 2007;5(9):e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis Raquel, Zhou Qi, Bachtrog Dorish. Sex-biased Transcriptome Evolution in Drosophila. Genome Biology and Evolution. 2012 Oct 23; doi: 10.1093/gbe/evs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Wolfner MF. Acp36DE Is Required for Uterine Conformational Changes in Mated Drosophila Females. Proceedings of the National Academy of Sciences. 2009 Sep 1;106(37):15796–15800. doi: 10.1073/pnas.0904029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA. Rapid evolution of genomic ACP complement in the melanogaster subgroup of Drosophila. Molecular Biology and Evolution. 2005 Oct;22(10):2010–2021. doi: 10.1093/molbev/msi201. [DOI] [PubMed] [Google Scholar]
- Bejerano Gill, Pheasant Michael, Makunin Igor, Stephen Stuart, Kent WJames, Mattick John S, Haussler David. Ultraconserved Elements in the Human Genome. Science (New York, N.Y.) 2004 May 28;304(5675):1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Blair SS. Imaginal Discs. In: Sullivan William, Ashburner Micheal, Hawley R Scott., editors. Drosophila Protocols. Cold Spring Harbor Laboratory Press; 2000. pp. 159–173. [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. De Novo Origination of a New Protein-Coding Gene in Saccharomyces Cerevisiae. Genetics. 2008 May 1;179(1):487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A-R, Rolland T, I Wapinski, MA Calderwood, MA Yildirim, et al. Proto- genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Bangham Jenny, Vinti Giovanna, Seifried Beth, Lung Oliver, Wolfner Mariana F, Smith Hazel K, Partridge Linda. The Sex Peptide of Drosophila Melanogaster: Female Post-mating Responses Analyzed by Using RNA Interference. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 19;100(17):9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Herndon LA, Heifetz Y, Partridge L, Wolfner MF. The Acp26Aa Seminal Fluid Protein Is a Modulator of Early Egg Hatchability in Drosophila Melanogaster. Proceedings. Biological Sciences / The Royal Society. 2001 Aug 22;268(1477):1647–1654. doi: 10.1098/rspb.2001.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. New Genes in Drosophila Quickly Become Essential. Science. 2010 Dec 16;330(6011):1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Yap VB, Miller W. Scoring Pairwise Genomic Sequence Alignments. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2002:115–126. doi: 10.1142/9789812799623_0012. [DOI] [PubMed] [Google Scholar]
- Chintapalli Venkateswara R, Wang Jing, Dow Julian A T. Using FlyAtlas to Identify Better Drosophila Melanogaster Models of Human Disease. Nature Genetics. 2007 Jun;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Clark Andrew G, Eisen Michael B, Smith Douglas R, Bergman Casey M, Oliver Brian, Markow Therese A, Kaufman Thomas C, et al. Evolution of Genes and Genomes on the Drosophila Phylogeny. Nature. 2007 Nov 8;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Daines Bryce, Wang Hui, Wang Liguo, Li Yumei, Han Yi, Emmert David, Gelbart William, et al. The Drosophila Melanogaster Transcriptome by Paired-end RNA Sequencing. Genome Research. 2011 Feb;21(2):315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling Aaron CE, Mau Bob, Blattner Frederick R, Perna Nicole T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Research. 2004 Jul;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling Aaron E, Mau Bob, Perna Nicole T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. In: Stajich Jason E., editor. PLoS ONE. 6. Vol. 5. 2010. Jun 25, p. e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, Dickson BJ. fruitless Splicing Specifies Male Courtship Behavior in Drosophila. Cell. 2005 Jun 3;151(5):785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Dietzl Georg, Chen Doris, Schnorrer Frank, Su Kuan-Chung, Barinova Yulia, Fellner Michaela, Gasser Beate, et al. A Genome-wide Transgenic RNAi Library for Conditional Gene Inactivation in Drosophila. Nature. 2007 Jul 12;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Ding Yun, Zhao Li, Yang Shuang, Jiang Yu, Chen Yuan, Zhao Ruoping, Zhang Yue, et al. A Young Drosophila Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-linked Male Fertility Genes. PLoS Genetics. 2010;6(12):e1001255. doi: 10.1371/journal.pgen.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Da-Fei, Doolittle Russel F. Progressive Sequence Alignment as a Prerequisite to Correct Phylogenetic Trees. Journal of Molecular Evolution. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Friedman Erin J, Temple Brenda RS, Hicks Stephanie N, Sondek John, Jones Corbin D, Jones Alan M. Prediction of Protein-protein Interfaces on G-protein Beta Subunits Reveals a Novel Phospholipase C Beta2 Binding Domain. Journal of Molecular Biology. 2009 Oct 2;392(4):1044–1054. doi: 10.1016/j.jmb.2009.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical Tests of Neutrality of Mutations. Genetics. 1993 Mar;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley Brenton R, Brooks Angela N, Carlson Joseph W, Duff Michael O, Landolin Jane M, Yang Li, Artieri Carlo G, et al. The Developmental Transcriptome of Drosophila Melanogaster. Nature. 2010 Dec 22;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty Wilfried, Jagadeeshan Santosh, Kulathinal Rob J, Wong Alex, Ram Kristipati Ravi, Sirot Laura K, Levesque Lisa, et al. Evolution in the Fast Lane: Rapidly Evolving Sex-related Genes in Drosophila. Genetics. 2007 Nov;177(3):1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen Tobias JAJ, Staubach Fabian, Häming Daniela, Tautz Diethard. Emergence of a new gene from an intergenic region. Current Biology. 2009;19:1527–1531. doi: 10.1016/j.cub.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Jiang Jianqiao, Benson Elizabeth, Bausek Nina, Doggett Karen, White-Cooper Helen. Tombola, a tesmin/TSO1-family Protein, Regulates Transcriptional Activation in the Drosophila Male Germline and Physically Interacts with Always Early. Development. 2007 Apr;134(8):1549–1559. doi: 10.1242/dev.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann Henrik. Origins, Evolution, and Phenotypic Impact of New Genes. Genome Research. 2010 Oct;20(10):1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman Sol, Kern Andrew D, Bejerano Gill, Fewell Ginger, Fulton Lucinda, Wilson Richard K, Salama Sofie R, Haussler David. Human Genome Ultraconserved Elements Are Ultraselected. Science. 2007 Aug 17;317(5840):915–915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Kleene Kenneth. A Possible Meiotic Function of the Peculiar Patterns of Gene Expression in Mammalian Spermatogenic Cells. Mechanisms of Development. 2001 Aug;106(1–2):3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- Kleene Kenneth. Sexual Selection, Genetic Conflict, Selfish Genes, and the Atypical Patterns of Gene Expression in Spermatogenic Cells. Developmental Biology. 2005 Jan 1;277(1):16–26. doi: 10.1016/j.ydbio.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Knowles David G, McLysaght Aoife. Recent De Novo Origin of Human Protein-coding Genes. Genome Research. 2009 Oct;109(1):1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley Charles H, Stevens Kristian, Cardeno Charis, Lee Yuh Chwen G, Schrider Daniel R, Pool John E, Langley Sasha A, et al. Genomic Variation in Natural Populations of Drosophila Melanogaster. Genetics. 2012 Jun 5;192(2):533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine Mia T, Jones Corbin D, Kern Andrew D, Lindfors Heather A, Begun David J. Novel Genes Derived from Noncoding DNA in Drosophila Melanogaster Are Frequently X-linked and Exhibit Testis-biased Expression. Proceedings of the National Academy of Sciences. 2006 Jun 27;103(26):9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Chuan-Yun, Zhang Yong, Wang Zhanbo, Zhang Yan, Cao Chunmei, Zhang Ping-Wu, Lu Shu-Juan, et al. A Human-specific De Novo Protein-coding Gene Associated with Human Brain Functions. PLoS Computational Biology. 2010 Mar;6(3):e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Dan, Dong Yang, Jiang Yu, Jiang Huifeng, Cai Jing, Wang Wen. A De Novo Originated Gene Depresses Budding Yeast Mating Pathway and Is Repressed by the Protein Encoded by Its Antisense Strand. Cell Research. 2010 Apr;20(4):408–420. doi: 10.1038/cr.2010.31. [DOI] [PubMed] [Google Scholar]
- Li Wen-Hsiung, Gojobori Takashi, Nei Masatoshi. Pseudogenes as a Paradigm of Neutral Evolution. Nature. 1981 Jul 16;292(5820):237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP V5: a Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics (Oxford, England) 2009 Jun 1;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Marks Debora S, Colwell Lucy J, Sheridan Robert, Hopf Thomas A, Pagnani Andrea, Zecchina Riccardo, Sander Chris. Protein 3D Structure Computed from Evolutionary Sequence Variation. In: Sali Andrej., editor. PLoS ONE. 12. Vol. 6. 2011. Dec 7, p. e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold Steven J, Leyland Paul C, Seal Ruth L, Goodman Joshua L, Thurmond Jim, Strelets Victor B, Wilson Robert J, consortium the FlyBase. FlyBase: Improvements to the Bibliography. Nucleic Acids Research. 2012 Nov 3;41(D1):D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald John H, Kreitman Martin. Adaptive Protein Evolution at the Adh Locus in Drosophila. Nature. 1991 Jun 20;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McGraw Lisa A, Gibson Greg, Clark Andrew G, Wolfner Mariana F. Genes Regulated by Mating, Sperm, or Seminal Proteins in Mated Female Drosophila Melanogaster. Current Biology: CB. 2004 Aug 24;14(16):1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Meiklejohn Colin D, Parsch John, Ranz Jose M, Hartl Daniel L. Rapid Evolution of Male-Biased Gene Expression in Drosophila. Proceedings of the National Academy of Sciences USA. 2003;17:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali Thilakam, Pacifico Svetlana, Yu Jingkai, Guest Stephen, Roberts George G, 3rd, FinleyL Russell L., Jr DroID 2011: a Comprehensive, Integrated Resource for Protein, Transcription Factor, RNA and Gene Interactions for Drosophila. Nucleic Acids Research. 2011 Jan;39:D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard Darren J, Welch John J, Kim Kang-Wook, Jiggins Francis M. Quantifying Adaptive Evolution in the Drosophila Immune System. PLoS Genetics. 2009 Oct;5(10):e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschel Matthias, Zhang Zhi, Parsch John. Widespread Adaptive Evolution of Drosophila Genes With Sex-Biased Expression. Genetics. 2006 Oct;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Peter, Longden Ian, Bleasby Alan. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics: TIG. 2000 Jun;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, BM Wanjiru, AT Brandt, P Saelao, DJ Begun, CD Jones. De Novo ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly From Previously Non-Coding Sequences. PLoS Genetics. In Press doi: 10.1371/journal.pgen.1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton Timothy B, Lazzaro Brian P, Schlenke Todd A, Evans Jay D, Hultmark Dan, Clark Andrew G. Dynamic Evolution of the Innate Immune System in Drosophila. Nature Genetics. 2007 Dec;39(12):1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Sun Sha, Ting Chau-Ti, Wu Chung-I. The Normal Function of a Speciation Gene, Odysseus, and Its Hybrid Sterility Effect. Science (New York, N.Y.) 2004 Jul 2;305(5680):81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics. 1989 Nov;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple Brenda RS, Jones Corbin D, Jones Alan M. Evolution of a Signaling Nexus Constrained by Protein Interfaces and Conformational States. PLoS Computational Biology. 2010;6(10):e1000962. doi: 10.1371/journal.pcbi.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson Julie D, Gibson Toby J, Higgins Des G. Multiple Sequence Alignment Using ClustalW and ClustalX. Current Protocols in Bioinformatics / Editoral Board, Andreas D. Baxevanis … [et Al.] 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2 August Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Toll-Riera Macarena, Bosch Nina, Bellora Nicolás, Castelo Robert, Armengol Lluis, Estivill Xavier, Alba MMar. Origin of Primate Orphan Genes: A Comparative Genomics Approach. Molecular Biology and Evolution. 2008 Dec 23;26(3):603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- Tsaur Shun-Chern, Ting Chau-Ti, Wu Chung-I. Positive Selection Driving the Evolution of a Gene of Male Reproduction, Acp26Aa, of Drosophila: II. Divergence Versus Polymorphism. Molecular Biology and Evolution. 1998 Aug;15(8):1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- Turner Leslie M, Hoekstra Hopi E. Adaptive Evolution of Fertilization Proteins Within a Genus: Variation in ZP2 and ZP3 in Deer Mice (Peromyscus) Molecular Biology and Evolution. 2006 Sep;23(9):1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- Tweedie Susan, Ashburner Michael, Falls Kathleen, Leyland Paul, McQuilton Peter, Marygold Steven, Millburn Gillian, et al. FlyBase: Enhancing Drosophila Gene Ontology Annotations. Nucleic Acids Research. 2009 Jan;37(Database issue):D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen Leigh. A New Evolutionary Law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- Wagstaff BJ Begun DJ. Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura . Mol Biol Evol. 2005a;22:818–832. doi: 10.1093/molbev/msi067. [DOI] [PubMed] [Google Scholar]
- Wagstaff BJ Begun DJ. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae . Genetics. 2005b;171:1083–1101. doi: 10.1534/genetics.105.043372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff BJ Begun DJ. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics. 2007;177:1023–1030. doi: 10.1534/genetics.107.077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse Robert M, Tegenfeldt Fredrik, Li Jia, Zdobnov EEvgeny M, Kriventseva Evgenia V. OrthoDB: a Hierarchical Catalog of Animal, Fungal and Bacterial Orthologs. Nucleic Acids Research. 2012 Nov 24;41(D1):D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper Helen, Bausek Nina. Evolution and Spermatogenesis. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010 Apr 19;365(1546):1465–1480. doi: 10.1098/rstb.2009.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Alex, Albright Shannon N, Wolfner Mariana F. Evidence for Structural Constraint on Ovulin, a Rapidly Evolving Drosophila Melanogaster Seminal Protein. Proceedings of the National Academy of Sciences of the United States of America. 2006 Dec 5;103(49):18644–18649. doi: 10.1073/pnas.0601849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Alex, Christopher Adam B, Buehner Norene A, Wolfner Mariana F. Immortal Coils: Conserved Dimerization Motifs of the Drosophila Ovulation Prohormone Ovulin. Insect Biochemistry and Molecular Biology. 2010 Apr;40(4):303–310. doi: 10.1016/j.ibmb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Chen, Zhang Yong E, Chen Jia-Yu, Liu Chu-Jun, Zhou Wei-Zhen, Li Ying, Zhang Mao, Zhang Rongli, Wei Liping. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genetics. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Ziheng. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007 Aug;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang Zhi, Hambuch Tina M, Parsch John. Molecular Evolution of Sex-Biased Genes in Drosophila. Molecular Biology and Evolution. 2004;21(11):2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
- Zhao Jun, Klyne Graham, Benson Elizabeth, Gudmannsdottir Elin, White-Cooper Helen, Shotton David. FlyTED: The Drosophila Testis Gene Expression Database. Nucleic Acids Research. 2010 Jan;38(Database issue):D710–715. doi: 10.1093/nar/gkp1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Qi, Zhang Guo-jie, Zhang Yue, Xu Shi-yu, Zhao Ruo-ping, Zhan Zubing, Li Xin, Ding Yun, Yang Shuang, Wang Wen. On the origin of new genes in Drosophila. Genome Research. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.