Abstract
Aims: The purpose of this study was to determine whether 3′-5′-cyclic adenosine monophosphate (cAMP)-protein kinase A (PKA) and Sirtuin-1 (SIRT1) dependent mechanisms modulate ATP-binding Cassette (ABC) transport protein expression. ABC transport proteins (ABCC2–4) are essential for chemical elimination from hepatocytes and biliary excretion. Nuclear factor-E2 related-factor 2 (NRF2) is a transcription factor that mediates ABCC induction in response to chemical inducers and liver injury. However, a role for NRF2 in the regulation of transporter expression in nonchemical models of liver perturbation is largely undescribed. Results: Here we show that fasting increased NRF2 target gene expression through NRF2- and SIRT1–dependent mechanisms. In intact mouse liver, fasting induces NRF2 target gene expression by at least 1.5 to 5-fold. In mouse and human hepatocytes, treatment with 8-Bromoadenosine-cAMP, a cAMP analogue, increased NRF2 target gene expression and antioxidant response element activity, which was decreased by the PKA inhibitor, H-89. Moreover, fasting induced NRF2 target gene expression was decreased in liver and hepatocytes of SIRT1 liver-specific null mice and NRF2-null mice. Lastly, NRF2 and SIRT1 were recruited to MAREs and Antioxidant Response Elements (AREs) in the human ABCC2 promoter. Innovation: Oxidative stress mediated NRF2 activation is well described, yet the influence of basic metabolic processes on NRF2 activation is just emerging. Conclusion: The current data point toward a novel role of nutrient status in regulation of NRF2 activity and the antioxidant response, and indicates that cAMP/PKA and SIRT1 are upstream regulators for fasting-induced activation of the NRF2-ARE pathway. Antioxid. Redox Signal. 20, 15–30.
Introduction
Members of the ATP-binding cassette (ABC) superfamily, efflux organic anions from hepatocytes into bile (e.g., Abcc2) and blood (e.g., Abcc3 and 4). Increased Abcc2 expression increases biliary excretion of organic anion (e.g., disulfobromophthalein [DBSP]), whereas increased Abcc3 expression reverses vectorial excretion of glucuronide conjugates into blood (25). Genetic and pharmacological nuclear factor-E2 related factor 2 (NRF2) activation can upregulate Abcc2–4 expression in liver (25, 36).
NRF2 belongs to the basic leucine zipper family of transcription factors, present in the cytoplasm as a complex with Kelch like ECH-associated protein-1 (Keap1), Cul3 and ubiquitin-E3 ligase and undergoes continual proteasomal degradation (27, 50). Upon activation, NRF2 dissociates from the Keap1-Cul3-Ubiquitin E3 complex, translocates to nucleus and activates gene transcription by binding to Antioxidant Response Element (ARE). NRF2 is responsive to cellular redox status alterations (39) and is largely described to mediate induction of xenobiotic metabolism and cytoprotection genes, such as Nadph:quinone oxidoreductase (Nqo1) and glutamate-cysteine ligase, catalytic subunit, in response to oxidative stress and change in cellular redox status (23).
Innovation.
The present data demonstrate a regulatory role of nuclear factor-E2 related factor 2 in contributing to Abcc2–4 upon fasting, which corresponds to endogenous metabolite alterations. These observations also link the Sirtuin-1-PPAR gamma coactivator alpha pathway to mechanisms that dictate hepatic clearance and implicate other mechanisms, such as acetylation, are involved. Use of Antioxidant response element (ARE)-hPAP mice to study hepatic regulation of ARE activity has not been described. Detection of disulfobromophthalein in gallbladder bile to indirectly quantify Abcc2 activity in mice has not been described, which will help the transporter field be able to assess Abcc2 function in mice, which is challenging to do by cannulation surgery.
Nutrient homeostasis in liver relies on redox status. During fasting, decreased glucose and increased pyruvate concentrations increase the NAD+/NADH ratio, initiating a cascade of events that activate gluconeogenesis (44). Fasting increases glucagon, which increases intracellular 3′-5′-cyclic adenosine monophosphate (cAMP) content (30), activates protein kinase A (PKA), and modulates transcription of multiple genes in liver that maintain glucose homeostasis, metabolism, and transport (9). cAMP/PKA cascade activates downstream expression of gluconeogenic genes, restoring blood glucose levels to normal (5). Short-term fasting induces gluconeogenesis via mTOR/TORC2 pathway, which is inhibited during longer fasting intervals by Sirtuin1 (NAD-dependent deacetylase, SIRT1) switching to the use of free fatty acids as precursors for glucose production via deacetylation and activation of PPAR gamma coactivator alpha (Pgc-1α) and Forkhead Box O1 (Foxo1). (17, 34, 44).
Along with gluconeogenic genes, fasting induces transcription of some genes involved in biotransformation (e.g., Cyp2b10, CYP2C11, CYP2E1, CYP2B1/2) and transport (e.g., Slco1b2) in coordination with nuclear receptors (6, 9). However, little is known about effect of nutritional status on biliary clearance mechanisms or bile flow. Few studies exist regarding how fasting affects transporter or the nuclear receptor-regulated pathways which regulate expression. Fasting effects on the NRF2-Keap1 signaling pathway, which is responsive to oxidative stress and altered redox status, have not been elucidated. Some evidence suggests a possible role for NRF2 in lipid and glucose homeostasis (3, 40), very little is known about how fasting affects the NRF2-Keap1 pathway. To date, no study has addressed how SIRT1 regulates NRF2 activity and subsequent gene expression in vivo.
The purpose of this study was to identify whether (i) fasting increases Abcc expression through NRF2- and SIRT1-dependent mechanisms and (ii) cAMP/PKA activators increase ARE activation via NRF2- and SIRT1-dependent mechanisms in vitro. The current study determined whether fasting and pharmacological activation of the fasting response pathway increased NRF2 activity and upregulated Abcc2–4 expression through cAMP/PKA-SIRT1-dependent mechanisms. Data herein present an undescribed role for NRF2 as a “nutrient sensitive” responsive transcription factor that is regulated via SIRT1-dependent mechanisms in vivo, and illustrate hepatic clearance mechanisms are differentially affected by fasting.
Results
Fasting increases Abcc2–4 expression in liver and hepatocytes
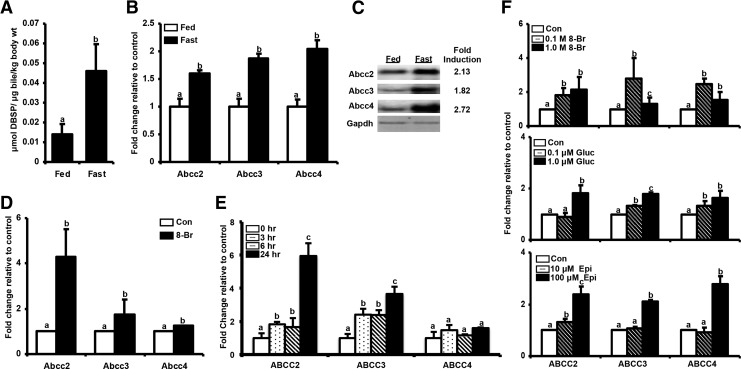
First, it was determined whether fasting modulates biliary clearance via induction of Abcc transporters expression and activity. The fasting model displayed a typical hepatic fasting response, with induction of Pgc-1α and Sult2a1 mRNA expression in liver (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). Fasting increased excretion of DBSP, a prototypical organic anion that undergoes biliary excretion via Abcc2 (25), into gallbladder bile by ∼3-fold compared to fed mice (Fig. 1A). Fasting correspondingly increased liver Abcc2–4 mRNA levels (Fig. 1B) and Abcc2–4 protein expression (Fig. 1C) compared to mice fed ad libitum. 8-Br-cAMP, a cAMP analogue that activates cAMP/PKA pathway, increased expression of Abcc2–4 by 6, 1.75 and 1.5-fold in primary mouse hepatocytes (Fig. 1D). Additionally, 8-Br-cAMP induced ABCC2 and 3 expression by 5, and 2-fold, in human Huh-7 hepatocarcinoma cells (Fig. 1E); indicating that fasting induced activation of NRF2 was conserved in human-derived cells. In line with these data, several upstream activators of fasting pathway, glucagon, epinephrine, and 8-Br-cAMP, increased ABCC2–4 expression in primary human hepatocytes (Fig. 1F).
FIG. 1.
Fasting induces Abcc2–4 expression in vitro and in vivo. (A) Increased biliary disulfobromophthalein (DBSP) excretion upon fasting. DBSP (150 μmol/kg/5 ml, intraperitoneal injection) was injected into C57BL/6 mice (n=5 per group) either fed ad libitum or fasted for 18 h. 45 min after injection, gallbladders were collected and biliary DBSP content was determined spectrophotometrically after appropriate dilution and alkalinization of bile with 0.1 N NaOH. The data are represented as average±SEM μmol of DBSP/μl bile/kg body weight. (B) Induction of Abcc 2–4 expression in mouse liver after a 24 h fast-Total RNA was isolated from livers of C57BL/6 mice either fed ad libitum or fasted for 24 h. Relative Abcc 2–4 mRNA expression was quantified by real-time quantitative PCR. The data are represented as average±SEM fold change over the fed controls. All data have been normalized to 18S rRNA. (C) Induction of Abcc2–4 protein expression in mouse liver after 24 h fasting-Crude membrane proteins were isolated from livers of C57BL/6 mice either fed ad libitum or fasted for 24 h. Abcc2–4 protein expression was quantified by western blotting. (D) Induction of Abcc 2–4 mRNA expression in primary mouse hepatocytes by 8-Bromoadenosine-3′,5′-Cyclic Adenosine Monophosphate (8-Br-cAMP)-Primary mouse hepatocytes were treated with DMSO or 8-Br-cAMP (1 mM) for 24 h. Total RNA was isolated 24 h after treatment and Abcc2–4 mRNA expression was quantified using real time quantitative PCR. The data are represented as average±SEM (n=3) fold change over the fed controls. All data are normalized to 18S rRNA. Groups without a common letter are significantly different. (E) Time dependent expression of ABCC 2–4 mRNA in Huh-7 cells by 8-Br-cAMP- Huh-7 cells were treated with 1.0 mM 8-Br-cAMP for 0, 3, 6, 12, 24, and 48 h with media as the control (n=3). Total RNA was isolated and ABCC 2–4 mRNA expression was quantified using the Branched DNA Signal Amplification assay. The data are represented as average Relative Light Units (RLU) per 10 μg total RNA±SEM (n=3). (F) Induction of ABCC 2–4 mRNA expression in primary human hepatocytes by 8-Brc-AMP-Primary human hepatocytes were treated with media, DMSO, 8-Br-cAMP (0.1 and 1 mM), epinephrine (10 and 100 μM) or glucagon (0.1 and 1 μM) for 24 h. At the end of 24 h cells were lysed using Lysis Mixture™. mRNA levels of ABCC 2–4 were quantified using Branched DNA signal amplification assay. The data are represented as relative mRNA expression per 25 μl lysate normalized to beta actin from three human hepatocyte donors.
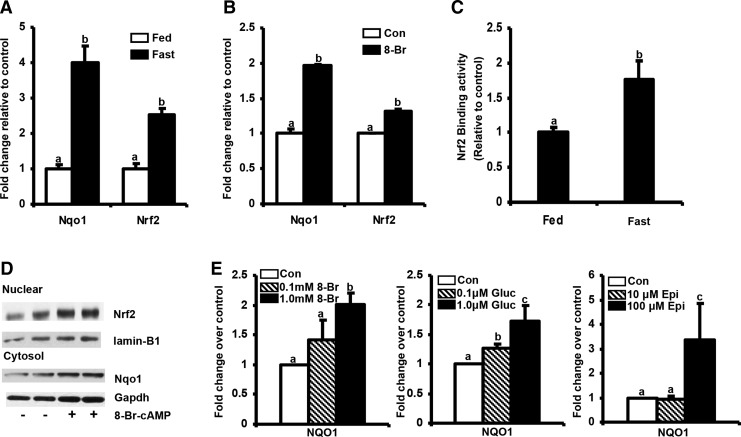
Nqo1 is considered to be a prototypical NRF2 target gene in mouse and human (26). Our previous work in demonstrated that food deprivation for 24 h increased Nrf2 and Nqo1 mRNA expression in livers of mice (55). Similar to the latter work, the cohort used herein also had increased Nqo1 and Nrf2 mRNA levels in liver compared to fed controls (Fig. 2A) fasting slightly increased Keap1 mRNA expression by 25% (Supplementary Fig. S2). Moreover, 8-Br-cAMP, a PKA activator, increased Nqo1 expression by 2-fold in primary mouse hepatocytes (Fig. 2B). This increase was accompanied by an increased Nrf2 binding to consensus ARE sequence in liver nuclear extracts from Nrf2 wild-type (WT) fasted mice (Fig. 2C) as determined by Procarta TF plex assay. To further explore the molecular mechanism underlying the fasting induced activation of NQO1 and ABCC2–4, we examined nuclear NRF2 levels before and after 8-Br-cAMP treatment. As shown in Figure 2D, 8-Br-cAMP increased nuclear NRF2 levels 1 h post treatment in Huh-7 cells along with increased cytosolic Nqo1 expression as determined by western blot (Fig. 2D). Additionally, PKA activators increased NQO1 expression in primary human hepatocytes (Fig. 2E). The observations indicate that activation of fasting pathways induces Nrf2, Nqo1, Abcc2–4 mRNA, and protein expression in mouse and human in vitro and in vivo models. Gsta-1, another Nrf2-regulated gene, was not induced in livers of mice after fasting, but was induced in human hepatocytes treated with 8-Br-cAMP (Supplementary Fig. S3).
FIG. 2.
Fasting induces nuclear factor-E2 related factor 2 (NRF2) target gene expression in vitro and in vivo. (A) Induction of Nadph:quinone oxidoreductase (Nqo1) and Nrf2 mRNA expression in mouse liver after 24 h fast- Total RNA was isolated from livers of C57BL/6 mice either fed ad libitum or fasted for 24 h. Gene expression of Nqo1 was quantified by real-time quantitative PCR. The data are represented as average±SEM fold change over the fed controls. All data have been normalized to 18S rRNA. (B) Induction of Nqo1 and NRF2 mRNA expression in primary mouse hepatocytes by 8-Br-cAMP-Primary mouse hepatocytes were treated with DMSO or 8-Br-cAMP (1 mM) for 24 h. Total RNA was isolated 24 h after treatment and Nqo1 and NRF2 mRNA expression was quantified using real time quantitative PCR. The data are represented as average±SEM (n=3) fold change over the fed controls. All data are normalized to 18S rRNA. Groups without a common letter are significantly different. (C) Fasting increases binding of NRF2 to its consensus binding sequence-Liver nuclear extracts were prepared from control and fasted C57BL/6 mice using Procarta TF nuclear extraction kit and analysis for transcription factor binding was determined using a Procarta TF binding assay. The results obtained are represented as data normalized to Transcription Factor II D. Data are expressed as Fluorescence Intensity/10 μg total liver nuclear extract normalized to C57BL/6 fed controls. p<0.05 was considered statistically significant. (D) Time dependent increase in NQO1 and NRF2 protein expression in Huh-7 cells by 8-Br-cAMP- Huh-7 cells were treated with1.0 mM 8-Br-cAMP for 0, 3, 6, 12, 24 h with media as the control (n=3). mRNA levels of NQO1 was quantified using Branched DNA signal amplification assay. The data are represented as fold change in NQO1 mRNA expression (n=3). NQO1 protein levels were determined by western blotting with Gapdh as the loading control. Nuclear NRF2 protein levels were measured by western blotting with laminB1 as loading control. (E) Induction of NQO1 mRNA expression in primary human hepatocytes by 8-Brc-AMP- Primary human hepatocytes were treated with media, DMSO, 8-Br-cAMP (0.1 and 1 mM), epinephrine (10 and 100 μM) or glucagon (0.1 and 1 μM) for 24 h. At the end of 24 h cells were lysed using Lysis Mixture. mRNA levels of NQO1 was quantified using Branched DNA signal amplification assay. The data are represented as relative mRNA expression per 25 μl lysate normalized to beta actin from three human hepatocyte donors.
Fasting induces Nrf2 activation and Abcc2–4, and Nqo1 expression in WT in part through Nrf2-dependent mechanisms
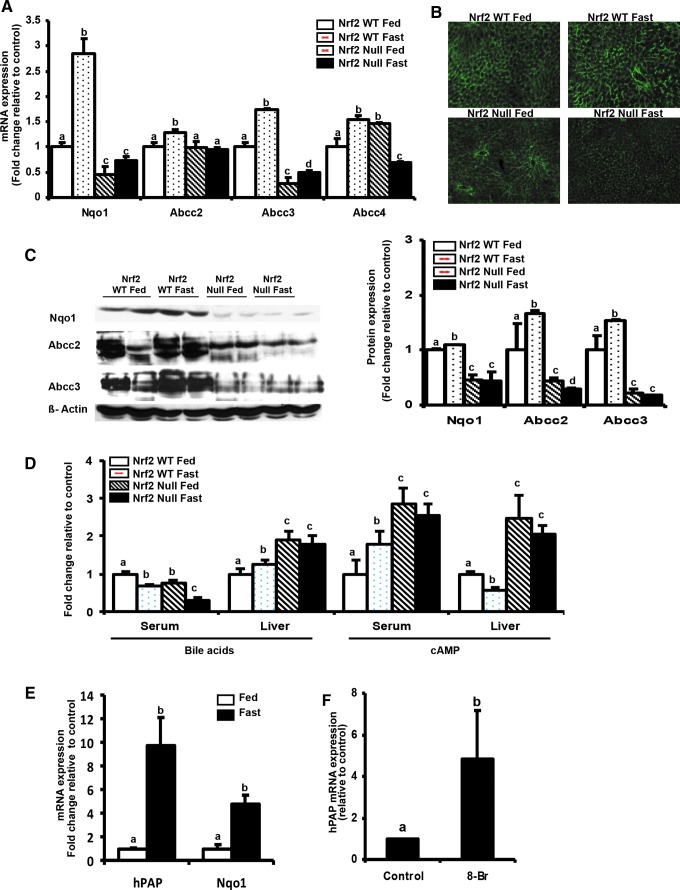
To determine whether fasting induces expression of Nrf2 target genes through Nrf2-dependent mechanisms, relative Nqo1 and Abcc expression in WT and Nrf2-null fasted mice was quantified. Increased Nqo1, Abcc2–4 expression at 24 h in Nrf2 WT mice, whereas expression in livers of Nrf2-null mice was significantly lower (Fig. 3A). The relative mRNA fold increase in Nqo1 mRNA was higher than Abcc2–4, which is consistent with previous observations (2, 57). Fasting also increased Abcc2 protein staining in livers of C57BL/6 mice, but not in livers of Nrf2 null mice (Fig. 3B). Further, fasting increased Abcc2, Abcc3, Nqo1 protein expression (Fig. 3C). To identify whether changes in mRNA and protein expression of Abccs during fasting contributed to changes in metabolite and substrate concentrations, serum and liver bile acids were quantified (Fig. 3D). Fasting increased bile acid levels by 23% in Nrf2 WT mice, while decreasing the serum bile acid content by ∼40%, which is consistent with a previous observation (32). Nrf2-null mice had lower serum bile acid levels in the fed state (Fig. 3D) consistent with previously published data (52), but after fasting, liver bile acid levels in the Nrf2-null mouse livers were similar to levels in fed Nrf2-null livers (Fig. 3D), suggesting possible decreased export of bile acids into the bile. We quantified serum and liver cAMP concentrations because cAMP is a described substrate for Abcc4 (11, 31). Fasting increased serum cAMP levels in WT mice by 77%, whereas levels were unchanged in NRF2 null mouse serum (Fig. 3D). Liver cAMP levels in Nrf2 WT mice decreased by 48% upon 24 h fasting, but remained unchanged in the Nrf2 null mice (Fig. 3D), which corresponds to the observed lack of Abcc4 induction (Fig. 3A) and suggesting increased hepatic retention of cAMP.
FIG. 3.
Fasting increases Abcc2–4, Nqo1 expression in Nrf2 dependent manner. (A) Fasting for 24 h induces the expression of Nrf2 target genes in wild-type C57BL/6 mice livers but not in Nrf2 null mice livers-Total RNA was isolated from livers of C57BL/6 (n=6) and Nrf2-null (n=6) mice either fed ad libitum or fasted for 24 h. Nqo1and Abcc2–4 gene expression was quantified by real time quantitative PCR. The data are represented as average±SEM (n=6) fold change over the fed controls. (B) Immunohistochemical staining of Abcc2-Cryosections were incubated with anti-Abcc2 antibody followed by incubation with AlexaFluor-conjugated secondary antibodies (green). (C) Induction of Abcc2-3 protein expression in Nrf2-WT but not in Nrf2-null mouse liver after 24 h fasting-Crude membrane proteins were isolated from livers of C57BL/6 (n=2) and Nrf2 null (n=2) mice either fed ad libitum or fasted for 24 h. Nqo1, Abcc2, Abcc3 and beta actin protein expression was quantified by western blotting. The data are representative of three individual protein quantifications. (D) Fasting for 24 h decreases liver cAMP, while increasing serum cAMP levels in Nrf2 WT but not in Nrf2 null mice. Serum bile acids and serum and liver cAMP levels were determined from serum and livers obtained from C57BL/6 (n=4) and Nrf2 null (n=4) mice fasted for 24 h. Data are represented as average±SEM (n=4) fold change relative to control. (E) Fasting increases hPAP and Nqo1 mRNA in livers of ARE-hPAP mice-Total RNA was isolated from livers of C57BL/6 (n=3) and ARE-hPAP (n=3) mice either fed ad libitum or fasted for 24 h. Nqo1 and human alkaline phosphatase (hPAP) mRNA expression was quantified by real time quantitative PCR. The data are represented as average±SEM (n=3) fold change over the fed controls. All data have been normalized to 18S rRNA. (F) 8-Br-cAMP induces hPAP mRNA expression in primary mouse hepatocytes-ARE-hPAP mouse hepatocytes were treated with 1 mM 8-Br-cAMP for 24 h. Total RNA was isolated and ARE-hPAP mRNA expression was quantified using RT2-PCR. Target gene expression was normalized to 18s rRNA. Groups without a common letter are significantly different. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To further test the possibility that Nrf2 induces expression of its targets through ARE, ARE-hPAP reporter transgenic mice possessing a human alkaline phosphatase reporter downstream of ARE consensus sequence (22) were fasted hPAP mRNA expression was determined. Fasting increased hPAP and Nqo1 mRNA expression in livers of ARE-hPAP mice by 8 and 5-fold compared to feed controls (Fig. 3E). hPAP mRNA expression was somewhat more inducible than Nqo1 mRNA expression, which is consistent with a previous publication (54). 8-Br-cAMP increased hPAP mRNA expression by about 2.5-fold in primary mouse hepatocytes isolated from ARE-hPAP mice (Fig. 3F). These results indicate that fasting induces Nrf2, Nqo1 and Abcc2–4 in part through Nrf2. Other transcription factors, such as CAR, PPARα also regulate expression of Abcc2–4 (15). However, Nrf2-null mice exhibited dampened induction of Abcc2–4 indicating that fasting induction of Abcc2–4 occurs via some Nrf2 dependent mechanisms.
8-Br-cAMP activates cAMP/PKA pathway upstream of NRF2-ARE activation
To identify a connection between cAMP/PKA signaling pathway and NRF2 activation, induction of ARE-driven luciferase reporter was analyzed in Huh-7 cells treated with 8-Br-cAMP, glucagon and epinephrine. As shown in Figure 4A and Supplementary Figure S4, all three treatments increased ARE-driven luciferase activity by about 1.5–2.5-fold. Primary ARE-hPAP mouse hepatocytes treated with 8-Br-cAMP showed increased hPAP expression, which was decreased upon treatment with H89 (Fig. 4B). 8-Br-cAMP treatment increased Nrf2, Nqo1, Abcc2, and 4 mRNA expression in primary hepatocytes isolated from Nrf2-WT, but not in Nrf2-null hepatocytes (Fig. 4C, D). Induction of Nrf2, Nqo1, Abcc2, and Abcc4 expression by 8-Br-cAMP was blunted by H89 in Nrf2-WT hepatocytes, but not in Nrf2-null hepatocytes (Fig. 4C, D).
FIG. 4.
Protein kinase A (PKA) pathway acts as upstream regulator of NRF2-ARE pathway. (A) 8-Br-cAMP induces antioxidant response element (ARE) in Huh-7 cells-Huh-7 cells were cultured to 85% conflucence and transiently transfected with an ARE–luciferase reporter construct (100 ng) for 24 h. The cells were then treated with 0.1 and 1.0 mM 8-Br-cAMP for 24 h. Luciferase activity was measured by dual luciferase assay with renilla luciferase as a control. The data are expressed as average fold induction in activity of the ARE-luciferase construct±SEM (n=4). (B) H89 inhibits 8Br-cAMP induced mRNA expression of hPAP-ARE-hPAP mouse hepatocytes were treated with 1 mM 8-Br-cAMP with or without H89 (10 μM) for 24 h. Total RNA was isolated and hPAP, expression was quantified using RT2-PCR. Target gene expression was normalized to 18s rRNA. 8-Br-cAMP induced (C) NRF2, Nqo1 and (D) Abcc2 and 4 mRNA expression is muted by H89 cotreatment in NRF2 WT, but not NRF2-null hepatocytes- Primary mouse hepatocytes obtained from WT or Nrf2-null mice were treated with DMSO or 8-Br-cAMP (1 mM) with or without H89 (20 μM) for 24 h. Total RNA was isolated 24 h after treatment and Abcc2 mRNA expression was measured using real time quantitative PCR. The data are represented as average±SEM (n=3) fold change over the fed controls. All data are normalized to 18S rRNA. p<0.05 was considered statistically significant. Groups without a common letter are significantly different. (E) Fasting induced upregulation of hepatic NAD+/NADH levels occurs to the greater extent in NRF2 null mice-NAD+/NADH ratio was quantified in livers obtained from C57BL/6 mice fasted for 24 h according to manufacturer's protocol. The data are represented as average±SEM (n=4) fold change over the fed controls. (F) AMP-activated protein kinase (AMPK) activators do not induce expression of Nrf2 target genes-Primary mouse hepatocytes were treated with control, AICAR (0.5 mM), NAD+ (5 mM) or metformin (1 mM) for 24 h. Total RNA was isolated 6 h after treatment and mRNA expression of Nqo1 and Nrf2 was quantified using real time quantitative PCR. The data are represented as average±SEM (n=3) fold change over the fed controls. All data are normalized to 18S rRNA. Groups without a common letter are significantly different. (G) Total AMPKα and p-AMPKα is attenuated in Nrf2 WT and Nrf2 null mouse livers upon fasting-Proteins lysates were obtained from livers of C57BL/6 (n=2) and Nrf2 null (n=2) mice either fed ad libitum or fasted for 24 h. Total AMPK and p-AMPKα and Gapdh protein expression was quantified by western blotting.
Induction of NAD+/NADH is one mechanism that activates SIRT1. Liver NAD+/NADH levels were decreased by ∼50% in NRF2-null compared with Nrf2 WT mice (Fig. 4E). Fasting resulted in a greater fold increase in liver NAD+/NADH ratio in Nrf2-null mice as compared to the WT mice (Fig. 4E), but as seen from Figure 3D, Nrf2-null mice did not demonstrate the changes in cAMP levels as seen in the Nrf2 WT mice. Lack of Nrf2 did not affect Pgc-1α and Pepck expression in these mice (Supplementary Fig. S5), indicating that fasting-induced expression of gluconeogenic genes is not Nrf2 dependent. However, even though upstream pathways of fasting were activated in Nrf2 null mice, Abcc2–4 and Nqo1 expression was muted indicating that induction of these target genes occurs via Nrf2 dependent mechanisms.
In primary mouse hepatocytes, AMP-activated protein kinase (AMPK) activators AICAR, MET, and NAD+ failed to induce Nrf2 and Nqo1, further suggesting a role for cAMP/PKA instead of the AMPK pathway (Fig. 4F). Total AMPKα and p-AMPKα levels decreased in livers of WT mice, but were unchanged in Nrf2-null livers, further suggesting that AMPK activation was not upstream to Nrf2 activation (Fig. 4G). These observations indicate that cAMP/PKA induces expression of ARE-containing genes via Nrf2-dependent mechanisms.
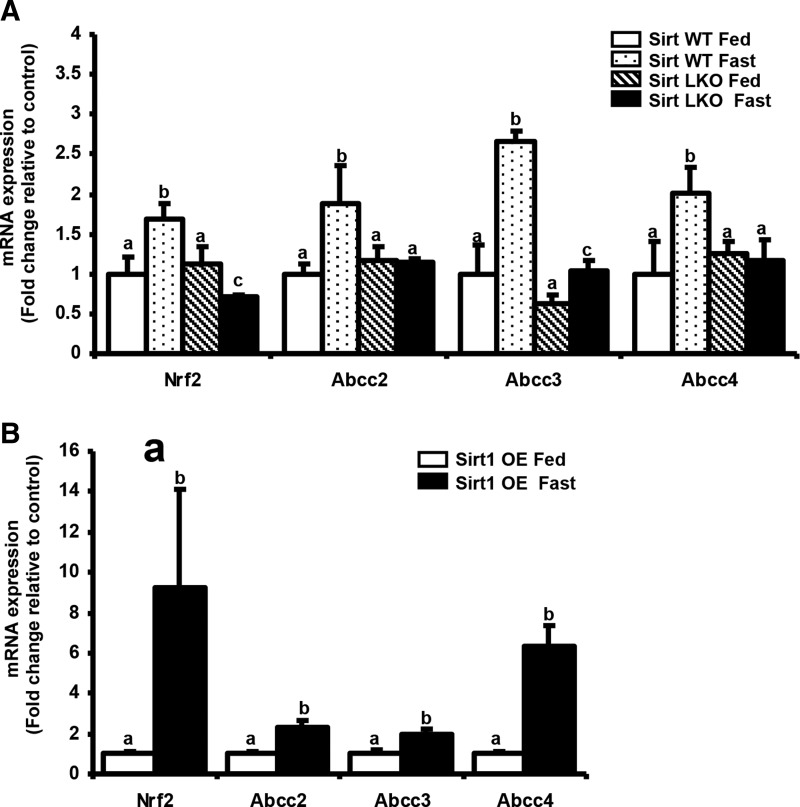
Fasting induced Abcc2–4 and NRF2-target gene expression is SIRT1-dependent in vivo
In addition to cAMP/PKA signaling pathway, previous studies have indicated that SIRT1 is a critical mediator of fasting-induced expression and activation of Pgc1-α (45), as well as other fasting physiologies, such as gluconeogenesis, fatty acid oxidation, and ketogenesis (7). To test whether SIRT1 plays a role in fasting induced activation of NRF2, liver-specific SIRT1-null mice were fasted and expression of Abcc2–4 determined. Absence of functional SIRT1 in liver minimally affected basal expression of genes measured (Fig. 5A). Upon fasting, Nrf2 and Abcc2–4 mRNA expression were induced in livers of control mice, but not in SIRT1LKO mice (Fig. 5A). Fasting increased Nqo1 expression to a similar degree in SIRT1-WT and SIRT1-LKO mice. Conversely, adenoviral over-expression of SIRT1 enhanced fasting induced Nrf2 and Abcc2–4 expression (Fig. 5B).
FIG. 5.
Changes in Nrf2-ARE pathway during fasting are Sirtuin-1 (SIRT1) dependent. (A) SIRT1LKO mice demonstrate resistance to effects of fasting-Total RNA was isolated from livers of Sirt WT (n=5) and SIRT1LKO (n=3) mice either fed ad libitum or fasted for 24 h. Gene expression of NRF2, Abcc 2–4 was quantified by real time quantitative PCR. The data are represented as average±SEM fold change over the fed controls. All data have been normalized to 18S rRNA. p<0.05 was considered statistically significant. Groups without a common letter are significantly different. (B) SIRT1OE mice demonstrate enhanced induction of Abcc2–4 and Nrf2 target genes upon fasting-Total RNA was isolated from livers of SIRT1OE (n=8) mice either fed ad libitum or fasted for 19 h. Gene expression of NRF2, Abcc 2–4 was quantified by real time quantitative PCR. The data are represented as average±SEM fold change over the fed controls. Groups without a common letter are significantly different. All data are normalized to 18S rRNA. p<0.05 was considered statistically significant. Groups without a common letter are significantly different.
NRF2 expression increases coordinately with increasing SIRT1 and Pgc-1α in vitro
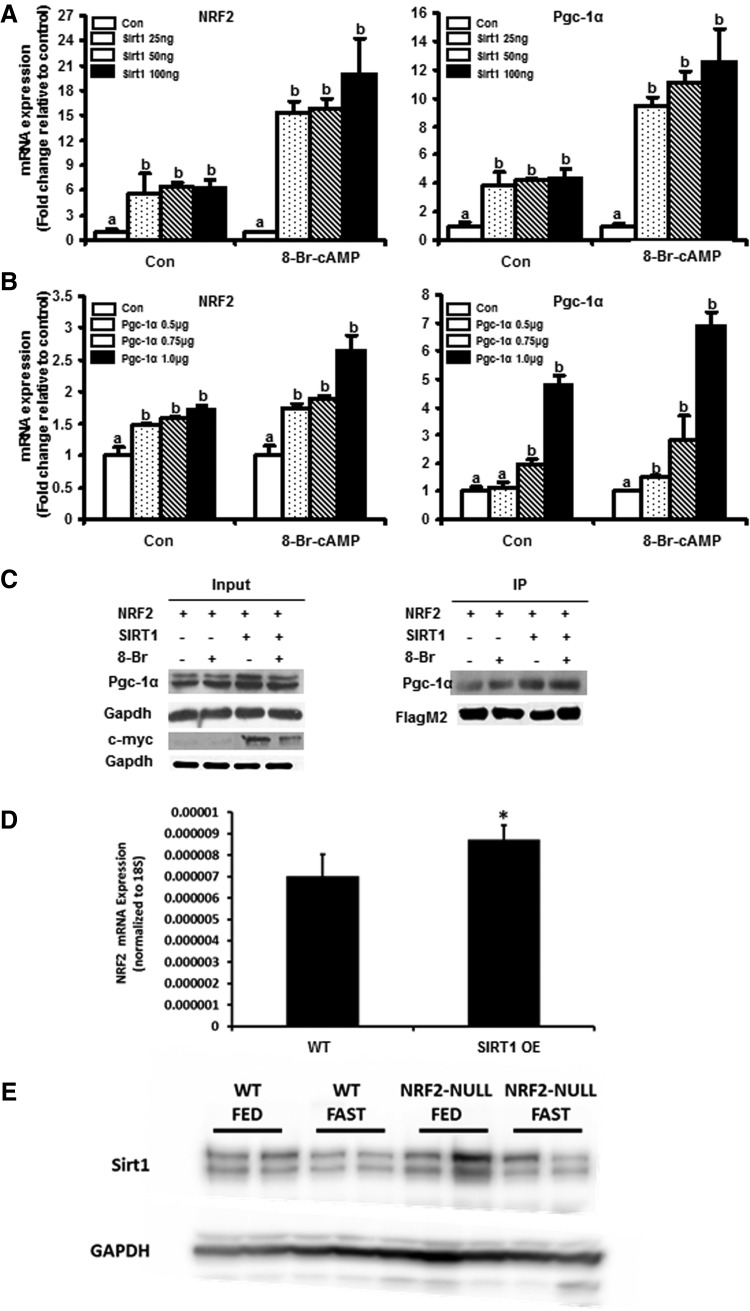
To identify whether the SIRT1-Pgc-1α cascade acts upstream of the NRF2-ARE pathway during fasting, NRF2 and Pgc-1α mRNA expression was quantified in Huh-7 cells transfected with either constant or varying amounts of NRF2, SIRT1 and Pgc-1α. As depicted in Figure 6C and D, NRF2 expression was coordinately regulated with Pgc-1α. NRF2 expression increased by 5–7-fold (Fig. 6C) and 1.5–2-fold (Fig. 6D) with increasing concentration of SIRT1 and Pgc-1α expression plasmid, respectively. 8-Br-cAMP increases this fold induction of NRF2 further in both cases (Fig. 6C, D). Thus, SIRT1-Pgc-1α cascade potentially acts upstream of NRF2-ARE pathway, increasing transcription of NRF2 and possibly its activity. Immunoprecipitation (IP) assays were performed using Huh-7 cells transfected with Flag-NRF2/cMyc-SIRT1 or Flag-NRF2 only and treated with 8-Br-cAMP to determine whether SIRT1 or Pgc-1α possibly could act as transcriptional activators. 8-Br-cAMP induced cMyc protein expression indicating and induction of SIRT1 protein (Supplementary Fig. S6), as well as FlagM2 protein in the immunoprecipitate indicating an increase in NRF2 expression (Fig. 6C). IP of Flag- tag by Anti-Flag M2 antibody resulted in co-IP of endogenous Pgc-1α along with Flag-NRF2 (Fig. 6C) indicating that Pgc-1α could be a coactivator of NRF2-ARE transcriptional pathway. It is also interesting to observe that this co-IP was less obvious in cells which were not transfected with SIRT1 expression plasmid (Fig. 6C). SIRT1 (c-Myc antibody) is also coimmunoprecipitated along with NRF2 (Fig. 6C). In addition, in livers from transgenic mice that moderately over express SIRT1 (19), had a slight elevation of NRF2 expression, further suggesting that increased SIRT1 expression can increase NRF2 expression (Fig. 6D). Lastly, fasting slightly decreased SIRT1 protein expression in WT and NRF2-null mice (Supplementary Fig. S2), suggesting that the observed induction of NRF2 expression may not be via increased SIRT1 protein expression. The above observations indicate that Pgc-1α acts as a positive regulator of NRF2 expression and NRF2 transcriptional pathway with activity of Pgc-1α partly dependent on presence of SIRT1.
FIG. 6.
NRF2 expression increases coordinately with PPAR gamma coactivator alpha (Pgc-1α) and SIRT1 expression. (A) NRF2 mRNA coordinately increases with increasing activation of SIRT1-Huh-7 cells were cultured to 85% confluence and transiently transfected (50–100 ng) Myc-SIRT1, 75 ng Flag-NRF2 and 0.5 μg Flag-Pgc-1α expression plasmids using Lipofectamine LTX-plus reagent (Invitrogen) for 24 h. The cells were then treated with 1.0 mM 8-Br-cAMP for 24 h. Endogenous NRF2 and Pgc-1α mRNA expression was quantified by real time quantitative PCR. The data are represented as average±SEM (n=3) over control All data have been normalized to 18S rRNA. (B) NRF2 mRNA expression coordinately increases with increasing Pgc-1α expression-Huh-7 cells were cultured to 85% confluence and transiently transfected with (0.5–1.0 μg) Flag-Pgc-1α, 50 ng Myc-SIRT1, 75 ng Flag-NRF2 and 0.5 μg Flag-Pgc-1α expression plasmids for 24 h. The cells were then treated with 1.0 mM 8-Br-cAMP for 24 h. NRF2 and Pgc-1α mRNA expression was quantified by real time quantitative PCR. The data are represented as average±SEM (n=3) over control All data have been normalized to 18S rRNA. (C) Pgc-1α coimmunoprecipitates with NRF2 in a SIRT1 dependent manner-Huh-7 cells were cultured to 85% confluence in 6 cm dishes and transiently transfected with 3.0 μg Flag-tagged NRF2, with or without 0.75 μg cMyc-tagged SIRT1 plasmid for 24 h. At the end of 24 h, cells were treated with 1.0 mM 8-Br-cAMP for 45 min, and then lysed using lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40). 1 mg of the protein lysate was incubated with Anti-FlagM2 magnetic beads overnight in lysis buffer and immunoprecipitates were separated on sodium dodecyl sulfate polyacrylamide gel electrophoresie (SDS-PAGE) and immunoblotted for Flag-M2 and endogenous Pgc-1α. Gapdh was used as protein loading control. (D) Moderate SIRT1 overexpression (OE) increased Nrf2 mRNA expression in mouse liver. Total RNA was isolated from SIRT1 WT (n=5) and SIRT1 OE (n=5) mouse livers and NRF2 mRNA expression quantified using real-time quantitative PCR. The data are presented as average±SEM (n=2–3) fold change in RNA expression over SIRT1 WT controls. (E) Fasting slightly decreases SIRT1 protein in mouse liver. Liver lysates in RIPA buffer were subjected to SDS-PAGE followed by western blot for SIRT1 and Gapdh.
NRF2 and SIRT1 are enriched at ARE sites in the 5′-flanking region of the ABCC2 gene
To identify whether activation of fasting pathways increased recruitment of NRF2 to the potential ABCC2 AREs, Chromatin IP (ChIP) assays were performed. These potential AREs in ABCC2 promoter region were identified using MatInspector algorithm. Although AREs have been described in human ABCC2 promoter by Stockel et al. (47), we analyzed the promoter region of ABCC2 again to identify any other potential ARE-like sites, due to differences in ABCC2 basal expression between HepG2 and Huh-7 cell lines.
Soluble chromatin fragments obtained from Huh-7 cells treated with 8-Br-cAMP were analyzed for increased NRF2 and SIRT1 enrichment at the MARE_ARE site identified by MatInspector. As seen in Figure 7A, 8-Br-cAMP increased NRF2 recruitment at the MARE_ARE site in ABCC2 promoter. A similar observation was not made with ARE-like element at ∼4467 bp (Supplementary Fig. S7) and hence, was not further analyzed. To identify whether 8-Br-cAMP could also cause increased NRF2 recruitment at the previously described ARE by (47), we used primers designed against the ARE like element (∼−335 from transcription start site) described to be important in regulation of basal ABCC2 expression along with a negative primer designed against a non ARE-containing site in the promoter. 8-Br-cAMP increased the recruitment of NRF2 to the ARE-like element described previously as well as MARE_ARE as identified by MatInspector algorithm, but does not show any changes when analyzed using negative primers (Fig. 7B). Similarly, IP of the soluble chromatin complexes with anti-SIRT1 antibody and control IgG antibody demonstrates that 8-Br-cAMP treatment causes an increase in recruitment of SIRT1 to the MARE_ARE as well as the previously described ABCC2-ARE (Fig. 7C) indicating that SIRT1 and NRF2 possibly coregulate ABCC2 expression. Whether SIRT1 binds directly to the DNA or regulates ABCC2 expression transactivation via NRF2 cannot be demonstrated.
FIG. 7.
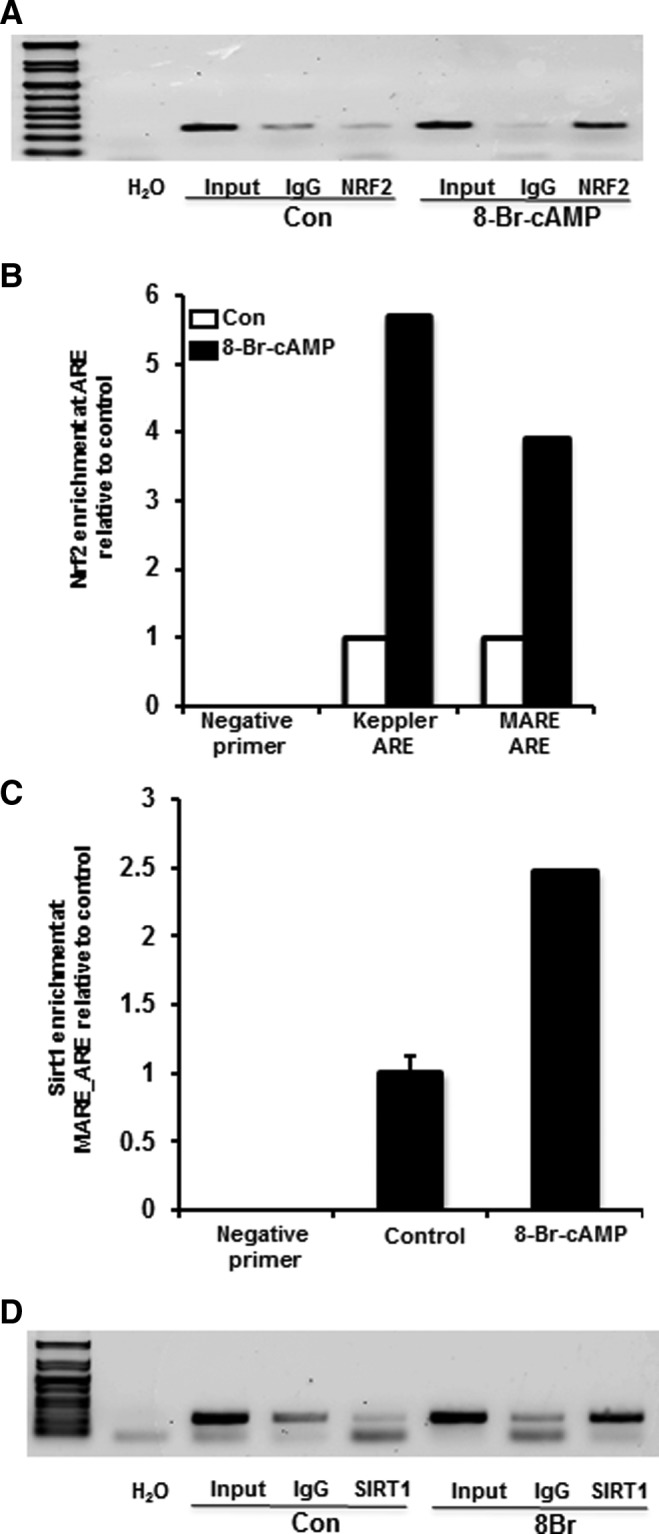
Activation of cAMP/PKA pathway increases NRF2 enrichment at ARE like elements in human ABCC2 promoter. (A) 8-Br-cAMP increases NRF2 enrichment at ARE-like sites in ABCC2 promoter. Huh-7 cells were treated with 8-Br-cAMP (0.1 mM) for 45 min and were washed and cross-linked for 10 min with 1% formaldehyde and crosslinking terminated by 125 mM glycine. The chromatin were sheared and prepared and incubated overnight with Protein G magnetic beads with Anti-NRF2 antibody or preimmune IgG. The antibody-bound chromatins and input DNA (1/10 dilution) were analyzed by PCR for the presence of DNA-fragments containing NRF2 bound-ARE-like element (MARE_ARE) with primers as indicated in Table 1. (B) 8-Br-cAMP increases NRF2 enrichment at other known ABCC2 AREs. Huh-7 cells were treated in a manner similar to (A) and the NRF2-antibody bound chromatin and input DNA were analyzed by RT2-PCR for the presence of DNA fragments containing MARE_ARE as well as ARE-like elements described by Stockel et al. (47) in ABCC2 promoter as well as negative primers directed at a non ARE-containing promoter region (Table 1). (C) 8-Br-cAMP increases SIRT1 enrichment at ABCC2 ARE-like elements in Huh-7 cells. Huh-7 cells were treated in a manner similar to (A), and the sheared chromatin fragments were incubated with anti-SIRT1 antibody and preimmune IgG overnight and the antibody-bound chromatins were analyzed by RT2-PCR for DNA fragments containing MARE_ARE (Table 1). (D) 8-Br-cAMP increases SIRT1 enrichment at the NQO1 ARE. Huh-7 cells treated in a manner similar to (A) and incubated with anti-SIRT1 antibody and preimmune IgG overnight and the antibody-bound chromatins were analyzed by PCR for DNA fragments containing ARE-like elements as described by Dhakshinamoorthy et al. (13) using primers indicated in Table 1.
8-Br-cAMP increases SIRT1 recruitment at NQO1 ARE
It is also important to determine whether SIRT1 is recruited (directly or indirectly) other known NRF2 targets as well. Hence, ChIP assays were performed on soluble chromatin obtained from Huh-7 cells treated with 8-Br-cAMP using anti-SIRT1antibody and analyzed for the presence of DNA containing NQO1 ARE described in (13) using primer sequences as mentioned by the authors. As seen in Figure 7D, not only does 8-Br-cAMP treatment increase SIRT1 recruitment at ABCC2-ARE, but also to a previously described ARE in 5′-flanking region of NQO1. This observation indicates that SIRT1 potentially regulates ABCC2 as well as NQO1.
Discussion
Studies herein demonstrate that fasting upregulates NRF2 activity and increases mouse and human ABCC2–4 expression in liver via cAMP/PKA-SIRT1 dependent mechanisms. Hepatic transport mechanisms are integral to liver function to maintain systemic hormone and second messenger levels. There are only a handful of published observations that document effects of fasting on hepatic clearance mechanisms.
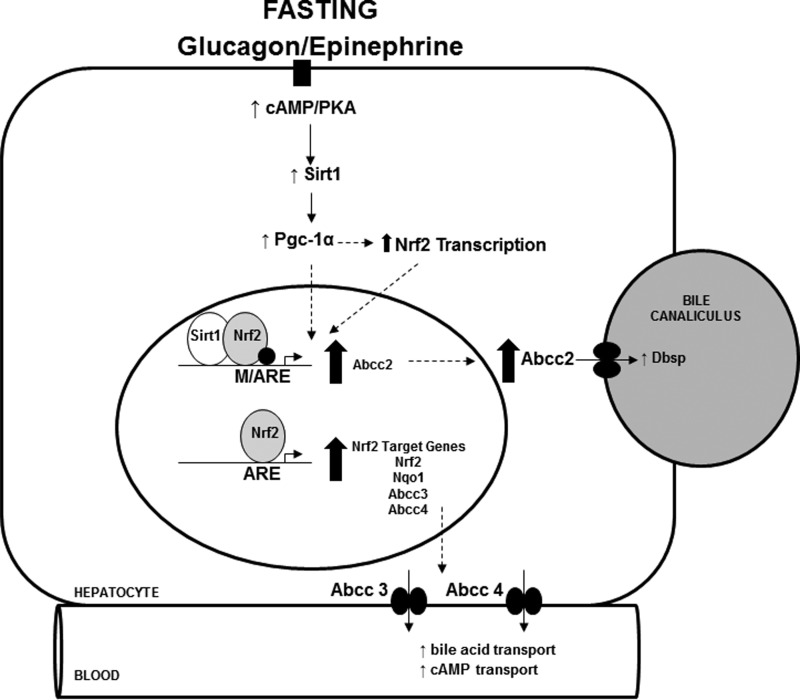
NRF2 is best described for regulating xenobiotic detoxification and clearance mechanisms, such as glutathione synthesis and conjugation enzymes, Phase-II biotransformation enzymes, and Abcc transporters (26). As an oxidative stress sensor; it is activated by reactive oxygen species, as well as antioxidants. A majority of studies demonstrate a classical function of NRF2 in mediating cytoprotective response in response to xenobiotic insult or disease, but few identify whether NRF2 participates in basic metabolic functions. Herein, we demonstrate that fasting response increased NRF2-ARE binding, upregulates NRF2-ARE activation, and induces subsequent target gene mRNA expression both mouse and human models. Moreover, the present data illustrate recruitment of NRF2 and SIRT1 to ARE elements in the human ABCC2 promoter. Figure 8 illustrates the hypothesized mechanism(s), by which activation of the fasting pathway may induce NRF2 expression, as well as ABCC2–4 induction.
FIG. 8.
Proposed model for fasting induced activation of NRF2-ARE pathway and expression of downstream target genes. Fasting increases circulating glucagon and epinephrine levels activating cAMP/PKA cascade, which eventually will increase Pgc-1α expression and also activate SIRT1. cAMP/PKA induces NRF2 binding to various AREs in a SIRT1-dependent manner leading to an increase in Nqo1, Abcc2–4 expression. Specifically, induction of Pgc-1α and SIRT1 can induce Nrf2 mRNA expression, which can lead to subsequent induction of Nrf2-regulated target genes, such as Nqo1 and Abcc2–4. In addition, activation of the PKA pathway increases NRF2 and SIRT1 recruitment to MARE_ARE elements in the ABCC2 promoter. It is hypothesized fasting will result in the production or accumulation of metabolic intermediates, which could be eliminated from hepatocytes via Abcc2–4. Fasting increases NAD+/NADH ratios, cAMP, bile acids, lipid peroxidation, glucuronide and glutathione (GSH) conjugation, and decreases GSH levels within the hepatocyte. GSH and glucuronide conjugates are substrates for Abcc2–4, some lipid peroxidation products are substrates for Abcc3. Abcc4 transports cyclic nucleotides, such as cAMP and cGMP. Overall, fasting could increase the hepatic efflux of metabolite conjugates and intermediates resulting from induction of the fasting response to maintain cellular homeostasis.
It is interesting to consider why NRF2 activation and hepatic clearance mechanisms are upregulated during food deprivation. Fasting causes a metabolism shift from lipid storage to lipid oxidation for energy generation, resulting in fat mobilization from adipose to liver, and consequently increasing lipid peroxidation products (1). Lipid breakdown in fasting preferentially increases arachidonic acid, stearic acid and docosahexanoic acid, which are probable substrates for Abcc3 (12, 29). A consequence of fasting is hepatic lipid accumulation and break down, as well as, increased bilirubin, bile acids, and cyclic nucleotides, which are all substrates for Abcc2–4. Fasting increases hepatic cAMP (30), cGMP (35), bilirubin (28), UDP-glucuronosyltransferase (UGT) expression (33), lipid peroxidation products, bile acids (43), bile flow (48), and forms glutathione conjugates, which depletes cellular glutathione (1). NRF2 and the ARE to enhance hepatic clearance mechanisms of endogenous metabolites through Abcc2–4 upregulation. For example, mouse and human ABCC2 are known to be high affinity transporters for bilirubin-glucuronide and contribute to bilirubin excretion to bile (21). Fasting increases bilirubin glucuronidation (51) and Abcc2 induction could be in response to increase bilirubin clearance. Second, as ABCC2–4 contributes to bile acid efflux from hepatocytes, induction might aid in decreasing the increased bile acid load that occurs with fasting. Third, cAMP and cGMP are substrates for Abcc4 (53), and our data herein illustrate that serum and liver cAMP levels corresponded with Abcc4 mRNA expression. Abcc4 induction during food deprivation may be a response to control intracellular cyclic nucleotide concentrations within liver to better regulate the fasting response. Fasting is known to increase NAD+/NADH and glutathione disulfide/glutathione (GSH) ratios within liver (17). Use of GSH in conjugating reactions potentially with lipid peroxide intermediates also increases GSH conjugates, which are also ABCC2–4 substrates. Moreover, upregulation of GSH synthesis restores redox balance and can enhance bile flow. Altered NAD+ and NADH levels in NRF2-null mice reflect the inability to cycle NADH to NAD+; required for glycolytic processes, which might be compensated by alternative pathways as indicated by a higher fold increased NAD+/NADH ratios upon fasting. This observation is consistent with a recent observation that PKA activates SIRT1 in NAD+ independent mechanisms (16). However, the role of NAD+ in activating SIRT1 cannot be overlooked. Lastly, fasting mobilizes fats from adipose tissue to liver resulting in fasting-induced steatosis. As NRF2 has been implicated in promoting steatosis, perhaps early NRF2 activation during fasting is one of the cellular responses to aid this process and promote energy conservation in the form of triglyceride (TG) storage (40).
Downstream regulation of Abcc2–4 by NRF2 is well described, yet upstream signaling pathways are less described. Insulin activates NRF2 via PI3K, mTOR and Akt pathways in diabetic models (38). However, pathways contributing to fasting- dependent NRF2 activation and subsequent target gene expression have not been described. To date, two in vitro studies illustrate that cAMP regulates Abcc3 and 4 expression (8, 46). Presently, cAMP/PKA pathway activators (e.g., epinephrine, glucagon, cAMP analogs, and food deprivation) increased ARE activity NRF2 target gene expression in primary human and mouse hepatocytes, as well as mouse liver. H89, a well described PKA inhibitor attenuated 8-Br-cAMP induction of ARE activation and NRF2 target gene upregulation, in vitro, further supporting a role for the cAMP/PKA pathway. Moreover, AMPK activators decreased NRF2 target gene expression and were not increased in fasting. Thus, the data support a role for cAMP/PKA cascade as a regulator of NRF2-ARE activation.
An additional aspect of the current study was to identify factors downstream of cAMP/PKA cascade, which mediate activation of NRF2- ARE pathway. The cAMP/PKA cascade activates cAMP response element-binding protein, which is also known to interact with NRF2, and synergistically increase NRF2 target gene transcription (49). Because Pgc-1α is a central transcriptional coactivator to multiple nuclear receptors (15), we hypothesized that fasting might also modulate NRF2 via SIRT1-Pgc-1α related mechanisms in vivo. Epigenetic mechanisms, such as acetylation/deacetylation by p300/cbp protein on lysine residues in the Neh1 domain of NRF2 as well as phosphorylation/dephosphorylation at serine 40 and tyrosine 568 modulate NRF2-ARE binding (23, 24, 49). SIRT1 can decrease NRF2 binding to ARE in vitro (24). However, to date, no studies have addressed how SIRT1 regulates NRF2 activity and subsequent gene expression in vivo. The data herein illustrate that fasting mediated NRF2 activation and Abcc upregulation is SIRT1 dependent. Moreover, we demonstrated that the PKA activator, 8-Br-cAMP is able to increase NRF2 recruitment to the ABCC2 promoter along with SIRT1.
SIRT1 and Pgc-1α act in concert to activate gluconeogenesis pathways in the hepatocytes (44). Mice expressing SIRT1 in liver induced NRF2 target gene mRNA expression upon fasting. However, fasting induction of NRF2 target gene expression (including Abcc2–4) was muted in livers of mice with hepatocyte-specific SIRT1 deletion. Moreover, fasting generally increased liver Abcc2–4 mRNA expression with SIRT1 overexpression. The above observations indicate toward a probable role of SIRT1 as an upstream regulator of NRF2-ARE pathway.
In summary, fasting and PKA activators increased mouse Abcc2–4 expression in liver by NRF2- and SIRT1-related mechanisms in mouse liver and hepatocytes. Additionally, 8-Br-cAMP and PKA activating compounds increased NRF2 target gene and ABCC2–4 mRNA expression in human hepatocytes. Our data illustrate an undescribed role for NRF2 as a “fasting” responsive transcription factor that is activated via cAMP/PKA-SIRT1 upstream mechanisms.
Materials and Methods
Animals and treatments
Adult male C57BL/6 (n=8/group), Nrf2-null (n=6/group) (20) and ARE-hPAP reporter mice (n=3/group) (22) were fed Harlan Teklad LM-485 Mouse/Rat Sterilizable Diet (Harlan Laboratories) ad libitum or were food withheld for 24 h. Adult male SIRT1-LKO (42) and Sirt-WT mice (n=3) were fed standard chow ad libitum or were food deprived (fasted) for 18 h. Blood and livers were collected between 9–11 am for all individual fasting experiments. Livers from SIRT1 overexpressing fed or fasted mice were obtained from J. Rodgers and P. Puigserver from a previously described study (45) and M. Serrano (19). All studies were carried out at the University of Rhode Island with institutional animal care and use committee approval. Serum glucose (Cayman Chemicals), triglycerides (Pointe Scientific) and free fatty acid (non-esterified fatty acids) concentrations (Wako Diagnostics), hepatic and serum cAMP (Cell Signaling Technologies), serum bile acids (Bioquant Kits) were determined according to manufacturer's instructions.
Determination of biliary excretion of DBSP
Adult, male, C57BL/6 (n=5 per group) were fed standard chow ad libitum or were food withheld for 18 h. DBSP, purchased from SERB Laboratories was injected intraperitoneal (150 μmol/kg/5 ml) and 45 min after injection gallbladders were collected. Bile DBSP content (μmol/μl bile/kg body weight) was determined spectrophotometrically at 580 nm after alkalinization with 0.1 N NaOH.
Cell lines and treatments
Huh-7 cells were treated with 0.5–2.0 mM 8-Bromoadenosine-cAMP (8-Br-cAMP) (Sigma Aldrich) or media alone for 24 h. Huh-7 cells were cultured to 85% confluence and transiently transfected with an ARE–luciferase (0.1 μg) (14) and pRL-CMV (0.01 μg) reporter constructs using lipofectamine-plus reagent (Invitrogen). Twenty-four hours after transfection, cells were treated with 0.1 mM or 1.0 mM 8-Br-cAMP for 24 h (15). Luciferase activity was measured using a Dual Luciferase Assay (Promega Corp.), and Firefly Luciferase Relative Light Units (RLUs) were normalized to Renilla Luciferase RLUs.
Plasmid transfections and endogenous gene expression analysis
All plasmids used were obtained from Addgene Plasmid Repository. Huh-7 cells were transiently transfected with (0.5–1.0 μg) Flag-Pgc-1α, 75 ng Flag-NRF2 and 50 ng Myc-SIRT1 expression plasmids or (50–100 ng) Myc-SIRT1, 75 ng Flag-NRF2 and 0.5 μg Flag-Pgc-1α expression plasmids using Lipofectamine LTX-plus reagent (Invitrogen) for 24 h. Cells were then treated with 1.0 mM 8-Br-cAMP for 24 h. NRF2 and Pgc-1α mRNA expression was measured using real time quantitative PCR, with expression being normalized to 18S rRNA expression.
IP assay
Huh-7 cells were cultured in 6 cm dishes and transiently transfected with 3.0 μg Flag-tagged NRF2, with or without 0.75 μg cMyc-tagged SIRT1 plasmid for 24 h using Lipofectamine LTX-plus reagent (Invitrogen). After 24 h, cells were treated with 1.0 mM 8-Br-cAMP for 45 min and lysed with buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40). 1 mg protein lysate was incubated with Anti-FlagM2 magnetic beads (Sigma Aldrich) overnight in lysis buffer. After incubation, beads were washed with Tris-Buffered Saline, pH 7.4, mixture boiled with sodium dodecyl sulfate (SDS) samples buffer, and immunoprecipitates were separated on a 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting against Pgc-1α (Abcam), FlagM2 and c-myc (Sigma Aldrich).
Primary hepatocyte isolation, culture, and treatment
Primary mouse hepatocytes were obtained from adult C57BL/6, ARE-hPAP and SirtLKO mice as described previously (55). Primary human hepatocytes (Cellzdirect™) were maintained in William's E Media supplemented with Cellzdirect recommended supplements. Hepatocytes were treated with DMSO vehicle or 1.0 mM 8-Br-cAMP dissolved in DMSO for 24 h. Human hepatocyte experiments were approved by the URI Institutional Review Board as an exemption.
mRNA quantification
Total RNA was isolated from tissue and cells using TRIzol reagent (Invitrogen). mRNA expression was quantified by Branched DNA signal amplification assay or real time quantitative PCR, with expression being normalized to 18S rRNA expression. At the end of treatment, primary human hepatocytes were lysed with Lysis Mixture™ (Panomics, Inc.) according to manufacturer's protocol and stored at −80°C until analysis. mRNA expression was measured using Quantigene® Discover Kit (Panomics, Inc.). Target gene expression was normalized to β-actin expression.
Western blot and immunohistochemistry
Previously described protocols were used (37). SIRT1 antibody (cat.WH0023411M1) was purchased from Sigma Aldrich. Proteins were electrophoretically separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Membranes were blocked with 5% Nonfat dry milk in phosphate-buffered saline (PBS) with 0.05% Tween-20 overnight, and then incubated with appropriate concentrations of primary and secondary antibodies. The blots were incubated in ECL-Plus western blot detection reagent. Bands were visualized using autoradiography film and quantified using Image Quant Software (GE Healthcare and Life Sciences). Details are described in (37) and Abcc2 staining immunostaining of liver sections was performed as described (4). In mouse liver lysates, Abcc2, 3, and 4 migrated to 180–190, 165, and 150–160 kDa, respectively. Nrf2 migrated to 110 kDa.
NRF2 binding assay
Nuclear extracts were isolated from livers using a TF Procarta nuclear extraction kit (Panomics, Inc.) and protein concentrations were measured by a BCA protein assay (Pierce). NRF2 binding to a prototypical ARE consensus sequence was quantified using a Procarta TF custom array (Affymetrix) according to manufacturer's instructions. Data were acquired by a Luminex Bio-plex™ 200 Array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex Data Manager Software Version 5.0 (Bio-Rad Laboratories). All data were normalized to Transcription Factor II D binding activity.
ABCC2 promoter analysis for ARE-like elements
To identify potential ARE-like elements in the 5′-flanking region of ABCC2, we used the Genomatix MatInspector Suite, (Genomatix Software GmbH) and identified potential MARE_AREs within −6000 to +100 bp from the transcriptional start site as detected by Transcriptional Regulatory Element Database (TRED) (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home). The generic transcription factor binding algorithm was used, which identified one MARE_ARE like element at ∼3272 bp and two NFE2-p45 like elements, one overlapping the MARE_ARE ∼3272 and another site at ∼4467 bp. To provide a positive control for the ChIP experiments, primers corresponding to previously identified ARE-like elements in human ABCC2 (47) were also included.
Chromatin IP
Huh-7 cells were treated with 8-Br-cAMP (0.1 mM) or control (media) for 45 min. The chromatin preparation and subsequent IP of the cells was performed using Active Motif ChIP-IT express kit (Active Motif) and optimized according to manufacturer's protocol. Briefly, the cells were washed, cross linked using 1% formaldehyde for 10 min and crosslinking was stopped by 125 mM glycine solution. The cells were scraped using 1×PBS solution containing protease inhibitors and pelleted by centrifugation. The pellet was then lysed using SDS-Lysis Buffer (provided) supplemented with protease inhibitors and sonicated using Branson Sonifier 250 (Branson Ultrasonics Corporation) thrice for 10 s with a rest period of 20–30 s at power level 2 on the machine. The shearing conditions were optimized according to manufacturer's guide to obtain chromatin fragments between 200–1200 bp as determined by agarose gel electrophoresis. Equal amounts of sheared chromatin were incubated overnight with Protein-G magnetic beads and ChIP validated rabbit anti-NRF2 antibody (sc-722×; Santa Cruz Biotechnologies) or rabbit Anti-SIRT1 antibody (07-131 Anti Sir2; EMD Millipore) along with preimmune Anti-Rabbit IgG antibody (Cell Signaling). Antibody bound chromatin was then eluted and the DNA reverse-cross linked and DNA purified by Proteinase K treatment. The DNA fragments were analyzed by PCR or RT2-PCR for the presence of NRF2-ARE like elements using primers as indicated in Table 1. The primers sequences for NQO1-ARE were obtained from (13).
Table 1.
Primers Used to Amplify ARE/ARE-Like Sequences in the Human ABCC2 Promoter
| Forward | Reverse | |
|---|---|---|
| PCR | ||
| MARE_ARE |
TTTAATCCGTGTGAGGAAGGAG |
CTGTTCTTGTTCACCCTCTAAGT |
| RT2-PCR | ||
| MARE_ARE |
TGAGAACCTCTTTGAATTCCTATGG |
GGCCCAGTGTAGATAAGTTTGAG |
| ARE | CTGCCCTTTGTGGGTCATA | TCTGTAGGAGTGGCCATACA |
NAD+/NADH assay
Tissue NAD+/NADH was measured using NAD/NADH assay kit (BioVision, Inc.) according to manufacturer's protocol.
Statistics
Statistical significance of differences was determined by Duncan's factorial ANOVA test. p<0.05 was considered statistically significant. Groups without a common letter are significantly different.
Supplementary Material
Abbreviations Used
- AMPK
AMP-activated protein kinase
- ARE
antioxidant response element
- 8-Br-cAMP
8-bromoadenosine-3′,5′-cyclic adenosine monophosphate
- cAMP
3′-5′-cyclic adenosine monophosphate
- ChIP
chromatin immunoprecipitation
- DBSP
disulfobromophthalein
- GSH
glutathione
- IP
immunoprecipitation
- Keap1
Kelch-like ECH-associated protein-1
- NRF2
nuclear factor-E2 related-factor 2
- Nqo1
Nadph:quinone oxidoreductase
- Pgc-1α
PPAR gamma coactivator-1 alpha
- SDS
sodium dodecyl sulfate
- SIRT1
sirtuin-1
- PAGE
polyacrylamide gel electrophoresis
- PKA
protein kinase A
- RLUs
relative light units
- TG
triglyceride
- WT
wild-type
Acknowledgments
The authors thank Drs. Joseph T. Rodgers and Pere Puigserver (Dana-Farber Cancer Institute, Harvard Medical School) for sharing liver tissue, Dr. Curtis D. Klaassen (University of Kansas Medical School) and Dr. Jeff Chan (UC-Irvine) for sharing NRF2-null mice. We thank Dr. Michael Goedken for histopathology consultation, as well as, Laura Armstrong, Vijay More, Prajakta Shimpi, and Maneesha Paranjpe for technical assistance. This work was supported by National Institutes of Health [5R01ES016042], and, in part, by Rhode Island IDeA Network of Biomedical Research Excellence [Award # P20RR016457-10] from the National Center for Research Resources, National Institutes of Health, National Institutes of Health [5K22ES013782], and the Rhode Island Foundation.
Author Disclosure Statement
No competing financial interests exists.
References
- 1.Abdelmegeed MA, Moon KH, Hardwick JP, Gonzalez FJ, and Song BJ. Role of peroxisome proliferator-activated receptor-alpha in fasting-mediated oxidative stress. Free Radic Biol Med 47: 767–778, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleksunes LM. and Klaassen CD. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab Dispos 40: 1366–1379, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aleksunes LM, Reisman SA, Yeager RL, Goedken MJ, and Klaassen CD. Nuclear factor erythroid 2-related factor 2 deletion impairs glucose tolerance and exacerbates hyperglycemia in type 1 diabetic mice. J Pharmacol Exp Ther 333: 140–151, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aleksunes LM, Scheffer GL, Jakowski AB, Pruimboom-Brees IM, and Manautou JE. Coordinated expression of multidrug resistance-associated proteins (Mrps) in mouse liver during toxicant-induced injury. Toxicol Sci 89: 370–379, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Altarejos JY. and Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12: 141–151, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buler M, Aatsinki SM, Skoumal R, and Hakkola J. Energy sensing factors PGC-1alpha and SIRT1 modulate PXR expression and function. Biochem Pharmacol 82: 2008–2015, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Canto C. and Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol 20: 98–105, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra P, Zhang P, and Brouwer KL. Short-term regulation of multidrug resistance-associated protein 3 in rat and human hepatocytes. Am J Physiol Gastrointest Liver Physiol 288: G1252–G1258, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Cheng X, Dieter MZ, Tanaka Y, and Klaassen CD. Activation of cAMP-dependent signaling pathway induces mouse organic anion transporting polypeptide 2 expression. Mol Pharmacol 71: 1159–1164, 2007 [DOI] [PubMed] [Google Scholar]
- 10.This reference has been deleted.
- 11.Chen ZS, Lee K, and Kruh GD. Transport of cyclic nucleotides and estradiol 17-beta-D-glucuronide by multidrug resistance protein 4. Resistance to 6-mercaptopurine and 6-thioguanine. J Biol Chem 276: 33747–33754, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Chen ZY. and Cunnane SC. Preferential retention of linoleic acid-enriched triacylglycerols in liver and serum during fasting. Am J Physiol 263: R233–R239, 1992 [DOI] [PubMed] [Google Scholar]
- 13.Dhakshinamoorthy S, Jain AK, Bloom DA, and Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem 280: 16891–16900, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Dieter MZ, Freshwater SL, Solis WA, Nebert DW, and Dalton TP. Tyrphostin [correction of Tryphostin] AG879, a tyrosine kinase inhibitor: prevention of transcriptional activation of the electrophile and the aromatic hydrocarbon response elements. Biochem Pharmacol 61: 215–225, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Ding X, Lichti K, Kim I, Gonzalez FJ, and Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem 281: 26540–26551, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerhart-Hines Z, Dominy JE, Jr., Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, and Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 44: 851–863, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashida S, Arimoto A, Kuramoto Y, Kozako T, Honda S, Shimeno H, and Soeda S. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol Cell Biochem 339: 285–292, 2010 [DOI] [PubMed] [Google Scholar]
- 18.This reference has been deleted.
- 19.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, and Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 1: 3, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Jedlitschky G, Hoffmann U, and Kroemer HK. Structure and function of the MRP2 (ABCC2) protein and its role in drug disposition. Expert Opin Drug Metab Toxicol 2: 351–366, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Johnson DA, Andrews GK, Xu W, and Johnson JA. Activation of the antioxidant response element in primary cortical neuronal cultures derived from transgenic reporter mice. J Neurochem 81: 1233–1241, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Kaspar JW, Niture SK, and Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med 47: 1304–1309, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai Y, Garduno L, Theodore M, Yang J, and Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem 286: 7629–7640, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaassen CD. and Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62: 1–96, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klaassen CD. and Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol 244: 57–65, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi M. and Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal 7: 385–394, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Koh YW, Choi EC, Kang SU, Hwang HS, Lee MH, Pyun J, Park R, Lee Y, and Kim CH. Green tea (-)-epigallocatechin-3-gallate inhibits HGF-induced progression in oral cavity cancer through suppression of HGF/c-Met. J Nutr Biochem 22: 1074–1083, 2011 [DOI] [PubMed] [Google Scholar]
- 29.Kubo K, Sekine S, and Saito M. Induction of multidrug resistance-associated protein MRP3 in the liver of rats fed with docosahexaenoic acid. Biosci Biotechnol Biochem 70: 1672–1680, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Lavine RL, Voyles N, Perrino PV, and Recant L. The effect of fasting on tissue cyclic cAMP and plasma glucagon in the obese hyperglycemic mouse. Endocrinology 97: 615–620, 1975 [DOI] [PubMed] [Google Scholar]
- 31.Li C, Krishnamurthy PC, Penmatsa H, Marrs KL, Wang XQ, Zaccolo M, Jalink K, Li M, Nelson DJ, Schuetz JD, and Naren AP. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell 131: 940–951, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, and Chiang JY. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem 287: 1861–1873, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, Gao B, Wierzbicki M, Verbeuren TJ, Shaw RJ, Cohen RA, and Zang M. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, and Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456: 269–273, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz SZ, Hennige AM, Feil S, Peter A, Gerling A, Machann J, Krober SM, Rath M, Schurmann A, Weigert C, Haring HU, and Feil R. Genetic ablation of cGMP-dependent protein kinase type I causes liver inflammation and fasting hyperglycemia. Diabetes 60: 1566–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maher JM, Cheng X, Slitt AL, Dieter MZ, and Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos 33: 956–962, 2005 [DOI] [PubMed] [Google Scholar]
- 37.More VR. and Slitt AL. Alteration of hepatic but not renal transporter expression in diet-induced obese mice. Drug Metab Dispos 39: 992–999, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okouchi M, Okayama N, Alexander JS, and Aw TY. NRF2-dependent glutamate-L-cysteine ligase catalytic subunit expression mediates insulin protection against hyperglycemia- induced brain endothelial cell apoptosis. Curr Neurovasc Res 3: 249–261, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Osburn WO. and Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res 659: 31–39, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, Yehuda-Shnaidman E, Lee C, Lau J, Kurtz TW, and Chan JY. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem 285: 9292–9300, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.This reference has been deleted.
- 42.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, and Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9: 327–338, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson DG, Ruepp SU, Stryker SA, Hnatyshyn SY, Shipkova PA, Aranibar N, McNaney CA, Fiehn O, and Reily MD. Metabolomic and transcriptomic changes induced by overnight (16 h) fasting in male and female Sprague-Dawley rats. Chem Res Toxicol 24: 481–487, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Rodgers JT, Lerin C, Gerhart-Hines Z, and Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett 582: 46–53, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodgers JT. and Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A 104: 12861–12866, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sassi Y, Lipskaia L, Vandecasteele G, Nikolaev VO, Hatem SN, Cohen Aubart F, Russel FG, Mougenot N, Vrignaud C, Lechat P, Lompre AM, and Hulot JS. Multidrug resistance-associated protein 4 regulates cAMP-dependent signaling pathways and controls human and rat SMC proliferation. J Clin Invest 118: 2747–2757, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stockel B, Konig J, Nies AT, Cui Y, Brom M, and Keppler D. Characterization of the 5'-flanking region of the human multidrug resistance protein 2 (MRP2) gene and its regulation in comparison with the multidrug resistance protein 3 (MRP3) gene. Eur J Biochem 267: 1347–1358, 2000 [DOI] [PubMed] [Google Scholar]
- 48.Strasberg SM, Siminovitch KA, and Ilson RG. Bile production in fasted and fed primates. Ann Surg 180: 356–363, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Z, Chin YE, and Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol 29: 2658–2672, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taguchi K, Motohashi H, and Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140, 2011 [DOI] [PubMed] [Google Scholar]
- 51.Visser TJ, van Haasteren GA, Linkels E, Kaptein E, van Toor H, and de Greef WJ. Gender-specific changes in thyroid hormone-glucuronidating enzymes in rat liver during short-term fasting and long-term food restriction. Eur J Endocrinol 135: 489–497, 1996 [DOI] [PubMed] [Google Scholar]
- 52.Weerachayaphorn J, Mennone A, Soroka CJ, Harry K, Hagey LR, Kensler TW, and Boyer JL. Nuclear factor-E2-related factor 2 is a major determinant of bile acid homeostasis in the liver and intestine. Am J Physiol Gastrointest Liver Physiol 302: G925–G936, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wielinga PR, van der Heijden I, Reid G, Beijnen JH, Wijnholds J, and Borst P. Characterization of the MRP4- and MRP5-mediated transport of cyclic nucleotides from intact cells. J Biol Chem 278: 17664–17671, 2003 [DOI] [PubMed] [Google Scholar]
- 54.Williamson TP, Johnson DA, and Johnson JA. Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology 33: 272–279, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J, Kulkarni SR, Li L, and Slitt AL. UDP-glucuronosyltransferase expression in mouse liver is increased in obesity- and fasting-induced steatosis. Drug Metab Dispos 40: 259–266, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.This reference has been deleted.
- 57.Zhang Y, Csanaky IL, Cheng X, Lehman-McKeeman LD, and Klaassen CD. Organic anion transporting polypeptide 1a1 null mice are sensitive to cholestatic liver injury. Toxicol Sci 127: 451–462, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.