Abstract
The Y chromosome is thought to be important for male reproduction. We have previously shown that with the use of assisted reproduction, live offspring can be obtained from mice lacking the entire Y chromosome long arm. Here, we demonstrated that live mouse progeny can also be generated using germ cells from males with the Y chromosome contribution limited to only two genes, the testis determinant factor Sry and the spermatogonial proliferation factor Eif2s3y. Sry is believed to function primarily in sex determination during fetal life. Eif2s3y may be the only Y chromosome gene required to drive mouse spermatogenesis allowing formation of haploid germ cells that are functional in assisted reproduction. Our findings are relevant but not directly translatable to human male infertility cases.
The mammalian Y chromosome, once thought to be a genetic wasteland (1), is now known to encode a battery of genes, many of which are thought to be involved in male reproduction (2). A substantial amount of work has been done to define which genes are important for maintaining sperm function under normal, in vivo, conditions. In the era of assisted reproduction technologies (ART) it is now possible to bypass several steps of normal human fertilization using immotile, non-viable, or even immature sperm. We have shown that infertile male mice lacking the entire Y chromosome long arm can generate live offspring when their severely morphologically abnormal sperm are delivered into oocytes via intracytoplasmic sperm injection (ICSI) (3). In these mice the Y chromosome is reduced from 78Mb to ~2Mb, and encodes only 7 genes and 3 gene families (Fig. S1, XY*XSxra).
In most mammals, including humans and mice, testis determination is regulated by Sry, which directs developing gonads to male differentiation (4–6). Upon transgenic addition of Sry, mice with one X chromosome (XOSry) develop testes which are populated with spermatogonia, male germ cells that have the potential to undergo differentiation and initiate spermatogenesis. In the absence of other Y chromosome genes these spermatogonia undergo proliferation arrest, and meiotic and post-meiotic stages of spermatogenesis are absent (7). Transgenic addition of individual missing Y genes led to the identification of Eif2s3y as the gene that restored normal spermatogonial proliferation (7). In XOSry males transgenic for Eif2s3y spermatogenesis was shown to complete meiotic prophase and the first meiotic division before the cells arrested as secondary spermatocytes, with the occasional production of spermatid-like cells (7, 8). Here, we tested whether these spermatid-like cells were functional in assisted reproduction, and what other components of the Y chromosome help to increase development of functional gametes.
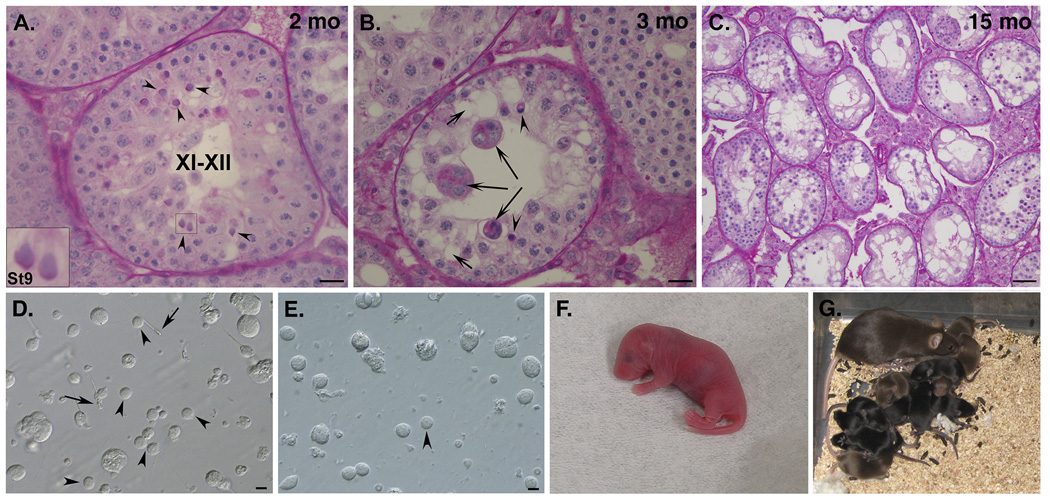
We first examined mice with the Y gene complement limited to two transgenically derived genes, autosomally located Sry and X chromosome located Eif2s3y (Fig. S1, XEOSry)(9). These mice had testes smaller than wild-type XY males (Fig. S2) but populated with germ cells. Analysis of testicular sections confirmed that spermatogenesis was ongoing allowing development of germ cells with spermatid-like morphology (Fig. 1A arrowheads), similar to those observed earlier (7, 10). The occurrence of these spermatids was low and their development was restricted to steps 5–7 of spermatid development, with the occasional presence of step 8–9 spermatids (Fig. 1A(inset)&S3). We also observed secondary changes to seminiferous tubules, with increased incidence of dying cells, multinucleate bodies, and vacuoles (Fig. 1B). These changes increased progressively as the males aged, ultimately leading to Sertoli cell only (SCO) syndrome tubules (Fig. 1C).
Fig. 1.
Testis histology, round spermatids and live offspring. (A) Males with only 2 Y genes, Eif2s3y and Sry, have meiotic and post-meiotic arrests that only occasionally allow formation of spermatids (arrowheads) that are frequently delayed and do not develop beyond St8/9. (B) Tubule degeneration with formation of 'multinucleate bodies' (long arrows), vacuoles (short arrows) and dying/apoptotic germ cells (arrowheads) is frequently observed. (C) Sertoli cell only-like phenotype in an old male. (D) Testicular suspension from wild-type males contains testicular sperm (arrows) and many round spermatids (arrowheads) with clear morphological features. (E) Males with 2 Y genes have significantly less germ cells in testicular cell suspension, with no sperm and very few round spermatids (arrowhead). These spermatids are functional in ROSI. (F) ROSI pup obtained after transfer of embryos generated with spermatids from a male with only two Y genes. (G) an adult female developed from the pup shown in (G) with her own litter. Roman and arabic numerals in (A–C) are tubule stages and steps of spermatid development (St), respectively (see Fig. S3). Scale is 50 µm (A–C) and 10 µm (D–F); insets = x3 magnification; mo = months of age.
We next tested the function of the spermatid-like cells from XEOSry in assisted reproduction. Round, spermatid-like cells could be found in testicular cell suspensions from all males used in ART trials although these cells were rare and their morphology was often slightly abnormal (increased size, less pronounced nuclei, rough rather than smooth surface), when compared to spermatids from control XY males (Fig. 1D&E). Nevertheless, when we performed round spermatid injection (ROSI), the oocytes were successfully fertilized as evidenced by the development of two pronuclei and extrusion of the second polar body, and subsequent cleavage (Fig. S4). When the developed 2-cell embryos were transferred into the oviducts of recipient females, live offspring were obtained (Table 1 and Fig. 1F). Three out of four males, which provided spermatids for injections, successfully sired offspring. The efficiency of ROSI with XEOSry males was significantly lower than with XY controls (Table 1, 9% vs. 26%). All the progeny had the genotypes as expected when derived from XEOSry fathers and were healthy; those bred were fertile (Fig. 1G,S5 and supplementary online text).
Table 1.
The results of round spermatid injection (ROSI) with spermatids from males with limited Y gene complement.
| Male genotype | Y gene contribution (see Fig. S1) |
Live offspring % (no)# |
Implants % (no)# |
|---|---|---|---|
| XEOSry | Eif2s3y & Sry | 9.1 (12/132)a | 29.5 (39/132)b |
| XEY*XSry | Eif2s3y & Sry | 5.7 (13/227)b | 27.3 (62/227)b |
| XESxrbO | Eif2s3y & Sxrb | 20.0 (24/120) | 45.8 (55/120)a |
| XESxrbY*X | Eif2s3y & Sxrb | 16.0 (24/150) | 37.3 (56/150)b |
| XYRIII control | intact Y | 26.0 (19/73) | 69.9 (51/73) |
Percentage calculated from embryos transferred. Male age ranged 63–229 days.
Statistical significance (Fisher's Exact Test):
P<0.01,
P<0.001 vs. XYRIII control.
Sxrb Sry, Zfy2/1, H2al2y, Rbmy cluster (reduced)
An unpaired sex chromosome leads to meiotic arrest and apoptosis (11) so partial meiotic failure in XEOSry males was not unexpected. Few spermatids that could be found in the testes could be the cells that 'leaked' through the meiotic arrest, i.e. finished meiosis and were haploid. Alternatively, these could be the cells that developed spermatid-like morphology without undergoing the second meiotic division (8, 10).
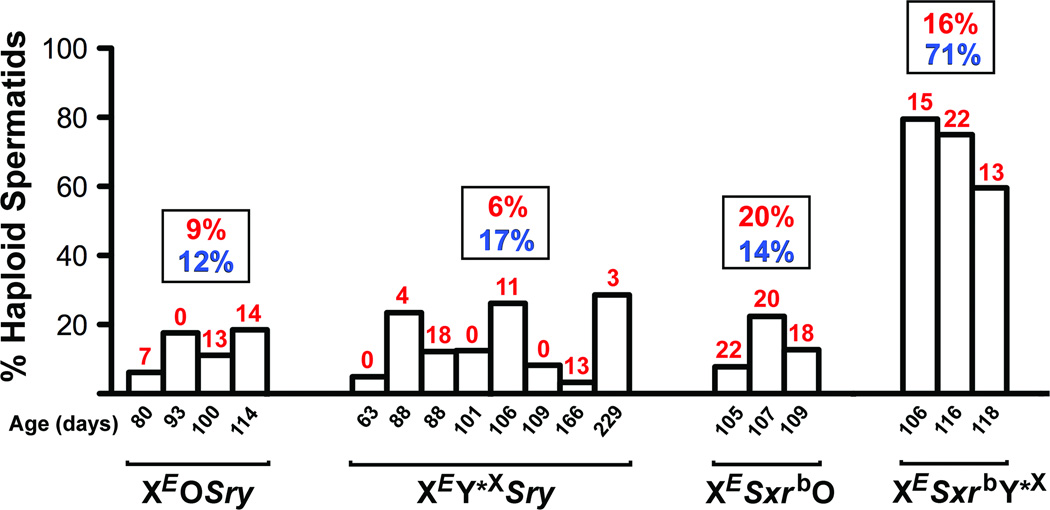
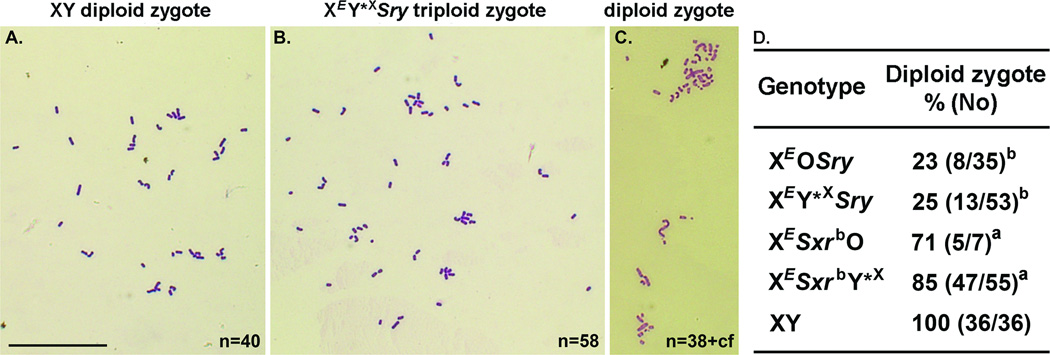
Spermatid nuclear DNA content (Fig. 2&S6) and zygotic chromosome analyses (Fig. 3) revealed that the great majority of spermatids from XEOSry males were diploid and yielded triploid zygotes, which would explain the poor ROSI success. In order to overcome the problem of meiotic block arising from X chromosome univalence we generated males, in which a minute Y*X chromosome (Fig. S1, Y*X) was added to provide a second pairing region (PAR) for PAR-PAR chromosome synapsis (12). In the resulting XEY*XSry males (Fig. S1), successful pairing of Y*X and X was observed in 85% of pachytene spermatocytes (Fig. S8). However, the testicular phenotype did not improve (Fig. S2&S7A–C). The proportion of live offspring obtained after injection was similarly low as with XEOSry males (Table 1) and only five out of eight males that provided cells for ROSI sired offspring. We therefore tested for ploidy and demonstrated that most of the XEY*XSry spermatids were diploid (Fig. 2&S6) and most zygotes after ROSI were triploid (Fig. 3). Thus, overcoming X chromosome univalence in XEY*XSry males did not allow overcoming meiotic arrest and increasing ROSI success (see supplementary online text).
Fig. 2.
Incidence of haploid spermatids. Each graph bar represents an individual male providing testicular cells for analysis; the numerals above show the percentage of pups obtained after ROSI. The data in rectangular boxes represent average percentage of haploid spermatids (blue) and ROSI offspring (red) for each genotype. Control male with intact Y chromosome had 97% (67/69) of spermatids haploid. XESxrbY*X males had higher average incidence of haploid spermatids than other genotypes (P<0.01).
Fig. 3.
Zygote chromosome analysis after ROSI. (A) Normal diploid mouse zygote generated after ROSI with XY male contains 40 chromosomes. (B) Triploid zygote obtained after ROSI with spermatids from a male with two Y genes (~60 chromosomes). (C) Diploid zygote after ROSI with spermatids from male of the same genotype (~40 chromosomes). (D) Incidences of diploid zygotes generated after ROSI. cf=chromosome fragments. n= chromosome number. a P<0.05, b P<0.001 vs. XY control. Bar=50 µm.
We next asked whether other Y chromosome genes may be beneficial for spermatid function in assisted reproduction. To address this, we produced males in which the Sry transgene driving sex determination was replaced with the sex reversal factor Sxrb (Fig. S1, XESxrbO & XESxrbY*X). In these males the X chromosome carries an Eif2s3y transgene necessary for spermatogonial proliferation (7) together with the Sxrb encoding for Zfy2/1, Sry, H2al2y, and the Rbmy gene cluster.
The testes from XESxrbO and XESxrbY*X males were larger than from XEOSry and XEY*XSry but smaller than in wild-type XY males (Fig. S2). The incidence of round spermatids increased only in XESxrbY*X (Fig. S3). In both male types spermatid development was more advanced with clear elongation up to step 10, occasional presence of step 11–12 spermatids, and even sporadic development to mature testicular sperm (Fig. S7D&G). There were secondary changes to the seminiferous epithelium (Fig. S7E,F,H&I), albeit in XESxrbY*X males less pronounced than in the other genotypes. Zygote chromosome analysis showed that most of the zygotes after ROSI were diploid (71–85%, Fig. 3), and the ROSI outcome was significantly improved, with all tested males (3 per genotype) yielding live offspring with rate now reaching 20% and 16% (Table 1). Interestingly, the frequency of haploid spermatids in XESxrbO males remained low (Fig. 2&S6). The discrepancy between the ploidy of the spermatids and zygotes raised the possibility that the second meiotic division took place in the oocytes. This suggests that Sxrb encodes a gene(s) that promotes the second meiotic division when all chromosomes are paired, and enables overcoming meiotic arrest of the diploid spermatid in the oocyte when an X chromosome pairing partner is missing (see supplementary online text).
We have shown that only two mouse Y chromosome genes, Sry and Eif2s3y, are necessary for the development of male haploid germ cells that are sufficient for successful reproduction yielding live offspring. This minimal Y gene complement must be enough to ensure correct male-specific methylation and proper formation of any other epigenetic modifications that are required for embryogenesis in the paternal genome.
That Sry is one of the two genes is not surprising as it drives testis determination (4–6). Mouse Sry expression and tramslation occurs very briefly during fetal development. In adult testes Sry transcripts are thought to be aberrant and not translatable (13, 14) although their role as epigenetic regulator cannot be excluded (15). Nevertheless, it is reasonable to conclude that Sry plays a role primarily during sex determination.
This suggests that the second Y gene, Eif2s3y, is the only gene necessary to drive spermatogenesis through the first meiotic division and with the occasional meiotic progression to form haploid round spermatids. Eif2s3y is a Y encoded subunit 3 of the eukaryotic translation initiation factor 2. It is ubiquitously expressed and in the testis plays a role in spermatogonial proliferation (7). The Eif2s3y gene has been conserved on the Y chromosome during eutherian evolution but there is no Y-linked copy of Eif2s3 detected in any of the simian primates including humans (16).
In men, spermatogonial proliferation arrest results most often from deletions of AZFa (azoospermia; Sertoli-cell-only syndrome) and it has been related to a loss of DDX3Y (17–19). In both men and mice the Eif2s3 and Ddx3 genes belong to the family of widely expressed genes encoding proteins involved in initiation of mRNA translation at the ribosome. These genes have X homologues that escape X-inactivation, and the Y and X copies are suspected to be at least in part interchangeable, with the Y copy conserved to provide two doses of gene product in both male and female. The loss of the mouse Y encoded Ddx3y is not detrimental for spermatogonesis because of the compensation provided by an X copy retrotransposed on an autosome (20). Analogically, in men, the presence of a retroposed X copy of EIF2S3X, in addition to EIF2S3X itself, explains why the loss of the Y copy was still permissive for spermatogenesis. Although there is no human copy of Eif2s3y, men have a Y encoded copy of the translation factor EIF1A (eukaryotic translation initiation factor 1A, Y-linked) (21), which likely acts as part of a multi-protein complex that includes EIF2S3X as well as other EIF family members. Interestingly, EIF1AY is found in the AZFb region, and its diminished expression sporadically contributes to azoospermia (22).
At present, our findings in mice do not translate directly to humans. ROSI is still considered experimental in human ART due to concerns regarding the safety of injecting immature germ cells and technical difficulties (23). In spite of this, some children have already been born (24, 25) and those were healthy. As we learn more about the effects and improve technical aspects of ROSI, this method may become more acceptable. Indeed, studies on ROSI effects in mice have been encouraging (26). Thus our study may bear importance for clinicians working in ART clinics supporting the possibility that ROSI may be a viable option for overcoming infertility in men with non-obstructive azoospermia.
Considering that we have obtained live offspring using germ cells from males with only two Y chromosome genes one could question the importance of Y chromosome in male reproduction. We believe that the answer lies in defining the need. Human Y chromosome is not on the way to oblivion, as it has been implied in the past (27), and its genetic information is undoubtedly important for many aspects of reproduction involving the development of mature sperm and its function in normal fertilization (28). Most of the mouse Y chromosome genes are involved in spermiogenesis and sperm function and as such are necessary for normal fertilization (29, 30). However, when it comes to assisted reproduction, our mouse study proves that the Y chromosome contribution can be brought to a bare minimum consisting of Sry and Eif2s3y. Indeed, it may well be possible to eliminate mouse Y chromosome altogether if appropriate replacements are made for those two genes.
Supplementary Material
Acknowledgements
The work was supported by NIH HD072380, HD058059 and GM103457 (Project 2) and HCF 13ADVC-60314 grants to MAW. The authors thank Paul Burgoyne for providing paternal stock mice for breeding, overall support, and insightful discussions. The authors also thank N. Vernet from P. Burgoyne's group for sharing protocols and expertise on histological and DNA content analyses. We are grateful to numerous students who have helped with mouse genotyping. Histological sections were prepared by JABSOM Histopathology Core supported by NIH grants NIMHHD G12 MD007601 and the NIGMS P30 GM103341.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Stern C. Am J Hum Genet. 1957 Sep;9:147. [PMC free article] [PubMed] [Google Scholar]
- 2.Skaletsky H, et al. Nature. 2003 Jun 19;423:825. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 3.Yamauchi Y, et al. Biol Reprod. 2009 Aug;81:353. doi: 10.1095/biolreprod.109.076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubbay J, et al. Nature. 1990 Jul 19;346:245. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 5.Sinclair AH, et al. Nature. 1990 Jul 19;346:240. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 6.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Nature. 1991 May 9;351:117. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 7.Mazeyrat S, et al. Nat Genet. 2001 Sep;29:49. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 8.Vernet N, et al. Curr Biol. 2011 May 10;21:787. doi: 10.1016/j.cub.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Materials and methods and further details are available as supporting material on Science Online.
- 10.Vernet N, Mahadevaiah SK, Ellis PJ, de Rooij DG, Burgoyne PS. Reproduction. 2012 Oct;144:433. doi: 10.1530/REP-12-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odorisio T, Rodriguez TA, Evans EP, Clarke AR, Burgoyne PS. Nat Genet. 1998 Mar;18:257. doi: 10.1038/ng0398-257. [DOI] [PubMed] [Google Scholar]
- 12.Burgoyne PS, Mahadevaiah SK, Sutcliffe MJ, Palmer SJ. Cell. 1992 Oct 30;71:391. doi: 10.1016/0092-8674(92)90509-b. [DOI] [PubMed] [Google Scholar]
- 13.Capel B, et al. Cell. 1993 Jun 4;73:1019. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 14.Jeske YW, Bowles J, Greenfield A, Koopman P. Nat Genet. 1995 Aug;10:480. doi: 10.1038/ng0895-480. [DOI] [PubMed] [Google Scholar]
- 15.Hansen TB, et al. Nature. 2013 Mar 21;495:384. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Ehrmann IE, et al. Hum Mol Genet. 1998 Oct;7:1725. doi: 10.1093/hmg/7.11.1725. [DOI] [PubMed] [Google Scholar]
- 17.Foresta C, Ferlin A, Moro E. Hum Mol Genet. 2000 May 1;9:1161. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 18.Sargent CA, et al. J Med Genet. 1999 Sep;36:670. [PMC free article] [PubMed] [Google Scholar]
- 19.Sun C, et al. Nat Genet. 1999 Dec;23:429. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell MJ. Results Probl Cell Differ. 2000;28:233. doi: 10.1007/978-3-540-48461-5_10. [DOI] [PubMed] [Google Scholar]
- 21.Lahn BT, Page DC. Science. 1997 Oct 24;278:675. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 22.Kleiman SE, et al. Hum Reprod. 2007 Jan;22:151. doi: 10.1093/humrep/del341. [DOI] [PubMed] [Google Scholar]
- 23.Fertil Steril. 2008 Nov;90:S199. doi: 10.1016/j.fertnstert.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Gianaroli L, et al. Fertil Steril. 1999;72:539. doi: 10.1016/s0015-0282(99)00285-x. [DOI] [PubMed] [Google Scholar]
- 25.Saremi A, Esfandiari N, Salehi N, Saremi MR. Arch Androl. 2002 Jul-Aug;48:315. doi: 10.1080/01485010290031637. [DOI] [PubMed] [Google Scholar]
- 26.Tamashiro KL, Kimura Y, Blanchard RJ, Blanchard DC, Yanagimachi R. J Assist Reprod Genet. 1999;16:315. doi: 10.1023/A:1020406016312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graves JA. Cell. 2006 Mar 10;124:901. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Hughes JF, et al. Nature. 2010 Jan 28;463:536. [Google Scholar]
- 29.Riel JM, et al. J Cell Sci. 2012 Nov 23; [Google Scholar]
- 30.Yamauchi Y, Riel JM, Stoytcheva Z, Burgoyne PS, Ward MA. Genome Biol. 2010;11:R66. doi: 10.1186/gb-2010-11-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura Y, Yanagimachi R. Biol Reprod. 1995;52:709. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- 32.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. J Reprod Fertil. 1989;86:679. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- 33.Mahadevaiah SK, et al. Hum Mol Genet. 1998 Apr;7:715. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- 34.Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. Cytogenet Cell Genet. 1998;80:37. doi: 10.1159/000014954. [DOI] [PubMed] [Google Scholar]
- 35.Burgoyne PS, Levy ER, McLaren A. Nature. 1986 Mar 13–19;320:170. doi: 10.1038/320170a0. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe MJ, Burgoyne PS. Development. 1989 Oct;107:373. doi: 10.1242/dev.107.2.373. [DOI] [PubMed] [Google Scholar]
- 37.Eicher EM, et al. Cytogenet Cell Genet. 1991;57:221. doi: 10.1159/000133152. [DOI] [PubMed] [Google Scholar]
- 38.Lane PW, Davisson MT. J Hered. 1990 Jan-Feb;81:43. doi: 10.1093/oxfordjournals.jhered.a110923. [DOI] [PubMed] [Google Scholar]
- 39.Gubbay J, et al. Proc Natl Acad Sci U S A. 1992 Sep 1;89:7953. doi: 10.1073/pnas.89.17.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazeyrat S, et al. Hum Mol Genet. 1998 Oct;7:1713. doi: 10.1093/hmg/7.3.557. [DOI] [PubMed] [Google Scholar]
- 41.Simpson EM, Page DC. Genomics. 1991 Nov;11:601. doi: 10.1016/0888-7543(91)90067-o. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed EA, de Rooij DG. Methods Mol Biol. 2009;558:263. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 43.Rasband WS. Bethesda: U.S. National Institute of Health; 2007. [cited 22 June 2007]. Available from: http://rsb.info.nih.gov/ij/. [Google Scholar]
- 44.Burgoyne PS, Mitchell MJ. In: The Y Chromosome and Male Germ Cell Biology in Health and Diseases. Y L, C F, C a, Y W, editors. New Jersey: World Scientific Publishing Co. Inc; 2007. pp. 1–25. [Google Scholar]
- 45.Ferguson L, Ellis PJ, Affara NA. Mamm Genome. 2009 Apr;20:193. doi: 10.1007/s00335-009-9175-8. [DOI] [PubMed] [Google Scholar]
- 46.Decarpentrie F, et al. Hum Mol Genet. 2012 Jun 15;21:2631. doi: 10.1093/hmg/dds088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura Y, Yanagimachi R. Biology of Reproduction. 1995;53:855. doi: 10.1095/biolreprod53.4.855. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Burgoyne PS, Arnold AP. Hum Mol Genet. 2002 Jun 1;11:1409. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Watkins R, Arnold AP. Gene Expr Patterns. 2006 Jan;6:146. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. Histological and Histopathological Evaluation of the testis. St. Louis, MO: Cache River Press; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.