Abstract
RING ubiquitin ligases (E3s) recruit E2 thioesterified with Ub to facilitate Ub transfer to a target. Certain RING E3s dimerize to form active ligases but structural evidence on how this process promotes Ub transfer is lacking. Several members of the baculovirus inhibitor of apoptosis repeat-containing (BIRC) family of proteins function as dimeric RING E3s in the regulation of cell death. Here we report the structure of the human dimeric RING domain from BIRC7 in complex with the E2 UbcH5B covalently linked to Ub at its active site (UbcH5B~Ub). In addition to the known E2–RING contacts, the structure reveals extensive non-covalent donor Ub interactions with UbcH5B and both subunits of the RING domain dimer. Mutations that disrupt these non-covalent interactions or RING dimerization reduce UbcH5B~Ub binding affinity and ubiquitination activity. Moreover, NMR analyses demonstrate that BIRC7 binding to UbcH5B~Ub induces peak shift perturbations in the donor Ub consistent with the crystallographically-observed BIRC7–Ub interactions. Our results provide structural insights into how dimeric RING E3s recruit E2~Ub and optimize the donor Ub configuration for transfer.
Keywords: Inhibitor of apoptosis, BIRC7, E3, E2 ubiquitin conjugate, RING dimerization
Post-translational modification by Ub is an important regulatory mechanism for a wide variety of cellular processes1,2. Ub is activated and conjugated to substrate by the consecutive actions of Ub-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub-ligase (E3)1. E1 activates and forms a covalent thioester intermediate with the C-terminus of Ub in an ATP-dependent manner. Ub is then transferred to E2, resulting in a covalent thioester linkage between E2’s catalytic cysteine and Ub’s C-terminus (E2~Ub). E3s promote transfer of Ub from E2 to the amino group of a substrate lysine and generally contain E2 and substrate-binding domains to facilitate this process. Several classes of E3s have been defined based on the mechanism of transfer: HECT, RING, U-box and RING-in-between-RING3,4. For RING E3s, the largest class5, Ub is transferred directly from E2~Ub to substrate without the formation of a covalent E3~Ub intermediate.
How RING E3s promote Ub transfer is unclear. In the absence of an E3, E2~Ub is able to catalyze the transfer of donor Ub (conjugated Ub) to an acceptor Ub, free amine, or substrate4,6-11, but addition of a RING E3 massively enhances the rate4,8,9,11. In several structures of RING E3–E2 complexes, the RING domain binds E2 at a region distal to the E2’s active site, precluding direct modification of the active site by the RING E3 (refs. 12,13). Only biochemical analyses provide some insight into how RING E3s promote Ub transfer. Recent studies have shown that interactions between the Ile44 surface of the donor Ub and E2 α3’s residues are important for donor Ub transfer in the absence and presence of RING E3s9,10. Interestingly, mutations on the surface of the E2 UbcH5B, which are distal from both the RING domain-binding site and active site, influence the ability of RING E3s to stimulate Ub transfer8,14. In addition, mutational analyses of several E2s have identified highly conserved key residues near E2’s active site that are critical for catalysis: an asparagine side chain may stabilize the oxyanion transition state intermediate15 and an aspartate side chain may suppress the pKa of lysine on a substrate16. These findings have led to the proposal that RING E3 binding may organize the E2’s active site through allosteric effects to enhance substrate ubiquitination8,9.
Several RING E3s must homo- or heterodimerize to form active ligases. For example, BIRC family RING E3s, Rad18, RNF4, TRAF6 and IDOL form homodimers, whereas others, like MDM2, BRCA1 and RNF2, heterodimerize with inactive RING domains, namely MDMX, BARD1 and BMI1, respectively (reviewed in13). Although the structures of dimeric RINGs differ slightly, dimerization is achieved via RING–RING interactions and residues flanking N- and C-terminal regions of the RING domain. Mutations that disrupt RING dimerization reduce ubiquitination activity17-20. Recent studies on BIRC family RING E3s and RNF4 show that RING dimerization is required for E2~Ub binding18,21. For RNF4, one subunit of the RING domain dimer is proposed to bind E2 and the C-terminal tail of the second to interact with the Ile44 surface of the corresponding donor Ub to facilitate Ub transfer18. The importance of the C-terminal tail of the RING dimer is supported by a previous study on MDM2–MDMX, where inactive MDM2 C-terminal tail mutants can be rescued by heterodimerzation with wild-type MDMX22. Several models describing how RING E3s promote Ub transfer have been proposed based on a wealth of biochemical data, but there is currently no structure of a RING E3–E2~Ub complex.
BIRC proteins play a major role as negative regulators of cell death and several members of this protein family, including BIRC2 and BIRC3, contain a C-terminal RING domain that homodimerizes to form an active E3 (refs. 17,21). The E3 activity is required for ubiquitination and subsequent degradation of critical components in the apoptotic pathways such as caspases and second mitochondrial activator of caspases (SMAC), for activation of NF-κB signaling and for degradation of other proteins (reviewed in23). Using BIRC RING E3s as a model system, we determined the crystal structure of the dimeric BIRC7 (also known as melanoma inhibitor of apoptosis protein and Livin) RING domain in complex with UbcH5B covalently linked to Ub’s C-terminus to gain insights into the role of RING dimerization and to understand how RING E3s interact with E2~Ub to promote Ub transfer. The structural and biochemical data reveal that combined interactions between UbcH5B, donor Ub and BIRC7 RING dimer are crucial for Ub transfer. These results elucidate the roles of catalytically important residues outside the E2’s active and RING-binding sites, and identify non-canonical interactions between the Ile36 surface of Ub and the BIRC7 RING dimer, highlighting the importance of RING dimerization. In addition, NMR chemical shift analyses show that BIRC7 binding induces peak shift perturbations in the donor Ub consistent with the observed crystallographic interactions. As suggested for several RING E3s8,9,18, we propose BIRC7 promotes activity by stabilizing an optimal E2~donor Ub arrangement for Ub transfer. Here, we show the first detailed structural evidence supporting this hypothesis.
Results
Characterization of the BIRC7 RING–UbcH5B~Ub complex
Several homodimeric RING E3s, such as RNF4 and BIRC3, have distinct binding preferences for E2~Ub compared to E2 as demonstrated by pull-down assays18,21. To determine if BIRC7 has similar preferences, we quantitatively measured binding affinities of the BIRC7 and BIRC3 RING domains for UbcH5B~Ub and UbcH5B using surface plasmon resonance (SPR) analysis. Because E2 thioesterified with Ub rapidly dissociates into E2 and Ub in the presence of a RING E3, UbcH5B’s catalytic Cys85 was mutated to Ser to generate a stable covalent ester linkage (UbcH5BS~Ub), which mimics the thioesterified Ub linkage. In the SPR analysis, the contact time between E3 and UbcH5BS~Ub is less than 30 seconds and UbcH5BS~Ub is stable under these conditions. Like BIRC3, BIRC7 showed a higher binding affinity for UbcH5BS~Ub than UbcH5B (Table 1 and Supplementary Fig. 1), suggesting the donor Ub contributes to the RING E3–E2~Ub interaction.
Table 1.
Dissociation constants (KD) for interactions between RING E3 variants, UbcH5B, and UbcH5BS~Ub variants.
| GST-Immobilized Protein | Analyte | KD (μM)1 |
|---|---|---|
|
| ||
| BIRC3541–C | UbcH5B | 300 ± 7 |
| BIRC3541–C | UbcH5BS~Ub | 5 ± 4 |
| BIRC7239–C | UbcH5B | N.M. |
| BIRC7239–C | UbcH5BS~Ub | 136 ± 2 |
| BIRC7FL | UbcH5B | N.M. |
| BIRC7FL | UbcH5BS~Ub | 181 ± 29 |
| BIRC7FL | UbcH5BS I88A~Ub | N.M. |
| BIRC7FL | UbcH5BS S108R~Ub | N.M. |
| BIRC7FL | UbcH5BS D117A~Ub | 191 ± 18 |
| BIRC7FL | UbcH5BS~Ub L8D | N.M. |
| BIRC7FL | UbcH5BS~Ub G35E | N.M. |
| BIRC7FL | UbcH5BS~Ub I36A | N.M. |
| BIRC7FL | UbcH5BS~Ub I44A | N.M. |
| BIRC7FL V254A | UbcH5BS~Ub | N.M. |
| BIRC7FL V263R | UbcH5BS~Ub | N.M. |
| BIRC7FL I284A | UbcH5BS~Ub | N.M. |
| BIRC7FL R286A | UbcH5BS~Ub | N.M. |
| BIRC7FL F296H | UbcH5BS~Ub | N.M. |
| BIRC7FL Δ296-298 | UbcH5BS~Ub | N.M. |
| MDM2428–C–MDMX428–C | UbcH5B | N.M. |
| MDM2428–C–MDMX428–C | UbcH5BS~Ub | 130 ± 31 |
| RNF21–114–BMI11–109 | UbcH5B | 91 ± 2 |
| RNF21-114–BMI11–109 | UbcH5BS~Ub | 90 ± 2 |
s.e.m. are indicated. Number of replicates, representative sensorgrams and binding curves are shown in Supplementary Fig. 1. Subscripts denote the range of residues incorporated in the variant. FL indicates full-length protein was used. N.M. indicates KD is not measurable.
Due to the weak binding, we could not saturate UbcH5B or UbcH5BS~Ub variants concentrations for some of these measurements. The KD values were estimated by fitting data obtained from 0–180 μM UbcH5B or UbcH5BS~Ub variants using steady state affinity analyses.
To investigate the donor Ub role in binding, we attempted to crystallize UbcH5BS~Ub bound to the BIRC7 RING domain. Unfortunately, UbcH5BS~Ub was not suitable for crystallization because it dissociated into UbcH5B and Ub after 1–3 days in the presence of a RING E3. The highly conserved E2 HPN motif is proposed to stabilize the oxyanion intermediate during RING E3-mediated Ub transfer, and mutation of the Asn to Ala within this motif greatly hinders RING-E3 mediated Ub transfer15. Hence, we incorporated this mutation and found that UbcH5BS N77A~Ub was stable for several days at 4°C in the presence of our RING E3s. To prevent the UbcH5 family canonical “backside” binding with Ub’s Ile44 patch14, we also mutated UbcH5B’s Ser22 to Arg. Following incorporation of the aforementioned mutations, we successfully crystallized and determined the structure of the BIRC7 RING domain (BIRC7239–C) bound to UbcH5BS S22R N77A~Ub (UbcH5BRAS~Ub) to 2.18 Å (Fig. 1 and Table 2).
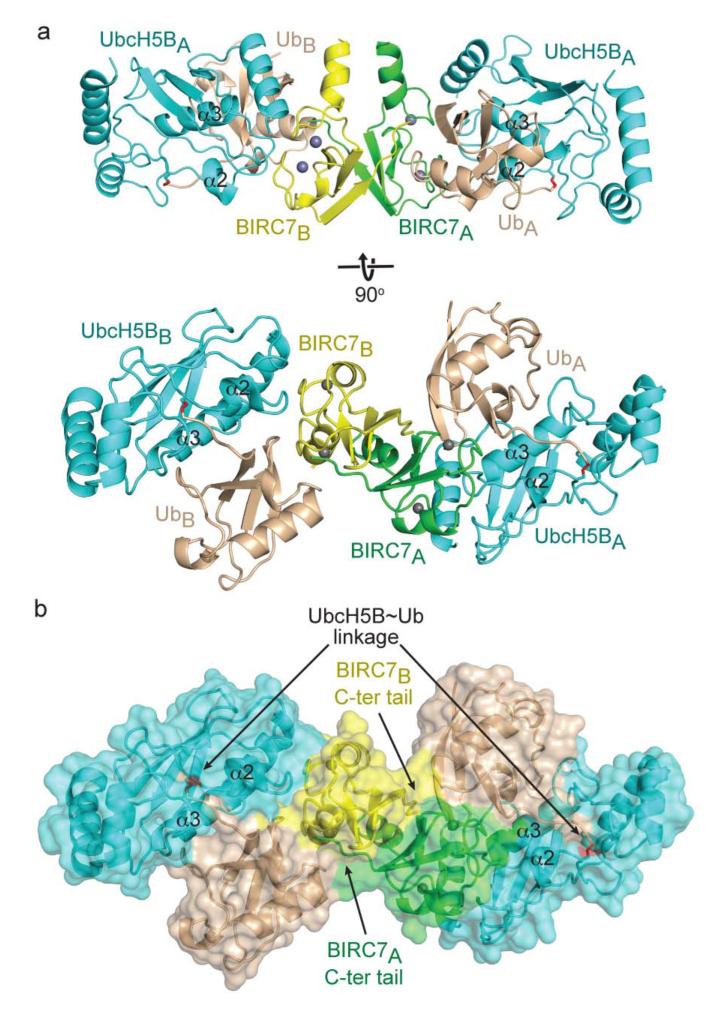
Figure 1.
Structure of BIRC7239–C–UbcH5BRAS~Ub. (a) Cartoon representation of the complex with the two crystallographic heterotrimers differentiated with A or B subscripts. Top and bottom panels are related by 90° rotation about the x-axis. Zn2+ atoms are depicted as grey spheres, Ub is colored wheat, BIRC7A green, BIRC7B yellow, and UbcH5B cyan with the covalent linkage highlighted in red. α2 and α3 from UbcH5B are labeled. (b) Surface representation of the complex, colored and oriented as in a bottom panel. UbcH5B~Ub linkages and the BIRC7 dimer C-terminal (C-ter) tails are indicated with arrows.
Table 2.
Data collection and refinement statistics
| BIRC7239–C–UbcH5B~Ub | |
|---|---|
| Data collection | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 100.6, 100.6, 123.9 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 30–2.18(2.29–2.18)1 |
| Rsym or Rmerge | 0.057(0.621) |
| I / σI | 16.5(3.3) |
| Completeness (%) | 99.8(99.2) |
| Redundancy | 6.7(6.6) |
| Refinement | |
| Resolution (Å) | 30–2.18 |
| No. reflections | 33958 |
| Rwork / Rfree | 0.202/0.249 |
| No. atoms | |
| Protein | 4232 |
| Ligand/ion | 4 |
| Water | 153 |
| B-factors | |
| Protein | 63.9 |
| Ligand/ion | 35.1 |
| Water | 50.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.24 |
Values in parentheses are for highest-resolution shell.
The complex has a compact overall structure and there are two copies of the BIRC7239–C–UbcH5BRAS~Ub heterotrimer in the asymmetric unit which superimpose with an r.m.s.d. of 0.43 Å for Cα atoms. For simplicity of discussion, we differentiate molecules from the two heterotrimers with A or B subscripts (e.g. BIRC7A or UbB, Fig. 1). The structures of UbcH5BRAS and Ub resemble prior UbcH5B and Ub structures, having an r.m.s.d. of 0.43 and 0.42 Å, respectively, for Cα atoms when compared to the models used for molecular replacement24. The structure of BIRC7239–C is similar to the BIRC3 RING domain17 (r.m.s.d. of 1.13 Å for Cα atoms) where an α-helix precedes the RING domain and the C-terminal tails of BIRC7 from both subunits participate in RING dimerization (Fig. 1).
The canonical RING–E2 interactions described previously12,17 are conserved in our BIRC7239–C–UbcH5BRAS~Ub complex. Ub nestles along UbcH5B’s α3 and the BIRC7 RING domain within each subunit of the asymmetric unit. Notably, the C-terminal tail of BIRC7B packs against UbA and the C-terminal tail of BIRC7A packs against UbB (Fig. 1). Ub’s Ile44 and Ile36 patches make extensive contacts with UbcH5B and the BIRC7 dimer, respectively. The C-terminal tail of Ub lies along the interface between UbcH5B’s α2 and α3, making numerous contacts with UbcH5B, the globular Ub body and the RING domain (Fig. 1). The quality of the electron density was better for subunit A of the BIRC7239–C–UbcH5BRAS~Ub complex, so our discussion focuses on this subunit and the C-terminal tail of BIRC7B (Fig. 1).
To assess the importance of the interactions observed in our structure (see below), we performed in vitro ubiquitination assays using full-length BIRC7 to test the effects of mutations on Ub transfer. For these assays, pulse-chase conditions were used to eliminate any mutational effects arising from E1-catalyzed UbcH5B~Ub formation. Under pulse-chase conditions, E2 is initially charged by incubation with Ub, E1, Mg2+ and ATP; subsequently, apyrase and EDTA are added prior to E3 to remove Mg2+–ATP and prevent further charging. Charging times were varied so that the initial concentrations of E2~Ub were similar in each assay.
We performed single turnover lysine discharge assays4 to exclusively monitor the effects on donor Ub transfer, thereby eliminating defects associated with Ub acting as an acceptor. Di-Ub formation is an alternative for exclusively monitoring donor Ub transfer, but under our reaction conditions, full-length BIRC7 has strong preferences for autoubiquitination (Supplementary Fig. 2a). We opted not to use K0-Ub (where all Lys are mutated to Arg) because Lys11 and Lys48 are within the Ub binding interface (Fig. 2a and Fig. 3a). BIRC7 contains an N-terminal BIR domain that binds SMAC, so, to investigate the mutational effects on poly-Ub formation, we employed BIRC7 autoubiquitination and SMAC ubiquitination assays. It is noteworthy that in the SMAC ubiquitination assay, BIRC7 autoubiquitination occurs concurrently (Supplementary Fig. 2b).
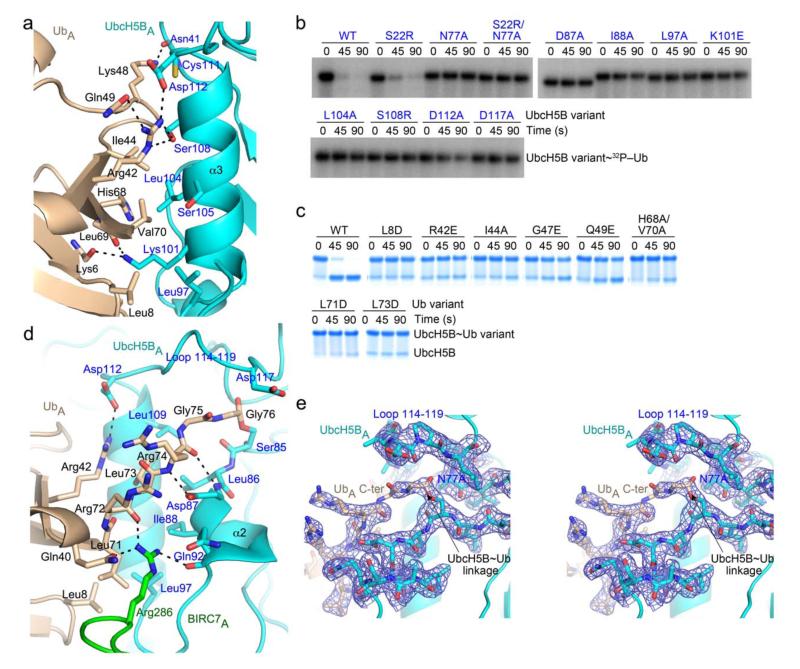
Figure 2.
UbcH5B–Ub interactions within the complex. (a) Close-up view of donor UbA Ile44 patch–UbcH5BA α3 interactions. (b) Non-reduced autoradiograms of pulse-chase reactions showing the disappearance of UbcH5B~32P-Ub with full-length BIRC7, L-lysine, and UbcH5B variants over time. (c) As in b but for Ub variants and visualized by SDS-PAGE without radiolabeling. (d) Close-up view of UbcH5BA active site–UbA tail interactions. (e) Stereo view of the UbcH5BA~UbA linkage, UbcH5BA loop 114–119 and UbA tail–UbcH5BA interactions with key residues shown as sticks and labelled. 2Fo-Fc electron density (blue) contoured at 1.0 σ is shown. For a, d and e, key residues are shown as sticks and coloring is as in Fig. 1 with N atoms blue, O atoms red, and S atoms yellow. Putative hydrogen bonds are shown as dashed lines.
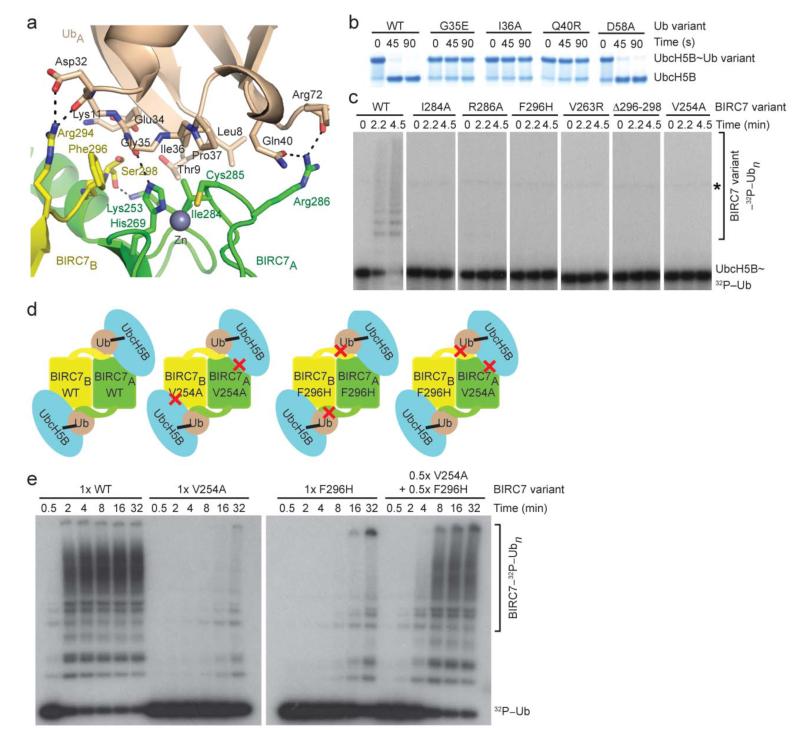
Figure 3.
Donor Ub–RING interactions are required for BIRC7-catalyzed reactions. (a) Close-up view of donor UbA Ile36 patch–BIRC7 dimer interactions. (b) Non-reduced SDS-PAGE of pulse-chase reactions showing the disappearance of UbcH5B~Ub with full-length BIRC7, L-lysine, and Ub variants over time. (c) Non-reduced autoradiograms of pulse-chase reactions showing the simultaneous formation of 32P-Ub products and disappearance of UbcH5B~32P-Ub with full-length BIRC7 variants over time. An asterisk indicates the E1~Ub band. (d) Experimental design to validate the cross-dimer–donor Ub interactions. Based on the structure, we designed BIRC7 mutants expected to disrupt the BIRC7 RING–UbcH5B interaction (V254A) or the BIRC7 tail–Ub interaction (F296H), as indicated by red crosses. A heterodimer of the RING and tail mutants should be more active than a homodimer of either mutant. (e) Reduced autoradiograms showing the formation of 32P-Ub products with wild-type, mutant homodimers, and the hetero-mutant BIRC7 dimer over time.
Donor Ub–UbcH5B interactions
Our structure shows, for the first time, detailed molecular interactions between a donor Ub covalently linked to an E2 in the presence of a RING E3. Unlike other E2~Ub structures, our E2~Ub conjugate appears poised for transfer. The Ile44 patch of Ub makes extensive non-covalent interactions with the entire length of UbcH5B’s α3 (Fig. 2a). Recently, E2’s α3–Ub Ile44 surface interactions have been identified to be important for Ub transfer by the E2s Ube2S and Cdc34 (refs. 9,10); for Cdc34, an S129L mutation on α3 of the E2 combined with an I44A mutation on Ub rescues Ub transfer defects associated with the individual mutants. In our structure, Ser108 of UbcH5B (corresponding to Ser129 in Cdc34) interacts with Ile44 of Ub. Furthermore, mutations of key residues on UbcH5B’s α3 (K101E, L104A, S108R and D112A) and Ub’s Ile44 surface (L8D, R42E, I44A, G47E, Q49E and H68A/V70A) cause defects in all three of our BIRC7-mediated Ub transfer assays (Fig. 2b,c and Supplementary Fig. 2c–g). In addition, Ub’s L8D and I44A and UbcH5B’s S108R greatly reduce binding affinity when incorporated into UbcH5BS~Ub in our SPR analyses (Table 1).
The observed UbcH5B~Ub conformation precludes binding of the donor Ub to UbcH5B’s backside in the presence of BIRC7 because both interactions use the same Ub surface. To investigate the role of UbcH5B’s backside Ub binding, we tested UbcH5B S22R in BIRC7-catalyzed assays. UbcH5B S22R displayed similar activity as wild-type in lysine discharge assays but was strongly defective in polyubiquitinating BIRC7 and SMAC (Fig. 2b and Supplementary Fig. 2d,e), consistent with previous reports showing that backside Ub binding is required for polyubiquitination14,24.
Residues 71 –74 of the C-terminal tail of UbA are maintained by contacts with UbcH5BA’s α2, UbA’s Leu8, Leu71 and Leu73, and BIRC7A’s Arg286 (Fig. 2d,e). To probe the significance of the observed interactions, we tested the activity of several Ub tail-interacting mutants: UbcH5B’s D87A, I88A, and L97A and Ub’s L71D and L73D are defective in BIRC7-mediated Ub transfer (Fig. 2b,c and Supplementary Fig. 2d–g). Furthermore, incorporation of UbcH5B’s I88A mutation into UbcH5BS~Ub reduces BIRC7 binding affinity (Table 1) suggesting that the conformation of Ub’s C-terminal tail residues 71–74 is important in BIRC7-catalyzed Ub transfer.
When compared to other UbcH5~Ub structures (PDB 3A33 and 3UGB)24,25, there is a paucity of interactions between the extreme C-terminal tail of Ub (residues 75 and 76) and the UbcH5 loop comprising residues 114–119. In these other structures lacking the E2 N77A mutation, the sidechain of Asp117 interacts with the amide of Ub’s Gly76 and the E2 loop, in turn, is stabilized by hydrogen bonding interactions with Asn77. In BIRC7239–C–UbcH5BRAS~Ub, it is noteworthy that the average B factors for the two tail Ub residues (115 Å2) and the E2 loop (95.7 Å2) are high compared to the overall structure; no electron density is present for several of the sidechains in the E2 loop and electron density for the covalent ester linkage is visible but poor around Ub’s Gly75, preventing accurate positioning of the carbonyl group of this residue (Fig. 2e). Nonetheless, modeling of Asn77 from the UbcH5BS~Ub structure (PDB 3A33)24 onto our mutant N77A UbcH5BA suggests that the Asn Nδ is within hydrogen bonding distance of UbA’s Gly76 carbonyl oxygen although the angles may not reflect transition state geometries (Supplementary Fig. 3a).
Previously, mutational analyses have shown that loop 114–119 is crucial in UbcH5-mediated catalysis4,8,24. Structural alignments of the Cα atoms of UbcH5BRAS~Ub with available structures of UbcH5B and UbcH5B~Ub variants lacking the N77A mutation show the loop encompassing residues 114–119 of the E2 adopts multiple conformations (Supplementary Fig. 3b), suggesting the loop is flexible. Moreover, superposition of UbcH5B from our structure onto Ubc9 (the SUMO E2) in the SUMO–RanGAP1–Ubc9–Nup358 complex structure (r.m.s.d. of 1.12 Å for Cα atoms, Supplementary Fig. 3c–e) reveals a similar E2–Ub like protein (Ubl) tail conformation. In the SUMO E2, Asp127 (corresponding to UbcH5B’s Asp117) contacts the amino group of lysine on the substrate and has been shown to play a critical role in lysine pKa suppression during catalysis16. Mutation of this residue produces defects in BIRC7 (Fig. 2b and Supplementary Fig. 2d,e) and other RING E3-mediated Ub transfer4,8,24. Interestingly, UbcH5BS D117A~Ub and UbcH5BS~Ub display similar binding affinities for BIRC7 (Table 1) suggesting that Asp117 may not be involved in E2~Ub recruitment but has an important role in the subsequent steps of the reaction. It is evident from biochemical studies that UbcH5B loop 114-119 has a role in catalysis, but the E2 active site arrangement observed in our structure may not be optimized for transfer due to the N77A mutation. Further investigation is required to elucidate the role of the UbcH5B loop in Ub transfer.
Donor Ub Ile36 patch–BIRC7 interactions
In UbA, the Leu8–Thr9 loop and the patch surrounding Ile36 make extensive contacts with the BIRC7A RING domain and BIRC7B’s C-terminal tail (Fig. 3a), providing the first structural evidence of donor Ub–RING interactions. UbA’s Leu8, Thr9, Ile36, and Pro37 form hydrophobic contacts with BIRC7A’s Ile284 and Zn2+-binding ligands, His269 and Cys285. In addition, hydrogen bonds occur between the sidechains of BIRC7A’s Arg286 and UbA’s Gln40 and between BIRC7A’s His269 and the backbone oxygen of UbA’s Gly35. The extreme C-terminal tail of BIRC7B makes several contacts with the donor UbA: Arg294 forms a hydrogen bond with the backbone oxygen of UbA’s Asp32; Phe296 contacts UbA’s Gly35; and Ser298 sits in a pocket created by UbA’s Thr9, Lys11 and Glu34.
To probe the observed Ub–BIRC7 interactions, we generated BIRC7 mutants (I284A, R286A, and F296H), Ub mutants within the Ile36 patch (G35E, I36A, and Q40R) and a control Ub mutant, D58A, which opposes the Ile44 and Ile36 patches. Phe296 was mutated to His because for the corresponding residue in RNF4, this was the only mutation that did not disrupt dimerization but still significantly reduced activity18. Ub mutants harboring G35E, I36A or Q40R are defective in BIRC7-mediated Ub transfer assays, whereas Ub D58A had similar activity to WT Ub (Fig. 3b and Supplementary Fig. 2f,g). Likewise, BIRC7 I284A, R286A or F296H mutants, which exist as dimers in solution as shown by size exclusion chromatography (Supplementary Fig. 4a), are also defective in catalyzing autoubiquitination (Fig. 3c). To test the importance of BIRC7 RING domain dimerization, we introduced V263R or a C-terminal tail deletion (Δ296-298) to disrupt dimerization. These mutants exist as monomers and are inactive in catalyzing BIRC7 autoubiquitination (Fig. 3c and Supplementary Fig. 4). SPR analyses show that UbcH5BS~Ub G35E and UbcH5BS~Ub I36A have reduced binding affinity for BIRC7 and BIRC7 mutants have reduced binding affinity for UbcH5BS~Ub (Table 1).
The dimeric arrangement shows that E2~Ub can be recruited via both BIRC7 domains, but competent binding of a single UbcH5B~Ub complex requires both BIRC7 domains – the RING domain of one dimer subunit binds UbcH5B and donor Ub whereas the tail of the second supports Ub binding via a “cross”-dimer arrangement (Fig. 1 and Fig. 3d) as proposed for RNF4 (ref. 18). To probe our BIRC7A RING–BIRC7B tail–UbcH5BA~UbA arrangement, we hypothesized that a dimer comprising a subunit of BIRC7 with a tail mutation and a second subunit with a RING mutation should be active. For our RING mutation, we generated BIRC7 V254A, which greatly reduces binding affinity for UbcH5BS~Ub and is defective in BIRC7 autoubiquitination (Fig. 3e). Incubation of a mixture of BIRC7 V254A with BIRC7 F296H led to enhanced BIRC7 autoubiquitination activity (Fig. 3e). Thus, the cross-dimer BIRC7A RING–BIRC7B tail arrangement and ensuing interactions with donor Ub are required for UbcH5B~Ub recruitment and Ub transfer.
BIRC7 binding reorganizes the donor Ub conformation
How do RING E3s promote donor Ub transfer? Our structural and biochemical data show that BIRC7 promotes Ub transfer by stabilizing UbcH5B and the donor Ub in a conformation optimal for conjugation. To confirm whether the donor Ub forms non-covalent interactions with UbcH5B and BIRC7 in solution, we charged UbcH5BRAS with 15N-labeled Ub (UbcH5BRAS~15N-Ub) and acquired 1H–15N HSQC spectra in the absence and presence of BIRC7239–C. The UbcH5BRAS~15N-Ub spectrum showed significant chemical shift perturbations in Ub’s Ile44 patch and C-terminus and moderate perturbations in other regions of Ub when compared to the 15N-Ub spectrum alone (Fig. 4a and Supplementary Fig. 5a). We do not know how Ub contacts UbcH5BRAS in the UbcH5BRAS~Ub complex, but UbcH5BRAS precludes backside binding. Based on the shift perturbations, Ub may interact with UbcH5B’s α3 in a similar manner to that observed in Ubc1~Ub and Ube2S~Ub models10,26. Mapping of these perturbations onto Ub in our structure reveals that some perturbations radiate away from the UbcH5B–Ub binding interface (Fig. 4b) suggesting that interactions between donor Ub’s Ile44 patch and UbcH5B are preferred but donor Ub may populate other conformations in solution as previously observed in UbcH5C~Ub complexes14,27.
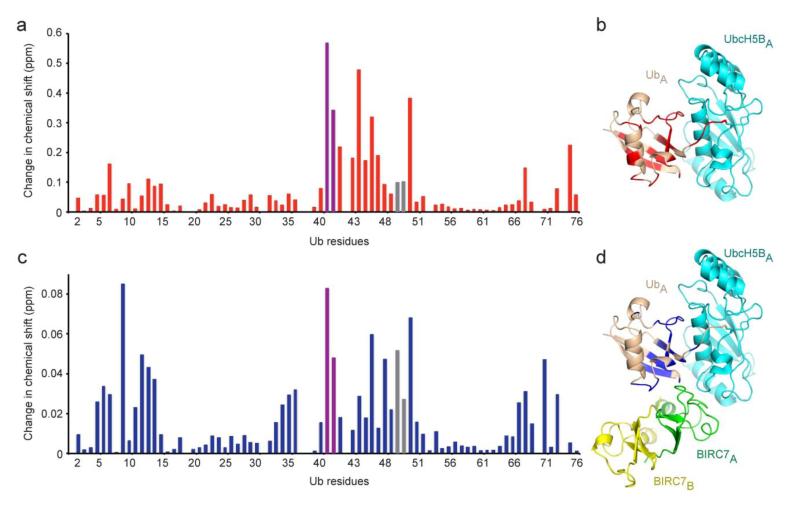
Figure 4.
Ub interaction surface in BIRC7239–C–UbcH5BRAS~Ub identified by NMR. (a) Changes in chemical shift per residue of 15N-Ub following covalent linkage to UbcH5BRAS determined by 1H–15N HSQC NMR. Changes were calculated according to the equation [(0.15 δN)2 + δH2]1/2. (b) Mapping of changes in a (> 1σ, 0.028 ppm) onto the Ub (colored red) in the UbcH5BA~UbA portion of BIRC7239–C–UbcH5BRAS~Ub. (c) Changes in chemical shift per residue of Ub in BIRC7239–C–UbcH5BRAS~15N-Ub compared to UbcH5BRAS~15N-Ub, calculated as in a. (d) Mapping of changes in c (> 1σ, 0.01 ppm) onto the Ub (colored blue) in the BIRC7 dimer– UbcH5BA~UbA portion of BIRC7239–C–UbcH5BRAS~Ub. For a and c, the violet and grey bars represent Gln40 and Gln49 sidechain chemical shift perturbations, respectively. For molecules, coloring is as described in Fig. 1.
Titration of an increasing molar ratio of BIRC7239–C to UbcH5BRAS~15N-Ub produced gradual chemical shift perturbations in the spectra (Supplementary Fig. 5b–d), indicative of changes in the donor Ub interaction network. To separate Ub peak shift perturbations induced by BIRC7 binding from conjugation of Ub to UbcH5BRAS, we compared the peak shifts from the BIRC7239–C–UbcH5BRAS~15N-Ub spectrum to the UbcH5BRAS~15N-Ub spectrum. These perturbations map to regions near the Leu8–Thr9 loop, Ile36 and Ile44 patches, and the C-terminal tail residues Leu71 and Leu73 (Fig. 4c and Supplementary Fig. 5e). These regions correspond to the donor Ub surfaces that are involved in non-covalent interactions with UbcH5B and BIRC7 in our structure (Fig. 4d) and are critical for activity. Together these results suggest that donor Ub has a preference to form non-covalent interactions with UbcH5B in the absence of E3, but BIRC7 binding further stabilizes the donor Ub interaction network.
Kinetic validation of the BIRC–E2~Ub model
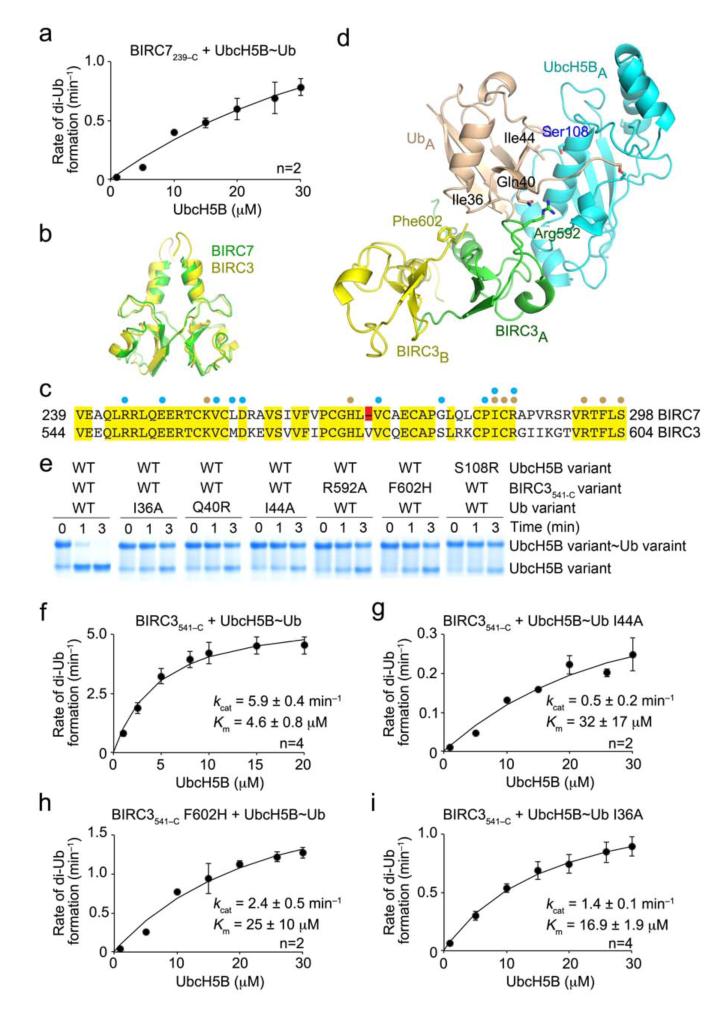
Our BIRC7239–C–UbcH5BRAS~Ub structure and supporting biochemical data suggest that E2–Ub and Ub–E3 interactions are required to optimally position the donor Ub for transfer. Disruption of either set of interactions is expected to hinder binding of E2~Ub and prevent proper positioning of the donor Ub for transfer – under steady-state conditions, an increase in Km and a decrease in kcat should be observed for any mutation that disrupts these interfaces. We attempted to assess mutational effects on kcat and Km using kinetic analyses on BIRC7-catalyzed di-Ub formation; to eliminate autoubiquitination, BIRC7239–C was used instead of full-length BIRC7. The donor Ub was labeled with 32P-ATP and we used a His-tagged Ub as the acceptor, which cannot be labeled. Reactions were carried out under pulse-chase conditions to avoid activating the acceptor Ub. At our highest achievable UbcH5B~Ub concentration (30 μM), our assay only approached saturation (Fig. 5a and Supplementary Fig. 6a) and was thus inappropriate for measuring mutational effects on kcat and Km.
Figure 5.
Effect of donor Ub interactions on the kinetics of Ub transfer. (a) Single turnover kinetics of di-Ub formation catalyzed by BIRC7239–C. The rate of di-Ub formation was plotted against UbcH5B concentration. (b) Superposition of the structure of BIRC3541–C dimer (yellow, PDB 3EB6)17 onto BIRC7239–C dimer (green) from BIRC7239–C–UbcH5BRAS~Ub. BIRC3541–C dimer was generated from a symmetry-related molecule. (c) ClustalW sequence alignment of BIRC7 and BIRC3 RING domains. Identical residues are highlighted in yellow and the Val deletion in BIRC7 in red. Cyan and wheat circles indicate residues involved in contacting UbcH5B and Ub, respectively, as observed in BIRC7239–C–UbcH5BRAS~Ub and PDB 3EB6. (d) Models of UbcH5B~Ub bound to BIRC3541–C dimer generated by superposing BIRC3541–C dimer onto BIRC7239–C–UbcH5BRAS~Ub. Key residues involved in donor Ub interactions are indicated. Coloring is as described in Figs. 1 and 2. (e) Non-reduced SDS-PAGE of pulse-chase lysine discharge reactions showing the disappearance of UbcH5B~Ub with variants of BIRC3541–C, UbcH5B or Ub over time. (f–i) As in a but performed with BIRC3541–C. The rate of di-Ub formation was plotted against UbcH5B concentration for WT reaction (f), 32P-Ub I44A (g), BIRC3541–C F602H (h) and 32P-Ub I36A (i). Kinetic parameters and number of replicates, n, are indicated. Error bars indicate standard errors.
Because the BIRC3 RING domain has ~25-fold higher binding affinity for UbcH5BS~Ub than BIRC7 in our SPR analyses (Table 1), we hypothesized it might be suitable for mutant kinetic assays to validate our model. The RING domain of BIRC3 resembles BIRC7 (r.ms.d. 1.13 Å) and the sequences are 74% identical (Fig. 5b,c). In the sequence alignment, the BIRC7 residues that interact with Ub in our structure are identical to those in BIRC3. All of the BIRC7–Ub interactions observed in our BIRC7239–C–UbcH5BRAS~Ub structure are present in a model of a BIRC3–UbcH5B~Ub complex built by superposing the RING domain of BIRC3 onto BIRC7 in our complex (Fig. 5d). Interestingly, BIRC7 has a Val deletion within a RING hydrophobic patch that is involved in E2 binding which may account for the observed weaker binding affinities (Fig. 5c and Supplementary Fig. 6b,c). To confirm that the BIRC3 and BIRC7 Ub transfer mechanisms are alike, we performed lysine discharge assays using the same sets of mutants tested in the BIRC7 assays. The activity profiles were identical – all of the mutants were defective in Ub transfer except UbcH5B’s S22R and Ub’s D58A (Fig. 5e and Supplementary Fig. 6d,e).
We first measured the kcat and Km for di-Ub formation using wild-type Ub, UbcH5B and BIRC3 RING domain to determine if saturating conditions were achievable. Indeed, the Km was well below the highest achievable UbcH5B~Ub concentration (Fig. 5f). We then selected three mutants for our kinetic assays to validate our model: Ub’s I44A and I36A and BIRC3’s F602H (corresponding to BIRC7’s Phe296). Ub’s Ile44 interacts with Ser108 on UbcH5B’s α3, and Ub’s Ile36 and BIRC3’s Phe602 comprise part of the E3–Ub interface. Each of these mutants increased Km and decreased kcat as predicted by our model (Fig. 5g–i and Supplementary Fig. 6f). Thus, donor Ub interactions with UbcH5B and the RING domain are crucial for optimal Ub transfer.
Discussion
The BIRC7239–C–UbcH5BRAS~Ub structure and biochemical data presented here provide insights into how a RING E3 facilitates Ub transfer and the role of RING dimerization in this process. The structure reveals how combined interactions between both subunits of the BIRC7 RING dimer, UbcH5B and donor Ub are vital for the stability of the BIRC7–UbcH5B~Ub complex and Ub transfer. Our NMR chemical shift experiments with Ub demonstrate that although the donor Ub can form non-covalent interactions with UbcH5B in the absence of BIRC7, further chemical shift perturbations are observed upon BIRC7 binding. These perturbations are consistent with interactions observed in the crystal structure.
Our results agree with a previous study on RNF4, where modeling and biochemical analyses predicted that the RING E3 binds directly to both E2 and the donor Ub, thereby optimizing the E2~Ub conformation and positioning the thioester linkage for transfer18. The RNF4 study demonstrated that dimerization is required for E3 ligase activity and that both subunits of the dimer are crucial for recruiting E2~Ub as we observed for BIRC7. In the RNF4–E2~Ub model, the Ile44 surface of Ub was predicted to bind to residues at the dimer interface, precluding interactions with E2’s α3 as observed in our structure. Instead, Ub’s Ile36 surface contacts the dimer interface to position the donor Ub for transfer.
Specific surfaces on Ub like the Ile44 patch are frequently observed to be involved in non-covalent interactions crucial for Ub transfer. Like BIRC7, Cullin-RING ligase-mediated Ub transfer also requires interactions between donor Ub’s Ile44 patch and residues Ile128 and Ser129 on α3 of the E2 Cdc34, corresponding to Cys107 and Ser108 of UbcH5B9,10. Donor Ub Ile44 patch–E2 α3 interactions are also required for di-Ub formation by the E2s Cdc34 and Ube2S in the absence of an E39,10. For Ubc13–MMS2, which catalyzes Lys63 poly-Ub chain formation, MMS2 interacts with the Ile44 patch of the acceptor Ub to promote Ub transfer28. Ub’s Ile36 patch has emerged as a second hydrophobic surface that is critical for Ub transfer. In BIRC7, it is required for reactions between the donor Ub and the RING E3 dimers but it is also recognized by HECT E3s29,30, deubiquitinating enzymes31 and Ub-binding domains32,33.
The UbcH5 family also has the ability to bind Ub’s Ile44 surface via its backside (Ser22 surface). This ability is crucial for processivity of poly-Ub chain synthesis14,24, but the mechanism is unknown. Our results and a recent study on Cullin-RING ligases9 suggest that the donor Ub cannot interact with the backside of any UbcH5 molecule during transfer. Hence, backside binding does not promote poly-Ub chain formation by positioning the donor Ub for transfer. Although the Ile44 patch on the donor Ub is unavailable during transfer, the backside of the donor UbcH5 (refers to UbcH5~Ub that is bound to E3) is not occupied and may recruit free UbcH5~Ub to enable self-assembly14,24,25 or bind and guide the growing poly-Ub chain to promote poly-Ub chain formation14. E2s have been shown to collaborate to catalyze poly-Ub chain34-36, so UbcH5 may also recruit other E2~Ub via backside binding. Further studies are required to elucidate the role of backside Ub-binding in poly-Ub formation.
Optimizing non-covalent donor Ub–E2 interactions and positioning the thioester linkage are mechanisms shared by other types of E3-catalyzed Ub or Ubl transfers. In the NEDD4L–UbcH5B~Ub complex, the HECT E3 NEDD4L binds both UbcH5B and Ub to position the E2~Ub thioester linkage for transfer29. The crystal structure of the post-conjugated SUMO–RanGAP1–Ubc9–Nup358 complex shows SUMO E3 Nup358–RanBP2 binds both Ubc9 and SUMO to prevent non-productive Ubc9~SUMO conformations and to position the Ubc9~SUMO thioester linkage for transfer37, though a recent study suggests Ubc9 only plays a structural role in this multisubunit E3 complex38. For both E3s, the interactions between E3 and donor Ub or Ubl enhance the catalytic efficiency of the transfer reaction by improving kcat and reducing Km. SPR analyses of UbcH5BS~Ub variants showed that mutations which disrupt any binding interface increase KD for the BIRC7–UbcH5B~Ub complex, but we were unable saturate reaction conditions to obtain kinetic constants due to the weak BIRC7–UbcH5B~Ub binding affinity. We can only speculate that some of these mutations would also have kcat defects in BIRC7-mediated Ub transfer. BIRC3541–C resembles BIRC7239–C and putatively has identical Ub binding interactions based on sequence alignment and modeling. Using BIRC3541–C, we demonstrated that disruption of donor Ub–UbcH5B and donor Ub–RING interactions reduce kcat and increase Km for Ub transfer.
RING E3 binding is proposed to induce allosteric changes in E2’s active site to promote Ub transfer8,9. Our data show conjugation to Ub and binding to an E3 do not significantly alter the active site of the E2 compared to other structures of free UbcH5, UbcH5~Ub, and UbcH5–E3. There is a slight shift in the E2 loop encompassing residues 114–119 compared to UbcH5B~Ub (PDB 3A33), but whether this is due to E3 binding, our Asn77 mutation, or inherent flexibility is unclear. Our results support findings from previous mutational analyses, where UbcH5’s α2 was revealed to contribute to the allosteric communication induced by E3 binding, possibly through interactions with Ub: UbcH5B I88A reduced and UbcH5C D87E abolished E3-stimulated Ub transfer4,8. In our complex, Ub tail residues 71–74 interact with α2 of UbcH5B. BIRC7’s Arg286, which is strictly conserved in many RING E3s as Arg or Lys, not only binds UbcH5B’s α2 in our structure but also coordinates Ub’s tail residues 71-74 to α2 of the E2.
The cross-dimer arrangement observed here and in RNF4 (ref. 18) is required for donor Ub interactions and E2~Ub recruitment. When compared to other dimeric RING E3s that function with the UbcH5 family E2s, we find this dimer arrangement is also observed in the structures of MDM2–MDMX39 and IDOL40, but absent in the structures of BRCA1–BARD1 (ref. 41) and RNF2–BMI1 (refs. 19,42). Modeling of the UbcH5BRAS~Ub fragment of our structure onto MDM2–MDMX, reveals the presence of a complementary donor Ub–RING interface and a conserved Phe (Phe296 in BIRC7) on the C-terminal tail required for the cross-dimer donor Ub binding (Supplementary Fig. 7). In contrast, a similar model of RNF2–BMI1 reveals potential sidechain clashes and the absence of favorable cross-dimer donor Ub interactions (Supplementary Fig. 7). Indeed, our data (Table 1) and previous studies18,21 show that the BIRCs, MDM2–MDMX and RNF4 have binding preferences for E2~Ub rather than free E2, whereas RNF2–BMI1 has no preference. Clearly, not all dimeric RING E3s recruit E2~Ub in a similar fashion, but our results suggest that RING E3s that adopt conformations similar to the BIRC7 RING dimer may have similar mechanisms of E2~Ub recruitment and Ub transfer.
This mechanism may not be unique to dimeric RING E3s. U-box domains structurally resemble RING domains and some form active homodimers in a manner analogous to RING E3 dimers. The structure of PRP19 U-box dimer shows a similar U-box C-terminal tail configuration as BIRC7, but with variations in the C-terminal tail sequence43. Modeling of the UbcH5BRAS~Ub portion of our structure onto the PRP19 U-box structure reveals a complementary interface between donor Ub and U-box with slight differences in the mode of C-terminal tail interactions (Supplementary Fig. 7), suggesting a similar E2~Ub recruitment mechanism. Interestingly, several of monomeric RING domains also exhibit higher binding affinities for E2~Ub than E2 despite lacking the cross-dimer arrangement. Examples include Cullin-RING ligases44, E3α45 and c-CBL46. Common UbcH5B surface residues outside the E2’s active site and E3 binding site – for example Ile88 – are essential for Apc2/11- and CNOT4-mediated Ub transfer8. Based on these observations, we speculate that other RING E3s may utilize similar modes of E2~Ub recruitment and Ub transfer.
Methods
Methods for protein preparation, structural determination and single turnover kinetics of di-Ub formation are in the Supplementary Methods.
Generation of UbcH5BS~Ub variants
Ub variants with a C-terminal Gly-Gly motif were cloned into pGEX4T1 with an N-terminal GST-tag followed by a TEV cleavage site and a Gly-Gly-Ser linker. GST-Ub variants were purified by glutathione-affinity chromatography. Untagged mustard Uba1 was expressed from pET23d, charged with GST-Ub variants with 5 mM MgCl2 and 5 mM ATP for 2h at 4°C and purified by glutathione-affinity chromatography eluted with 20 mM DTT, followed by anion exchange chromatography. Untagged UbcH5B C85S variants were expressed from pRSF_1b and purified by cation exchange chromatography. All proteins were dialyzed into 25 mM HEPES (pH 8.0), 0.15 M NaCl. UbcH5BS~Ub variants were formed by mixing arabidopsis thaliana Uba1, UbcH5B C85S variants, GST-Ub variants and TEV protease in the presence of 10 mM MgCl2 and 10 mM ATP at 19°C for 1 day. UbcH5BS~Ub variants were then purified by cation exchange chromatography followed by gel filtration chromatography using a Hi-Load 16/60 SD75 column. All UbcH5BS~Ub variants were stored in 25 mM HEPES (pH 7.5), 0.2 M NaCl and 1 mM DTT. UbcH5BS~Ub variants concentrations were determined by measuring absorbance at 280 nm in the presence of 6 M guanidine HCl using calculated molar extinction coefficient (26,900 M−1 cm−1) for Biacore analyses.
Crystallization
Crystals were obtained by mixing the protein with an equal volume of reservoir solution and grown by hanging drop vapor diffusion at 4 °C. BIRC7239–C (4.7 mg ml−1) was mixed with UbcH5BRAS~Ub (25.4 mg ml−1) at a 1:1 molar ratio, and crystals were grown in conditions containing 100 mM HEPES, pH 7.0, 17–20% (w/v) PEG 3350 and 0.2 M tri-ammonium citrate. The crystals were flash-frozen in 100 mM HEPES, pH 7.0, 23% (w/v) PEG 3350, 0.2 M tri-ammonium citrate and 20% (v/v) glycerol. Data were collected at beamlines I04 and I24 at DLS.
In vitro pulse-chase assays
E2 (10 μl) was charged in a buffer containing 50 mM Tris-HCl (pH 7.6), 50 mM NaCl, 5 mM MgCl2, 5 mM ATP, 1 mM DTT, 0.3 U ml−1 inorganic pyrophosphatase, 0.3 U ml−1 creatine kinase, 5 mM creatine phosphate, 1 mg ml−1 bovine serum albumin (except for assays detected with SimplyBlue™ SafeStain (Invitrogen)) and typically containing mouse UBA1 (0.4 μM), Ub or 32P-Ub (40 μM) and UbcH5B (20 μM) for 15–30 min at 23 °C. Charging was stopped by incubating the reaction with 0.25 U apyrase (Sigma) and 30 mM EDTA for 5 min at 23 °C. The autoubiquitination, SMAC-ubiquitination, di-Ub and lysine discharge reactions were then respectively initiated by the addition of BIRC7 variants (2 μM), a mixture of BIRC7 (0.220 μM) and 32P-SMAC (5.6 μM), a mixture of BIRC7 (0.220 μM) and His-Ub (200 μM), or a mixture of BIRC7 (2 μM) and L-lysine (150 mM) or a mixture of BIRC3541–C (0.220 μM) and L-lysine (150 mM). The parentheses indicate the final concentration. The final UbcH5B~Ub concentration was ~7 μM for all reactions. Reactions were quenched with 2X SDS loading buffer (with DTT only for the diubiquitin assays) at the indicated time, resolved by SDS-PAGE, dried and visualized by autoradiography, stained with SimplyBlue™ SafeStain, or immunoblotted with Ub (P4D1-A11, Millipore) or His-probe (sc-803, Santa Cruz) antibodies.
E3–E2~Ub binding assays
We used identical methods as previously described46 to couple E3s as listed in Table 1 to CM-5 chips (Biacore Life Sciences). All chips were equilibrated in running buffer containing 25 mM Tris (pH 7.6), 150 mM NaCl, 0.1 mg ml−1 BSA, 1 mM DTT and 0.005% (v/v) Tween-20 and analytes were serially diluted in running buffer. Binding was measured between concentration ranges of 0–180 μM for analytes. Data reported are the difference in SPR signal between GST-E3 variants and GST alone. The data were analyzed by steady state affinity analysis using the Biacore T200 Evaluation software package (Biacore Life Sciences) and Scrubber2.0c (BioLogic Software).
NMR Spectroscopy
Data were recorded at 25°C on a Varian VNMRS 600 spectrometer using a 5 mm triple resonance (HCN) cold probe. All protein samples were prepared or dialyzed in 25 mM sodium phosphate, 0.15 M NaCl buffer at pH 7.0 containing 90/10% H2O/D2O. 1H–15N HSQC spectra were recorded for 15N-Ub (100 μM) and UbcH5BRAS~15N-Ub (100 μM). For BIRC7239–C titration experiments, 1H–15N HSQC spectra of BIRC7239–C–UbcH5BRAS~15N-Ub complex was recorded with a molar ratio of BIRC7239–C:UbcH5BRAS~15N-Ub at 100:100 μM and then at 300:200 μM. Ub chemical shifts are available in BioMagResBank under accession code 6457. Due to large chemical shift changes upon covalent linkage to UbcH5BRAS and resonance broadening, Gln31, Arg42, Val70 and Arg74 were not assigned in Fig. 4 and Supplementary Fig. 5. The chemical shift perturbations were estimated using the equation [(0.15 δN)2 + δH2]1/2 (ppm), where δN and δH represent changes in nitrogen and proton chemical shifts, respectively, and analyzed using CCPN47. 1σ was determined using an iterative procedure to calculate a corrected standard deviation to zero as described previously48 but stopped after 10% of data were excluded.
Supplementary Material
Acknowledgements
We would like to thank K. Vousden and A. Schuettelkopf for discussion. W. Clark and A. Keith for in-house DNA sequencing; Diamond Light Source (DLS) for access to beamlines I04 and I24 beamlines (mx6683) that contributed to the results presented here. This work was supported by Cancer Research UK.
Footnotes
Accession codes Coordinates and structure factors for the crystal structures of has RCSB accession code of 4AUQ.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–50. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–8. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–81. [PubMed] [Google Scholar]
- 7.Pickart CM, Vella AT. Ubiquitin carrier protein-catalyzed ubiquitin transfer to histones. Mechanism and specificity. J Biol Chem. 1988;263:15076–82. [PubMed] [Google Scholar]
- 8.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–5. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–81. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 13.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Wu PY, et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–50. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 17.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–40. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 18.Plechanovova A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–9. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmil and Ringlb. EMBO J. 2006;25:2465–74. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–66. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feltham R, et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem. 2011;286:17015–28. doi: 10.1074/jbc.M111.222919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–12. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–52. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 24.Sakata E, et al. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–47. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Page RC, Pruneda JN, Amick J, Klevit RE, Misra S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry. 2012;51:4175–87. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 27.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry. 2011;50:1624–33. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 29.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maspero E, et al. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011;12:342–9. doi: 10.1038/embor.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu M, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–54. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 32.Prag G, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–20. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 33.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–39. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 35.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46:287–98. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–8. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–74. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–7. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–9. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 43.Vander Kooi CW, et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121–30. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Haas AL. Protein interactions within the N-end rule ubiquitin ligation pathway. J Biol Chem. 2003;278:9448–57. doi: 10.1074/jbc.M211240200. [DOI] [PubMed] [Google Scholar]
- 46.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–92. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 48.Schumann FH, Riepl H, Maurer T, Gronwald W, Neidig KP, Kalbitzer HR. Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J Biomol NMR. 2007;39:275–89. doi: 10.1007/s10858-007-9197-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.