Abstract
The gene transcript profile responses to metal oxide nanoparticles was studied using human cell lines derived from the colon and skin tumors. Much of the research on nanoparticle toxicology has focused on models of inhalation and intact skin exposure, and effects of ingestion exposure and application to diseased skin are relatively unknown. Powders of nominally nanosized SiO2, TiO2, ZnO and Fe2O3 were chosen because these substances are widely used in consumer products. The four oxides were evaluated using colon-derived cell lines, RKO and CaCo-2, and ZnO and TiO2 were evaluated further using skin-derived cell lines HaCaT and SK Mel-28. ZnO induced the most notable gene transcription changes, even though this material was applied at the lowest concentration. Nano-sized and conventional ZnO induced similar responses suggesting common mechanisms of action. The results showed neither a non-specific response pattern common to all substances nor synergy of the particles with TNF-α cotreatment. The response to ZnO was not consistent with a pronounced proinflammatory signature, but involved changes in metal metabolism, chaperonin proteins, and protein folding genes. This response was observed in all cell lines when ZnO was in contact with the human cells. When the cells were exposed to soluble Zn, the genes involved in metal metabolism were induced but the genes involved in protein refoldling were unaffected. This provides some of the first data on the effects of commercial metal oxide nanoparticles on human colon-derived and skin-derived cells.
Introduction
The ongoing search for unique physical and chemical properties has motivated the development of numerous types of metal oxide nanomaterials, but concerns remain regarding biological effects of particles comparable in size to ultrafine air pollution (d <100 nm). Much of the nanoparticle toxicological research has been on inhalation and intact-skin dermal exposures; however, there is also exposure and potential toxicity in the digestive tract and through damaged skin and other tissue epithelial barriers. Ingestion exposure can come from nano-sized materials used as food and drug ingredients, from hand-to-mouth contact, and through drinking water contamination. Dermal exposure can occur from nano-sized materials in sunscreens and cosmetics.
Numerous hypotheses have been proposed for potential toxicological responses induced by nanoparticle exposure including both chemical-specific effects of particle core composition or surface chemistry, and non-specific effects associated with the small size and high specific surface area of nano-sized particles.1 Mechanistic hypotheses include particle-induced pro-inflammatory signaling responses, oxidative signaling responses, non-specific effects that are dependent primarily on available surface area and less on the chemical composition of the core substance, and size-dependent uptake of insoluble particles.2
There have been a number of geneome-wide transcriptional studies of metal oxide nanoparticles. Nano-TiO2 activates inflammatory response and cell adhesion, but not oxidative stress, genes in human keratinocyte cells.3 In mouse lungs nano-TiO2 exacerbates LPS-induced inflammation, but does not alter the gene expression pattern.4 A comparison of 10- and 500-nm SiO2 particles using RAW264.7 mouse macrophages showed high correlation in gene expression changes between the sizes, suggesting common genomic responses to nanosilica.5 Ultrafine NiO, but not C60 fullerene particles, activated genes for chemokines, oxidative stress, and matrix metalloproteinase 12 in rat lungs.6 An inhalation comparison of multiwalled carbon nanotubes and α-quartz in rats revealed many interesting patterns such as expression of genes likely playing a role in fibrosis that were qualitatively similar for the two materials.7 The promotion of oxidative stress and inflammation are not restricted to engineered materials but have been associated with environmental nanoparticles. For example, environmental particulate like soot and diesel exhaust, are complex mixtures of organic and inorganic compounds, induce oxidative stress and inflammation pathways leading to disease.8
There have been a number of studies reporting preferential uptake of nanoparticles compared to larger particles of low-solubility substances. An ingestion study performed in rats showed that polystyrene particles smaller than 100 nm were taken up by the rat intestinal mucosa and enter systemic circulation.9 Indeed, inflamed colon cells internalize nanoparticles at a much greater rate than normal colon cells, and this has been exploited by studies using nanoparticles for drug delivery.10 Human epithelial cells take up and translocate nano-sized silica particles to the nucleus where the particles induced formation of aberrant clusters that inhibited replication, transcription, and cell proliferation.11
The substances SiO2, Fe2O3, and TiO2 are all used in skin care and cosmetic formulations and are approved as food additives where they can be present at levels up to 1–2%, and current regulations do not restrict allowable particle size. ZnO is used in sunscreens, cosmetics, dietary supplements, and paints. Many sunscreens now contain nano-sized TiO2 and ZnO, and there has been citizen concern about whether these nanomaterials pose different risks that the conventional forms of these oxides.12 In addition to consumer uses, all these materials are widely used in industrial processes where occupational exposure and wastewater releases can occur. The U.S. Environmental Protection Agency has announced regulatory policies regarding certain nanomaterials, such as carbon nanotubes, as novel chemical entities, and is considering policies regarding nanoscale silver, TiO2, and ZnO; however, most nanomaterials are still regulated only by their nominal chemical composition.
Given the limited data on the effects of nanoparticle ingestion we designed a screening study to evaluate gene regulation changes in colon cells treated with a suite of nanoparticles. The limitations of submerged cell culture using immortalized cell lines are acknowledged, but similar in vitro models are widely accepted for both preliminary screening and mechanistic studies. Based on the hypothesis that inflammation may increase the uptake and/or toxicity of ultrafine particles, 13 we also evaluated the transcriptional responses to nanoparticulate with TNF-α pretreatment to mimic a pre-existing inflammatory state. Zinc oxide was found to be the most potent of tested nano-sized powders, so we extended the study to directly compare gene transcription responses in colon cells exposed to ZnO sold as nano-sized and conventional powders. Next, we expanded these studies to evaluate ZnO and TiO2–dependent alterations in gene expression on skin-derived cells and found that similar genes are altered in these cells and in the colon-derived cells. In our previous studies, we determined that commercial ZnO particulate required contact to promote cytoxicity; therefore, we also evaluated gene expression changes dependent on soluble Zn. We determined that particulate ZnO promotes the induction of genes related to protein folding; these genes are not altered when exposed to soluble Zn, and therefore may represent early markers of ZnO-mediated toxicity.
Methods
Materials
TNF-α, Advanced DMEM, Glutamax, and Superscript III reagents were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum was purchased from Hyclone (Logan, UT). The RKO and SK Mel-28 cell lines were purchased from American Tissue Type Culture Collection (Manassas, VA). CaCo-2 cells were a gift from Dr. John G. Lamb (University of Utah). HaCaT cells were a gift of Dr. Doug Grossman (University of Utah). Antibodies directed against the gene products GADD45B (sc-8775), GAPDH (sc-25778), DNAJA4 (sc-100714), HMOX1 (sc1797), and secondary anti-goat, -mouse, and -rabbit antibodies were purchased from Santa Cruz Biotechnology; HSPA1A (C29F3A-5) was purchased from Stressgen; ZFP36 (ab36558) was purchased from Abcam. RNeasy Minikits were purchased from Qiagen (Valencia, CA). Cell Counting Kit-8 (CCK-8) was from Dojindo (Gaithersburg, MD). Lightcycler 480 SYBR Green I master mix was purchased from Roche Diagnostics (Indianapolis, IN). Reagent samples of anatase TiO2, and amorphous SiO2 were obtained from Nanostructured and Amorphous Materials, Inc. (Houston, TX), hematite Fe2O3, and nominally nanosized and conventional ZnO were obtained from Alpha Aesar (Ward Hill, MA).
Characterization of the particulate matter
The particle surface area per mass was measured by nitrogen adsorption (BET single point method) using a Quantachrome Monosorb analyzer. The surface-equivalent or surface mean diameter was calculated from the measured area assuming spherical particles and the appropriate mineral density. The geometric size of the particulate matter was verified by transmission electron microscopy. Briefly, samples of the powders were suspended at approximately 0.1 mg/ml in water containing 1% of FL-70 surfactant (Fisher Scientific), vortexed, and then dispersed using an ultrasonic tissue disruptor for 1 minute. A 1 μl drop of the suspension was placed on a carbon grid, air dried, and examined on a Tecnai G2 transmission electron microscope at 120 kV.
Dynamic light scattering (DLS) was used to measure the size distribution of the agglomerates formed when the particles were added to either water or cell culture media. Suspensions were prepared at 0.1 mg/ml in water (for zeta potential and DLS measurement), or at 300 μg/ml of particles in Advanced DMEM with Glutamax and 2% serum. The media-based DLS particle samples were prepared in the same manner as used for the cell treatments, below, and the tested suspension concentration corresponds to the 100 μg/cm2 cell exposures.
Soluble elements were measured by suspending 1 mg/ml of particles in Advanced DMEM culture media and incubating overnight at 37°C without cells. Composition was determined by ion coupled plasma spectrometry on a Perkin Elmer Optima 3100 XL.
Cell culture
All cells were maintained in Advanced DMEM supplemented with 1% Glutamax and 2% fetal bovine serum so that alterations in gene expression among the cell lines would not be due to distinct media. For experiments that used particulate matter, the nanomaterial was sterilized using 70% EtOH and vacuum drying, and then added to medium, vortexed, and sonicated for 15 min in a Cole-Parmer 8890 water bath sonicator to disperse the particulate matter. For experiments with TNF-α stimulation, cells were pretreated by incubation with 100 ng/ml TNF-α to create a drug-induced inflammatory state or as a control, respectively, for 1 hr, then the medium was exchanged with medium containing the particulate matter. For electron microscopy of the RKO cells with the ZnO and TiO2 nanomaterials, cells were seeded on a plastic surface, treated with the nanomaterials, and then fixed, stained and sectioned for transmission electron microscopy using standard methods used at the University of Utah electron microscopy core.
Viability assay
Cellular viability was determined using a Cell Counting Kit-8 (CCK-8) which relies on tetrazolium salt reduction by NADH in viable cells.14 Briefly, cells were seeded into 48 well plates at 4 × 104 cells/well and allowed to recover overnight. Nanoparticles were evaluated at concentrations ranging from 1–100 μg/cm2. Cells were treated as described above (Cell culture) and assessed for viability after 24 hrs. Following the treatment period, medium was aspirated and replaced with 4% CCK-8 in cell culture medium. Absorbance at 460 nm and 650 nm was measured after incubation at 37°C until the reagent developed sufficiently for maximal reading using a Perkin-Elmer VictorV3 Multimode Microplate Reader.
Microarray expression analysis
Cells were incubated as described above (Cell culture) and total RNA was collected after 4 hrs exposure to the particulate matter. For the expression analysis, we used 50 μg/cm2 of SiO2, TiO2, 5 μg/cm2 of ZnO (due to high toxicity) and 10 μg/cm2 of Fe2O3 (due to interference with RNA collection at higher particle concentration) in the colon cancer cell lines; in the skin-derived cell lines, 1 or 5 μg/cm2 of ZnO was used, and 5 or 10 μg/cm2 of TiO2. 100 μM ZnCl2 was used for the soluble Zn treatment. RNA was collected and prepared for expression analysis from cells treated for 4 hrs. RNA concentration was determined with a Nanodrop spectrophotometer and sufficient total RNA was recovered using the Qiagen RNeasy minikit protocol for the analysis. The quality of the RNA was monitored using an Experion automated electrophoresis station (BioRad) with standard sensitivity RNA chips, all samples displayed RNA Quality Index (RQI) levels greater than 8. Agilent labeling kits were utilized to amplify and generate Cy-dye labeled cRNA for hybridization to Agilent oligonucleotide arrays. A minimum of four biological replicates were collected representing each condition, and samples were combined, in equal fractions, for the microarray analysis. For most sample types, each of the four replicates were mixed together as the input sample for the expression analysis. An exception was the TNF-α pretreatment samples where composed of two sets of three biological replicates were combined so that replicates of this condition would be represented by more than a single sample in the expression analysis.
Agilent 44K (human whole genome) oligonucleotide microarrays were processed on site in the Microarray Resource located within the Huntsman Cancer Institute. Transcript levels were assessed on each channel and quantified by Agilent Feature Extraction software. This software preprocesses the data was as follows. Local background is subtracted, irregular spots flagged, and global linear regression (lowess) normalization is performed, and this ratio is log transformed. We import the data into TIGR MEV 3.1 software for further analysis. Generally, we use a supervised strategy to identify the genes with the greatest significant differences between stimulated cells to unstimulated cells using multiclass SAM.15 In each SAM analysis, 500 iterations were used when evaluating the false discovery rate, and a conservative cutoff was used that gave a median false discovery rate of 0%. The gene expression profiles were hierarchically clustered for visualization. Gene Ontology analysis of the differentially expressed genes was accomplished with EASE.16
Immunoblot analysis
Cells were grown in 6-well plates and treated with ZnO, ZnCl2 or were untreated as the control. Media was aspirated and cells were then washed with 1 ml of cold 1× PBS and the PBS aspirated. Cell lysates were collected in 8 M urea, 10 mM EDTA, 0.1 M Tris, pH 8, and the samples were reduced with the addition of dithiothreitol to 100 mM prior to protein estimation. Protein concentrations were determined using Coomassie Plus Protein Reagent (Pierce). Absorbance at 595 nm was measured using a Perkin-Elmer Victor3V plate reader. Fifteeen micrograms of protein were separated on 4–12% NuPAGE Bis-Tris gels and transferred to PVDF membranes, transferred to PVDF membrane, blocked with 5% non-fat dry milk, incubated with primary antibody (1:200 for DNAJA4, GADD45B, HSPA1A, HMOX1, and ZFP36, and 1:400 for GAPDH) overnight at 4°C, washed 3×, incubated with secondary antibody (1:5000) for 45 min at 22°C, washed 3×, incubated with chemiluminesence reagents (SuperSignal West Dura, Thermo Scientific), and visualized on a Kodak ImageStation.
Results
Particle characterization
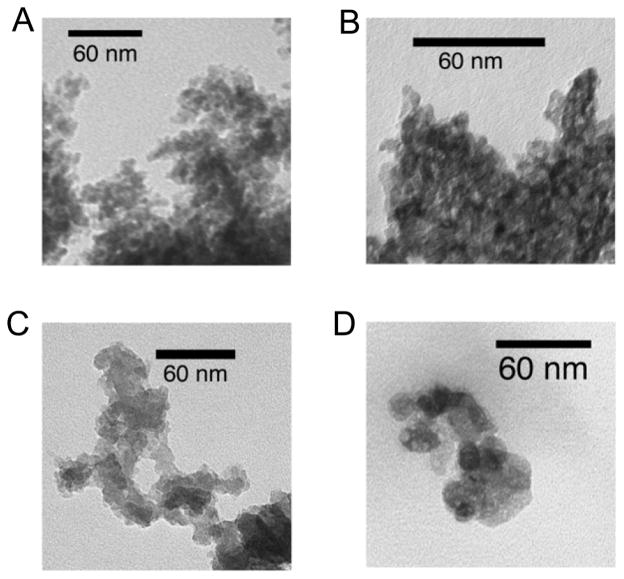
Documentation of particle characteristics is essential when reporting the toxicology of nanomaterials. Particle characteristics of the materials used in this study are summarized in Table 1. The TiO2, SiO2, and Fe2O3 nanomaterials were all over 60 m2/g by nitrogen absorption surface area measurement and had zeta potentials less than −40 mV. The ZnO nanoparticulate had specific surface of 24 m2/g and a zeta potential of −54 mV. Examination of the particles by TEM confirmed that all the nano-sized powders contained primary structures smaller than 100 nm (figure 1). The vendor’s nominal size, the surface-equivalent diameter determined by gas adsorption, and the geometric size observed by TEM were reasonably consistent. Agglomerates much larger than the nominal particle size formed when the particles were suspended either in water or in Advanced DMEM cell culture media with 2% serum (as observed previously for ZnO 17). The size distribution measured by dynamic light scattering was altered by water bath sonication and changed with time. The size of the major agglomerate mode in media after overnight incubation is listed in Table 1, and similar sized agglomerates were observed only minutes after sonication.
Table 1.
Particulate Matter Characterization
| Material and Abbreviation | Supplier’s Nominal Size | Surface Area (BET)a (m2/g) | Surface equivalent sphereb (nm) | Agglomerate mode: waterc (nm) | Agglomerate mode: mediad (nm) | Zeta Potentiale (mV) | Vendorf | Comment |
|---|---|---|---|---|---|---|---|---|
| Iron Oxide Fe2O3 | 3 nm | 222 | 5 | 557, >1000 (bimodal) | 1300 | −62 | AA | |
| Titanium Dioxide TiO2 | 5 nm | 240 | 6 | 749 | 1000 | −45 | NS | anatase |
| Silicon Dioxide SiO2 | 10 nm | 124 | 20 | 181 | 600 | −44 | NS | amorphous |
| Zinc Oxide, Nano ZnO | 8–10 nm | 24 | 40 | 161, 615 (bimodal) | 650 | −54 | AA | |
| Zinc Oxide, Conventional ZnO | −325 mesh (<44 μm) | 3.2 | 300 | ND | >5000 | ND | AA |
Surface area measured by nitrogen adsorption using a Quantachrome Monosorb analyzer.
Surface equivalent sphere is defined as the size of uniform spheres that have the same ratio of mass to surface area as the powder. Data were rounded to one figure due to uncertainties introduced by calculation assumptions.
Volume-weighted agglomerate mode in water at 0.1 mg/ml, vortexed, and sonicated immediately prior to DLS.
Volume-weighted agglomerate mode in cell culture media at 300 μg/ml as determined by DLS after 24 hr incubation.
Zeta potential were measured using a Malvern Zetasizer Nano ZS in water at 0.1 mg/ml, vortexed, and sonicated immediately prior analysis.
Vendor codes: NS, Nanostructured and Amorphous Materials; AA, Alpha Aesar.
ND = not done.
Fig. 1.
TEM images of nanoparticles suspended in water and dried on a grid; A) Fe2O3, B) TiO2, C) SiO2, and D) ZnO.
Soluble transition metals have been implicated as a cause of particle-induced oxidative stress, and trace metals can be a contaminant in insoluble materials. The concentration of trace elements, other than Zn, in cell culture media after overnight incubation with particles was near the blank media values. For example, based on an initial concentration of 1 mg/ml metal oxide concentration in media as a stock solution, soluble iron in the nano-Fe2O3 treated sample was 0.24 ppm compared to 0.17–0.19 ppm in blank media. Soluble zinc was 19 ppm in the nano-ZnO containing sample compared to 0.2 ppm in the blank media (~95× increase). Since there was no evidence that the particle suspensions, other than ZnO, resulted in soluble transition metals, this issue was not studied in greater detail herein.
Toxicity and comparisons with TNF-α in colon-derived cells
To determine if an induced inflammatory signaling condition altered the sensitivity of cells to the metal oxides, the effect of nanoparticulate matter on the viability of the RKO and CaCo-2 cells in the presence or absence of TNF-α pretreatment were assessed. Cells were treated with the particles in a dose-dependent manner ranging from 1 to 100 μg/cm2 using a formazan-style viability assay (Table 2). Initial experiments indicated that three of the metal oxide nanoparticles (TiO2, SiO2, and Fe2O3) had minimal toxicity below 100 μg/cm2 and ZnO displayed the most toxicity. TNF-α pretreatment was used to mimic an inflamed state;18 however, we did not observe dramatic differences in cell viability whether they were pretreated with TNF-α or BSA.
Table 2.
NanoParticulate LC50 (μg/cm2) in Colon-derived Cell Lines
| RKO | CaCo-2 | |||
|---|---|---|---|---|
| Material and Abbreviation | −TNF | +TNF | −TNF | +TNF |
| Iron Oxide Fe2O3 | >100 | >100 | >100 | >100 |
| Titanium Dioxide TiO2 | >100 | >100 | >100 | >100 |
| Silicon Dioxide SiO2, | >100 | >100 | >100 | >100 |
| Zinc Oxide, Nano ZnO | 27 ± 3.6 | 16 ± 2.6 | 28 ± 4.6 | 27 ± 1.8 |
Formazan-style viability assays were performed with each nanomaterial between 0–100 μg/cm2. In all cases, 100 μg/cm2 completely coated the bottom of the well. If a 50% loss of viability was not observed within this range the LD50 was designated >100 μg/cm2.
Effects due solely to high number concentration or due to high surface area of insoluble particles have been hypothesized. To evaluate if there was a consistent or non-specific transcriptional effect of the nanoparticles at an early (4 hr) timepoint we first compared BSA pretreated RKO or CaCo-2 colon cancer cells to cells treated with TNF-α, nanoparticulate only, or TNF-α plus nanoparticulate by whole genome expression analysis. If TNF-α had an effect on the cellular responsiveness to nanoparticles in these cell lines, we expected to observe potentiation of the metal oxide response. However, the most prominent response resulted from the TNF-α treatment, independent of particle type (all significant genes of interest are listed in supplemental tables). The induced genes that were statistically enriched in the molecular function or biological process gene ontology categories following EASE analysis were primarily involved in the regulation of apoptosis and cytokine pathways (figure S1), consistent with TNF-α-stimulation. The genes were consistent with NF-κB-mediated signaling pathways as these genes were primarily induced in the TNF-α-pretreatment groups but not in the BSA control group. No common set of genes modulated by the nano-sized TiO2, SiO2, Fe2O3, and ZnO was found, and this suggested that these materials did not induce a non-specific, nanoparticle-mediated response. Furthermore, co-treatment with TNF-α plus nanoparticles did not significantly alter the gene transcription response.
Particle-specific responses in colon-derived cells
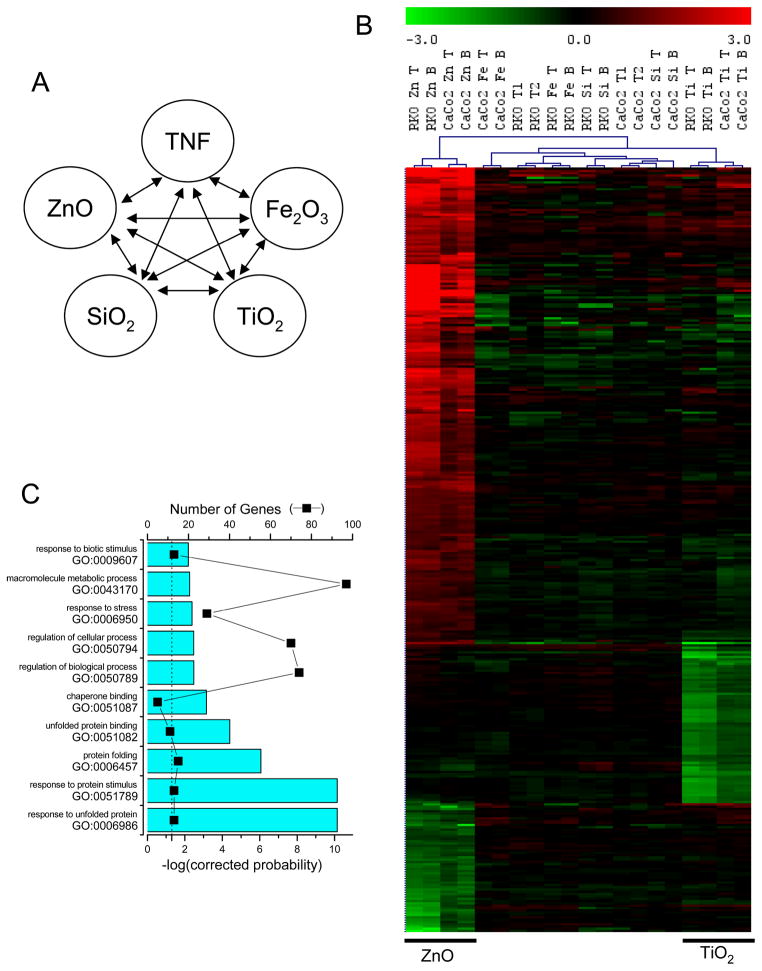
Next we evaluated the same dataset but grouped the expression data by nanoparticle type to determine if the particulate matter gave a chemical species-specific nanoparticle response (figure 2 and supplemental tables). This analysis indicated that ZnO and TiO2 displayed transcriptional effects with ZnO resulting in the largest number of gene expression changes. EASE annotation analysis indicated statistical enrichment of gene ontology categories related to unfolded protein, chaperon, and stress responses in ZnO-treated cells. Other enriched transcripts included genes whose products bind transition metals; many were Zn-finger containing proteins, so there was some overlap with the genes identified as transcriptional modulation as well as genes involved in metal metabolism. Genes involved in transcription regulation were also statistically enriched in the ZnO-treated cells, but many of these demonstrated decreased expression. In addition, decreased expression of genes involved in transcription were also identified in the TiO2 treatment condition.
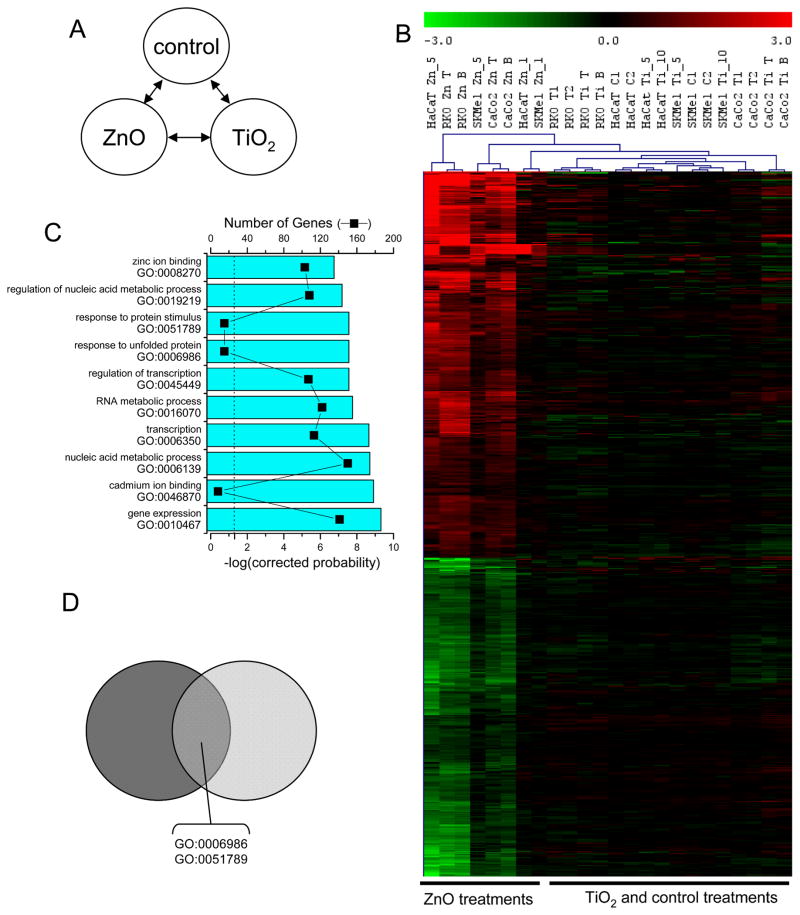
Fig. 2.
Gene expression changes of RKO and CaCo-2 cells following treatment with metal oxide nanomaterials. A) Multiclass SAM comparison indicated schematically. of nanoparticle-specific categories regardless of TNF-α pre-treatment was performed on the following categories (as represented schematically at the top of the figure): nanoparticle B) Hierarchical cluster visualization and heatmap of gene expression changes. nanoparticle-induced gene expression changes. Treatments are denoted: 1) Zn for ZnO; 2) Ti for TiO2; 3) Fe for Fe2O3; and 4) Si for SiO2, and 5) pre-treatments with T, T1 or T2 for TNF-α; samples are denoted with a B for BSA. 435 elements, representing 392 distinct probes, or 332 unique genes were identified as significant. ZnO treatment altered the expression of most of these genes, TiO2 inhibited the expression of a smaller set of genes, but Fe2O3 and SiO2 treatment did not generate coherent transcriptional responses. C) The top ten biological processes or molecular functions gene ontology (GO) categories as determined by EASE with the Bonferroni corrected probability (cyan bars), and the number of genes that identified the gene ontology category (■) displayed. The dashed line represents catagory significance at the p=0.05 level. The complete geneset and annotation analysis is provided in the microarray supplementary tables.
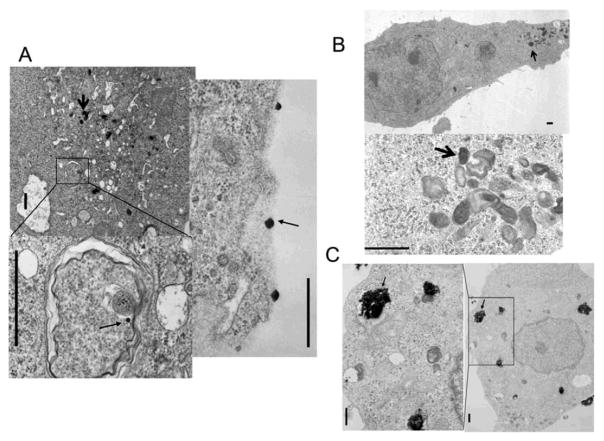
To evaluate whether ZnO and TiO2 transit across the plasma membrane to promote these transcriptional responses, we examined cells treated with the nanomaterials by electron microscopy (figure 3). TiO2 was clearly visible within RKO cells as large agglomerates while it was more challenging to identify the ZnO. The more pronounced ultrastructural features we found within the cells treated with ZnO were membrane whorls, a feature more consistent with late stage autophagasomes. Nevertheless, it did appear that the nanomaterials were entering the cells and ZnO-treatments demonstrated intracellular alterations consistent with autophagy (figures S3–S4).
Fig. 3.
TEM evaluation of ZnO and TiO2 nanoparticulate in RKO cells. A) ZnO; electron dense material was observed at the surface (light arrow right panel) and within membrane bound structures (heavy arrow lower panel). B) The major ultrastructural change following ZnO treatment was membrane-bound structures and whorls. C) TiO2; agglomerates were visible (light arrow) but did not always elicit membrane encapsulation. The solid bar on the images represents ~600 nm.
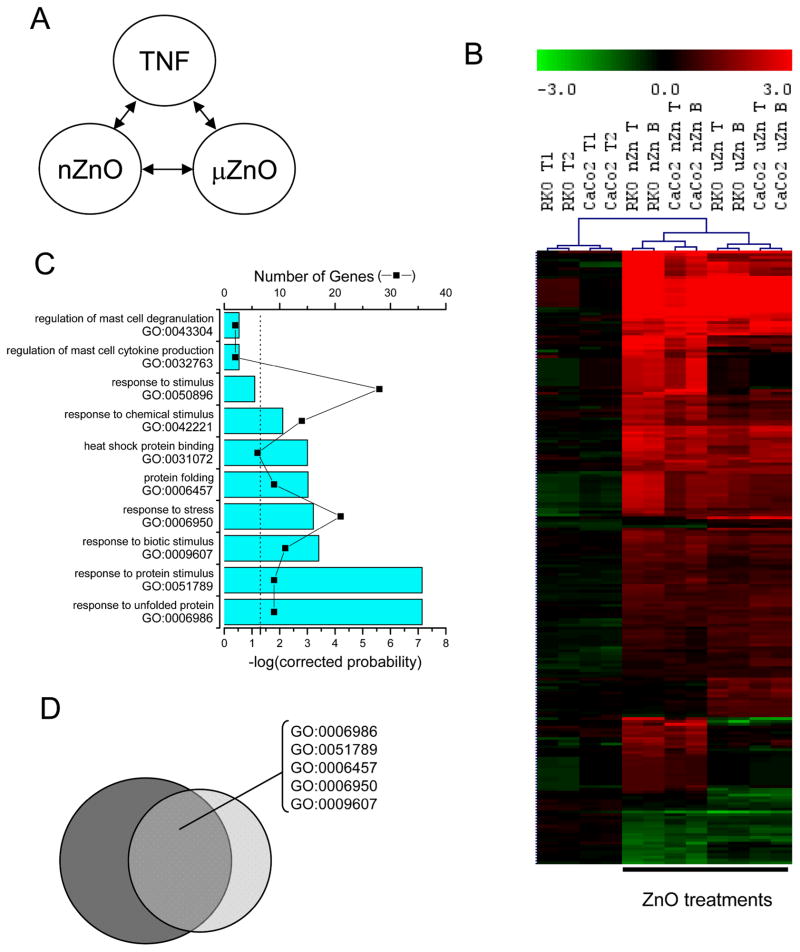
Whether novel nano-sized particle formulations induce different toxicological responses than conventional powders of the same nominal substance is a question of great interest. Since ZnO was the most cytotoxic and also induced the most prominent alterations in early transcriptional responses, experiments compared the nano-sized powder to a conventional ZnO powder that was nominally −325 mesh screen size (~44 μm) but actually contained a significant amount of submicron particles in agglomerates or aggregates (previously described 17). While there does appear to be some differences, we found that the conventional- and nano-sized ZnO induced similar changes in expression (figure 4 and supplemental tables). Again, the most pronounced sets of genes affected by both ZnO particle sizes were those involved in unfolded protein, chaperon, cellular stress responses, and metal metabolism.
Fig. 4.
Similar gene expression changes occur in RKO and CaCo-2 cells following treatment with nano-sized and micron-sized ZnO commercial powders. A) Schematic representation of the multiclass SAM performed on the following categories 1) TNF-α pre-treatment, 2) nanoZnO treatment, and 3) conventional ZnO treatment (designated by the μZnO). B) Hierarchical cluster visualization of ZnO particulate transcript profiles. 217 elements, representing 181 distinct probes, or 162 unique genes were identified as significantly different among these groups. C) The top ten gene ontology (GO) categories determined by EASE with the Bonferroni corrected probability (bars), and the number of genes that identified the gene ontology category (■) as described in figure 2. D) The Venn diagram represents the significant GO categories from figure 2C (dark grey) and this analysis (light grey) and shows that five of the top ten gene ontology categories were in common among the significantly enriched groups included the response to unfolded protein and response to protein stimulus categories.
We wished to specifically evaluate the involvement of inflammation-associated genes since particulate matter has demonstrated proinflammatory characteristics in other in vitro systems. We evaluated each cell line and ZnO treatment using multiclass significance analysis so that we could determine if inflammation-related genes were part of an early response to ZnO under any condition other than TNF-α cotreatment; however, a broad pro-inflammatory response was not observed (figure S5).
We validated the microarray results by quantifying transcription of specific genes that are often used as biomarkers of specific pathways using quantitative PCR (QPCR). We measured the transcriptional levels of BIRC3, ZFP36, PTGS2, SLC30A1, MT1F, and HSPA1A as biomarkers for inflammation, metal response, and chaperonin genes, respectively. For all of these genes, the QPCR validated the microarray analysis (figure S6).
Comparison of responses in colon- and skin-derived cells
Since our results demonstrated the greatest gene transcription responses for ZnO and TiO2, and these nanomaterials are frequently used for topical applications, we further evaluated these agents in HaCaT and SK Mel-28 cells as representative skin-derived keratinocytes and melanocytes. These cell lines displayed similar cytotoxic responsiveness to ZnO (table 3), and again, TiO2 was minimally toxic (LD50 >100 μg/cm2) in both cell lines. Next we evaluated the transcriptional response of these cell lines to ZnO and TiO2. With the additional gene expression experiments, we found a broader range of gene expression differences (figure 5). Still, the strongest changes in gene expression were related to metal metabolism, stress, and protein folding. There were also more modest changes in gene expression related to nucleic acid metabolism, and transcription (see gene expression supplemental tables). The genes that increased expression following exposure to ZnO were primarily those related to protein folding responses and stress-related transcriptional programs. The genes that were attenuated were related to nucleic acid metabolism.
Table 3.
NanoParticulate LC50 in Skin-derived Cell Lines
| Material and Abbreviation | HaCaT | SK Mel-28 |
|---|---|---|
| Titanium Dioxide TiO2 | >100 | >100 |
| Zinc Oxide, Nano ZnO | 11.8 ± 0.54 | 11.5 ± 2.6 |
Formazan-style viability assays were performed with each nanomaterial between 0–100 μg/cm2. In all cases, 100 μg/cm2 completely coated the bottom of the well. If a 50% loss of viability was not observed within this range the LD50 was designated >100 μg/cm2.
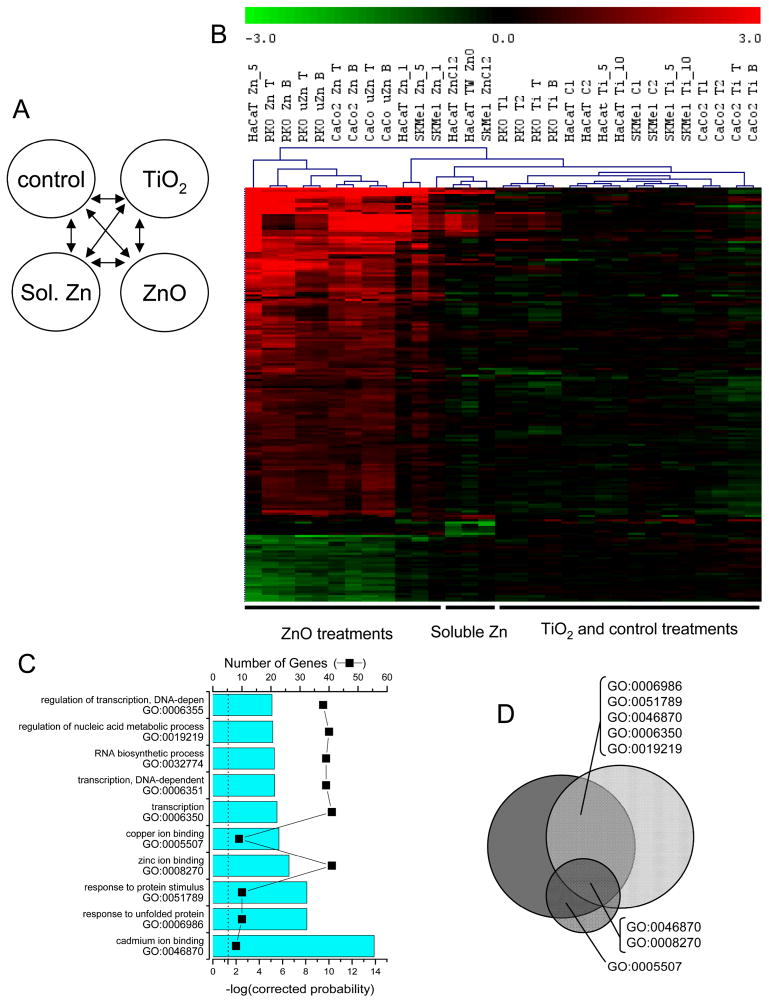
Fig. 5.
Gene expression in colon- and skin-derived cells. A) Schematic representation of the multiclass SAM performed on the following categories: 1) control samples (C1, C2) and TNF-α samples (T1, T2), 2) 5 μg/cm2 ZnO treatments, and 3) TiO2 treatments regardless of dose. B) Hierarchical cluster visualization of nanoparticle treatment transcript profiles. 917 elements, representing 818 distinct probes, or 690 unique genes were identified. C) The top ten gene ontology (GO) categories determined by EASE with the Bonferroni corrected probability (bars), and the number of genes that identified the gene ontology category (■) as described in figure 2. D) The Venn diagram represents the significant GO categories from figure 2C (dark grey) and this analysis (light grey) and depicts that the only GO categories that were consistent in the top ten categories were the protein stimulus and response to unfolding protein.
Our previous studies with these same nanomaterials have demonstrated that ZnO liberates Zn2+;17 therefore, we identified the gene expression changes among the cell lines with direct contact to the ZnO and TiO2 compared soluble Zn (liberated Zn from the nano-ZnO at 5 μg/cm2 was ~ 50 μM). To evaluate responses to soluble Zn, we measured the gene expression changes in the HaCaT cells treated with 100 μM ZnCl2, or ZnO (5 μg/cm2) that was separated from the cells with a Transwell insert, and SK Mel-28 cells treated with 100 μM ZnCl2. We then determined which genes are differentially expressed among these samples with soluble Zn compared to the ZnO direct contact samples. The genes associated with metal ion binding (copper and cadmium binding gene ontology categories; GO:0005507 and GO:0046870) are still induced by soluble Zn ions present in the media. However, the genes associated with protein turnover, cellular stress, nucleic acid metabolism, and transcription related pathways are the genes that no longer demonstrate changes in expression (figure 6 and S7).
Fig. 6.
ZnO contact dependence for gene expression modulation. A) Schematic of the multiclass SAM of skin-derived and colon-derived cells performed on the following categories: 1) control samples (C1, C2) and TNF-α samples (T1, T2), 2) all ZnO treatments, 3) all TiO2 treatments, and 4) skin-derived cells exposed to soluble Zn (ZnCl2) or ZnO separated from cells using a Transwell insert (TW). B) Hieararchical cluster visualization of nanoparticle treatment transcript profiles. 274 elements, representing 234 distinct probes, or 186 unique genes were identified. C) The top ten gene ontology (GO) categories determined by EASE with the Bonferroni corrected probability (bars), and the number of genes that identified the gene ontology category (■) as described in figure 2. D) The Venn diagram represents the significant GO categories from figure 5C (dark grey), this analysis (light grey), and the smaller circle represents the significant GO categories observed from analysis of soluble Zn samples. Two GO categories were in common among all these analyses (cadmium ion binding and zinc ion binding); categories that loose representation in the soluble Zn analysis are those relating to protein folding and transcriptional regulation.
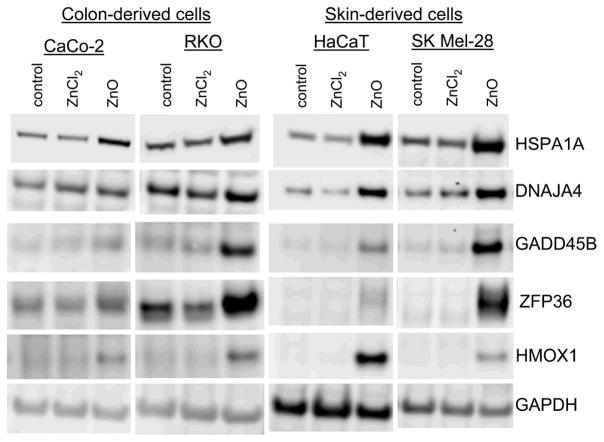
Finally, to validate that chaperon and stress response genes displayed differential expression between cells exposed to soluble Zn compared to ZnO, the gene products were confirmed and validated by immunoblot analysis (figure 7). The gene products of HSPA1A, DNAJA4, and GADD45B are representative of chaperon and stress pathways. Additional gene products that could represent oxidative stress (HMOX1) and inflammation (ZFP36) were also evaluated. However, since there is a paucity of other antioxidant and inflammatory genes, it is more likely that ZFP36 expression is responding to Zn levels, and that HMOX1 responds as a stress response gene under ZnO stimulation.
Fig. 7.
Immunoblot analysis of select gene products demonstrating ZnO-dependence of expression in RKO, CaCo-2, HaCaT, and SK Mel-28 cells. Protein levels in untreated cells or following 8 hrs of stimulation with 100 μM ZnCl2 or 5 μg/cm2 ZnO. Gene products representing protein folding and stress responses include; HSPA1A, DNAJ4A, GADD45B, ZFP36, and HMOX1; while GAPDH represents a control protein. Contact with ZnO demonstrates gene product modulation across all 4 cell lines derived from different tissues while soluble Zn did not elicit similar responses.
Discussion
The SiO2, Fe2O3, and TiO2 nanoparticulate displayed little toxicity and had limited effects on gene transcription in RKO and CaCo-2 colon cells, even at concentrations above likely physiological exposure. This adds to the growing evidence that low solubility inorganic particles do not have unique or unusual mechanisms of toxicity solely because of size by providing genome-wide expression profiling data for lesser-studied cell types. A TNF-α induced pro-inflammatory environment did not alter the responses of RKO and CaCo-2 cells to these metal oxide nanoparticles, refuting our initial hypothesis that inflammation would enhance response to nanoparticles. ZnO was the most toxic of the tested metal oxides and microarray evaluation of the whole human genome suggests that the cells produced a generalized stress response that affected the expression of Zn response proteins, heat shock proteins, early response proteins, and metal metabolism. Genomic analysis supplement studies that measure only a few protein expression biomarkers, as indicated by the observation that there was a limited pro-inflammatory response to the ZnO that involved a few genes, but not the entire pathway. Significantly, contact with nano-ZnO particles produced a different response than that resulting from an equivalent concentration of ZnCl2.
ZnO and TiO2 are common ingredients in topical consumer products. Therefore, we expanded our study to evaluate the cytotoxicity and transcriptional responses in skin-derived cells. The cytotoxicity was similar in these cells to the colon-derived cells. We also observed significant overlap in the gene expression from the ZnO treated cells indicating that ZnO likely has a common mechanism of action.
ZnO has displayed potential for numerous beneficial applications including food additives, sunscreens, and pigments as well as a broad range of engineering applications.19 ZnO has traditionally demonstrated utility by its antimicrobial activity.20 In addition, ZnO has anti-cancer activity, that may be associated with rapid cell division compared to non-maligant cells.21 Our studies utilized a variety of transformed cell lines and ZnO was cytotoxic in all. In addition, the cellular transcriptional response was similar in all cases. Therefore, ZnO may have a common mechanism of cytotoxocity among all the cells. Our previous observations, as well as those of other groups,17, 22 suggests that cytosolic Zn2+ liberated over time from ZnO taken up by cells promotes mitochondrial dysfunction, stress at the endoplasmic reticulum, and superoxide production that promote cell death and loss of membrane integrity.
Gene expression analysis provides insight into particular cellular response pathways in an unbiased manner. For example, the gene expression data strongly suggest that Zn ions are available to the cells exposed to ZnO. The upregulation of genes like the metallothioneins (MT1A, MT1B, MT1E, MT1F, MT1G, MT1H, MT1L, MT1M, MT1X, and MT2A) and the Zn efflux transporter (SLC30A1) are genes that are under the control of the metal transcription factor that requires Zn metal ions for transactivation.23 ZnO toxicity for some organisms is related to Zn solubility,24 and the development of ZnO tolerance appears to enlist genes that sequester Zn2+ like metallothionein.25 This early transcriptional response we observed is consistent with cells managing Zn but also showed additional transcription responses not related to the metal transcription factor.26 Since the metallothionein response was also observed when the cells were exposed to soluble Zn, this response appears to be an adaptive response to the increased Zn2+ in the cellular environment. Nevertheless, the induction of metallothioneins as well as additional stress response genes have been characterized as a gene expression response for toxic metals like cadmium.27
Besides the induction of metal responsive agents, the most characteristic cellular pathways influenced by ZnO were transcriptional regulation and protein folding responses. The repression of transcription and nucleic acid metabolism is consistent with Zn toxicity.28 At physiological doses, the transition metal, Zn, is critical for many proteins containing Zn-finger motifs; however, these protein motifs are subject to disruption by other transition metals, and even high concentrations of Zn.29 While it is unlikely that this is cell type specific response to nano-ZnO since others have shown Zn-dependent heat shock protein responses,30 this response was absent in experiments where the cells were exposed to soluble Zn instead of ZnO particles. Indeed, these stress responses appear to be common responses to metal toxicity,31 and suggests that the cytotoxicity of metals like Zn may be due to a broad disruption of protein function, analogous to thiol oxidation. The induction of a common set of genes involved in stress responses and protein folding (ATF3, BAG3, DNAJA1, DNAJA4, DNAJB1, DNAJB4, DNAJB6, ERRFI1, FOS, GADD454B, HSPA1A, HSPA1L, HSPA4L, HSPA6, and HSPH1) provides an opportunity to investigate the relationship of these gene products with Zn-mediated cytotoxicity since many heat shock proteins have roles in cell survival and apoptosis.32 Therefore, further studies are warrented to determine if the induction of these genes is consistent with disruption of the mitochondrion as previous studies have observed,17 as well as determining if impairment of protein translocation to the mitochondria33 are components of Zn-mediated cytotoxicity
The size-dependent variation in potency of nanomaterials is often explained in terms of surface area effects, as in a recent gene array comparison of 10 and 500 nm SiO2.5 In this study, we also observed similar genes responding to ZnO regardless of the primary particle size. However, the lack of stability of the nanomaterial dispursions, as defined by the agglomeration observed in aqueous solutions, limits our ability to explicitly assess the concentration-dependent interactions of the ZnO with the cells. It is possible that the agglomerates promote interactions with the cells due to settling. This hypothesis is supported by the conventional ZnO generating more pronounced responses by QPCR than the nano-sized ZnO. However, the conventional ZnO has been shown to be very polydisperse and contains a significant amount of submicron particles.17 Therefore, we cannot rule out that the smaller size particles from the conventional ZnO may contribute disporportionately to the cellular responses.
ZnO has demonstrated induction of oxidative stress responses and DNA damage in A431 cells,34 as does environmental particulate contaminated with Zn.35 The possibility of inhalation of Zn-containing compounds in environmental particulate has spurred an investigation of responsive genes in mice instilled with a Zn salt,36 and these investigators identified genes similar to those reported herein. Our gene expression data, evaluating early responses to ZnO, did not demonstrate widespread pro-inflammatory or oxidative stress responses gene expression induction. We did observe select gene modulation like ZFP36 and PTGS2 representing inflammation associated genes, and HMOX1 may be the most responsive gene, and could represent an oxidative response. However, there are few additional genes modulated by ZnO that were consistent with these functional categories. Indeed, the transcriptional regulation of HMOX1 by the transcriptional repressor, Bach1, has demonstrated cadmium responsiveness37 and HMOX1 may be the most responsive gene repressed by Bach1 in keratinocytes.38 Therefore, these genes are most likely responding to the ZnO explicitly and are not components of broader transcriptional responses.
The exposure to different nanomaterials via oral human consumption is an outstanding question as it is unknown whether dietary metal oxides nanomaterials have chronic effects on the colon. SiO2, and TiO2, are commonly used as food ingredients. They are used in cosmetics, drugs, and consumer products where exposure can result from hand-to-mouth contact and drinking water contamination. We have shown that colon cell responses to ZnO are mediated by cell-particle contact, and not by soluble Zn.17 Nanomaterial exposure to gastric acid may alter their properties, particularly since ZnO is more soluble in acidic solutions compared to other metal oxides. However, the ingestion of large amounts of ZnO has been reported to cause gastroduodenal corrosive injury in humans without systemic toxicity.39
Conclusions
This study has provided some of the first data on the effects of commercial metal oxide nanopartilces on human colon-derived as well as skin-derived cells. These gene transcription data are consistent with other recent work suggesting that low solubility metal oxide nanoparticles do not have unique mechanisms of action or orders-of-magnitude greater potency when compared to conventional powders of the same nominal substance.5 In this study we observed similar patterns of gene transcription in colon cells treated with materials marketed as nano-sized and conventional ZnO powders. Continued research on the environmental and health effects of new industrial chemical formulations, including smaller primary particle sizes and novel surface treatments, is consistent with the precautionary principle.
Supplementary Material
Acknowledgments
This work was supported by U.S. Environmental Protection Agency - Science to Achieve Results (STAR) program [grant number RD83333601 (PJM)] and in part by Huntsman Cancer Institute and Huntsman Cancer Foundation. We also acknowledge the use of core facilities (microarray, DNA synthesis, and sequencing cores) supported by a National Cancer Institute, Cancer Center Support Grant [grant number P30 CA042014] awarded to Huntsman Cancer Institute.
List of Abbreviations
- BIRC3
baculoviral IAP repeat-containing 3
- B2M
beta-2 microglobulin
- DLS
dynamic light scattering
- DNAJA4
DnaJ (Hsp40) homolog, subfamily A, member 4
- EASE
expression analysis systematic explorer
- Fe2O3
iron oxide
- GADD45B
growth arrest and DNA-damage-inducible, beta
- HSPA1A
inducible heat shock protein 70
- HMOX1
heme oxygenase 1
- MT1F
metallothionein 1F
- PTGS2
prostaglandin-endoperoxide synthase 2 (also known as COX-2, cyclooxygenase-2)
- SLC30A1
solute carrier family 30 (zinc transporter) member 1 (also known as ZnT1, zinc transporter 1)
- TIGR MEV
the institute for genomic research multiple experiment viewer
- TNF-α
tumor necrosis factor alpha
- SAM
significance analysis of microarrays
- SiO2
silicon dioxide
- TEM
transmission electron microscopy
- TiO2
titanium dioxide
- ZFP36
zinc finger protein 36 (also known as TTP, tristetraprolin)
- ZnO
zinc oxide
Footnotes
This article is published as part of a themed issue on Metal Toxicity, Guest Edited by Gregor Grass and Christopher Rensing.
Electronic Supplementary Information (ESI) available: The significant results for each assessment of the gene expression experiments, including the annotation, statistical information, and the expression data, as well as the annotation analysis (EASE) for each assessment is provided in an Excel workbook. Additional data and methods are included in a supplemental document. See DOI: 10.1039/b000000x/
References
- 1.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–7. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]; Veranth JM, Ghandehari H, Grainger DW. In: Comprehensive Toxicology. McQueen CA, editor. Vol. 8. Oxford: Academic Press; 2010. pp. 453–475. [Google Scholar]
- 2.Li JJ, Muralikrishnan S, Ng CT, Yung LY, Bay BH. Nanoparticle-induced pulmonary toxicity. Exp Biol Med (Maywood) 2010;235:1025–33. doi: 10.1258/ebm.2010.010021. [DOI] [PubMed] [Google Scholar]; Fubini B, Ghiazza M, Fenoglio I. Physico-chemical features of engineered nanoparticles relevant to their toxicity. Nanotoxicology. 2010;4:347–63. doi: 10.3109/17435390.2010.509519. [DOI] [PubMed] [Google Scholar]; Warheit DB, Donner EM. Rationale of genotoxicity testing of nanomaterials: regulatory requirements and appropriateness of available OECD test guidelines. Nanotoxicology. 2010;4:409–13. doi: 10.3109/17435390.2010.485704. [DOI] [PubMed] [Google Scholar]; Donaldson K, Poland CA, Schins RP. Possible genotoxic mechanisms of nanoparticles: criteria for improved test strategies. Nanotoxicology. 2010;4:414–20. doi: 10.3109/17435390.2010.482751. [DOI] [PubMed] [Google Scholar]; Napierska D, Thomassen LC, Lison D, Martens JA, Hoet PH. The nanosilica hazard: another variable entity. Part Fibre Toxicol. 2010;7:39. doi: 10.1186/1743-8977-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]; Menard A, Drobne D, Jemec A. Ecotoxicity of nanosized TiO2. Review of in vivo data. Environ Pollut. 2011;159:677–84. doi: 10.1016/j.envpol.2010.11.027. [DOI] [PubMed] [Google Scholar]; Terzano C, Di Stefano F, Conti V, Graziani E, Petroianni A. Air pollution ultrafine particles: toxicity beyond the lung. Eur Rev Med Pharmacol Sci. 2010;14:809–21. [PubMed] [Google Scholar]; Inoue K, Takano H. Aggravating impact of nanoparticles on immune-mediated pulmonary inflammation. ScientificWorldJournal. 2011;11:382–90. doi: 10.1100/tsw.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita K, Horie M, Kato H, Endoh S, Suzuki M, Nakamura A, Miyauchi A, Yamamoto K, Kinugasa S, Nishio K, Yoshida Y, Iwahashi H, Nakanishi J. Effects of ultrafine TiO2 particles on gene expression profile in human keratinocytes without illumination: involvement of extracellular matrix and cell adhesion. Toxicol Lett. 2009;191:109–17. doi: 10.1016/j.toxlet.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Inoue K, Takano H, Ohnuki M, Yanagisawa R, Sakurai M, Shimada A, Mizushima K, Yoshikawa T. Size effects of nanomaterials on lung inflammation and coagulatory disturbance. Int J Immunopathol Pharmacol. 2008;21:197–206. doi: 10.1177/039463200802100122. [DOI] [PubMed] [Google Scholar]
- 5.Waters KM, Masiello LM, Zangar RC, Tarasevich BJ, Karin NJ, Quesenberry RD, Bandyopadhyay S, Teeguarden JG, Pounds JG, Thrall BD. Macrophage responses to silica nanoparticles are highly conserved across particle sizes. Toxicol Sci. 2009;107:553–69. doi: 10.1093/toxsci/kfn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita K, Morimoto Y, Ogami A, Myojyo T, Tanaka I, Shimada M, Wang WN, Endoh S, Uchida K, Nakazato T, Yamamoto K, Fukui H, Horie M, Yoshida Y, Iwahashi H, Nakanishi J. Gene expression profiles in rat lung after inhalation exposure to C60 fullerene particles. Toxicology. 2009;258:47–55. doi: 10.1016/j.tox.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Ellinger-Ziegelbauer H, Pauluhn J. Pulmonary toxicity of multi-walled carbon nanotubes (Baytubes) relative to alpha-quartz following a single 6h inhalation exposure of rats and a 3 months post-exposure period. Toxicology. 2009;266:16–29. doi: 10.1016/j.tox.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Dobson J. Toxicological aspects and applications of nanoparticles in paediatric respiratory disease. Paediatr Respir Rev. 2007;8:62–6. doi: 10.1016/j.prrv.2007.02.005. [DOI] [PubMed] [Google Scholar]; Rouse RL, Murphy G, Boudreaux MJ, Paulsen DB, Penn AL. Soot nanoparticles promote biotransformation, oxidative stress, and inflammation in murine lungs. Am J Respir Cell Mol Biol. 2008;39:198–207. doi: 10.1165/rcmb.2008-0057OC. [DOI] [PubMed] [Google Scholar]; Pope CA, 3rd, Ezzati M, Dockery DW. Fine-particulate air pollution and life expectancy in the United States. N Engl J Med. 2009;360:376–86. doi: 10.1056/NEJMsa0805646. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krewski D, Jerrett M, Burnett RT, Ma R, Hughes E, Shi Y, Turner MC, Pope CA, 3rd, Thurston G, Calle EE, Thun MJ, Beckerman B, DeLuca P, Finkelstein N, Ito K, Moore DK, Newbold KB, Ramsay T, Ross Z, Shin H, Tempalski B. Extended follow-up and spatial analysis of the American Cancer Society study linking particulate air pollution and mortality. Res Rep Health Eff Inst. 2009:5–114. discussion 115–36. [PubMed] [Google Scholar]; Van Hee VC, Kaufman JD, Budinger GR, Mutlu GM. Update in environmental and occupational medicine 2009. Am J Respir Crit Care Med. 2010;181:1174–80. doi: 10.1164/rccm.201002-0183UP. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hesterberg TW, Long CM, Lapin CA, Hamade AK, Valberg PA. Diesel exhaust particulate (DEP) and nanoparticle exposures: what do DEP human clinical studies tell us about potential human health hazards of nanoparticles? Inhal Toxicol. 2010;22:679–94. doi: 10.3109/08958371003758823. [DOI] [PubMed] [Google Scholar]
- 9.Jani P, Halbert GW, Langridge J, Florence AT. Nanoparticle uptake by the rat gastrointestinal mucosa: quantitation and particle size dependency. J Pharm Pharmacol. 1990;42:821–6. doi: 10.1111/j.2042-7158.1990.tb07033.x. [DOI] [PubMed] [Google Scholar]
- 10.Lamprecht A, Yamamoto H, Takeuchi H, Kawashima Y. Nanoparticles enhance therapeutic efficiency by selectively increased local drug dose in experimental colitis in rats. J Pharmacol Exp Ther. 2005;315:196–202. doi: 10.1124/jpet.105.088146. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, von Mikecz A. Formation of nucleoplasmic protein aggregates impairs nuclear function in response to SiO2 nanoparticles. Exp Cell Res. 2005;305:51–62. doi: 10.1016/j.yexcr.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Faunce TA. Toxicological and public good considerations for the regulation of nanomaterial-containing medical products. Expert Opin Drug Saf. 2008;7:103–6. doi: 10.1517/14740338.7.2.103. [DOI] [PubMed] [Google Scholar]
- 13.Peters K, Unger RE, Gatti AM, Sabbioni E, Tsaryk R, Kirkpatrick CJ. Metallic nanoparticles exhibit paradoxical effects on oxidative stress and pro-inflammatory response in endothelial cells in vitro. Int J Immunopathol Pharmacol. 2007;20:685–95. doi: 10.1177/039463200702000404. [DOI] [PubMed] [Google Scholar]; Gojova A, Guo B, Kota RS, Rutledge JC, Kennedy IM, Barakat AI. Induction of inflammation in vascular endothelial cells by metal oxide nanoparticles: effect of particle composition. Environ Health Perspect. 2007;115:403–9. doi: 10.1289/ehp.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berridge MV, Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev. 2005;11:127–52. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moos PJ, Chung K, Woessner D, Honeggar M, Cutler NS, Veranth JM. ZnO particulate matter requires cell contact for toxicity in human colon cancer cells. Chem Res Toxicol. 2010;23:733–9. doi: 10.1021/tx900203v. [DOI] [PubMed] [Google Scholar]
- 18.Ma TY, Boivin MA, Ye D, Pedram A, Said HM. Mechanism of TNF-{alpha} modulation of Caco-2 intestinal epithelial tight junction barrier: role of myosin light-chain kinase protein expression. Am J Physiol Gastrointest Liver Physiol. 2005;288:G422–30. doi: 10.1152/ajpgi.00412.2004. [DOI] [PubMed] [Google Scholar]
- 19.Popov AP, Zvyagin AV, Lademann J, Roberts MS, Sanchez W, Priezzhev AV, Myllyla R. Designing inorganic light-protective skin nanotechnology products. J Biomed Nanotechnol. 2010;6:432–51. doi: 10.1166/jbn.2010.1144. [DOI] [PubMed] [Google Scholar]; Weintraub B, Zhou Z, Li Y, Deng Y. Solution synthesis of one-dimensional ZnO nanomaterials and their applications. Nanoscale. 2010;2:1573–87. doi: 10.1039/c0nr00047g. [DOI] [PubMed] [Google Scholar]
- 20.Sawai J. Quantitative evaluation of antibacterial activities of metallic oxide powders (ZnO, MgO and CaO) by conductimetric assay. J Microbiol Methods. 2003;54:177–82. doi: 10.1016/s0167-7012(03)00037-x. [DOI] [PubMed] [Google Scholar]; Li Q, Mahendra S, Lyon DY, Brunet L, Liga MV, Li D, Alvarez PJ. Antimicrobial nanomaterials for water disinfection and microbial control: potential applications and implications. Water Res. 2008;42:4591–602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]; Gajjar P, Pettee B, Britt DW, Huang W, Johnson WP, Anderson AJ. Antimicrobial activities of commercial nanoparticles against an environmental soil microbe, Pseudomonas putida KT2440. J Biol Eng. 2009;3:9. doi: 10.1186/1754-1611-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aydin Sevinc B, Hanley L. Antibacterial activity of dental composites containing zinc oxide nanoparticles. J Biomed Mater Res B Appl Biomater. 2010;94:22–31. doi: 10.1002/jbm.b.31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrovsky S, Kazimirsky G, Gedanken A, Brodie C. Selective Cytotoxic Effect of ZnO Nanoparticles on Glioma Cells. Nano Res. 2009;2:882–890. doi: 10.1007/s12274-009-9089-5. [DOI] [Google Scholar]; Hanley C, Layne J, Punnoose A, Reddy KM, Coombs I, Coombs A, Feris K, Wingett D. Preferential killing of cancer cells and activated human T cells using ZnO nanoparticles. Nanotechnology. 2008;19:295103. doi: 10.1088/0957-4484/19/29/295103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rasmussen JW, Martinez E, Louka P, Wingett DG. Zinc oxide nanoparticles for selective destruction of tumor cells and potential for drug delivery applications. Expert Opin Drug Deliv. 2010;7:1063–77. doi: 10.1517/17425247.2010.502560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CC, Aronstam RS, Chen DR, Huang YW. Oxidative stress, calcium homeostasis, and altered gene expression in human lung epithelial cells exposed to ZnO nanoparticles. Toxicol In Vitro. 2010;24:45–55. doi: 10.1016/j.tiv.2009.09.007. [DOI] [PubMed] [Google Scholar]; De Berardis B, Civitelli G, Condello M, Lista P, Pozzi R, Arancia G, Meschini S. Exposure to ZnO nanoparticles induces oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol Appl Pharmacol. 2010 doi: 10.1016/j.taap.2010.04.012. [DOI] [PubMed] [Google Scholar]; Xia T, Kovochich M, Liong M, Madler L, Gilbert B, Shi H, Yeh JI, Zink JI, Nel AE. Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano. 2008;2:2121–34. doi: 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. Embo J. 1994;13:2870–5. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Palmiter RD, Findley SD. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. Embo J. 1995;14:639–49. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin NM, Rogers NJ, Apte SC, Batley GE, Gadd GE, Casey PS. Comparative toxicity of nanoparticulate ZnO, bulk ZnO, and ZnCl2 to a freshwater microalga (Pseudokirchneriella subcapitata): the importance of particle solubility. Environ Sci Technol. 2007;41:8484–90. doi: 10.1021/es071445r. [DOI] [PubMed] [Google Scholar]
- 25.Wesselkamper SC, Chen LC, Gordon T. Development of pulmonary tolerance in mice exposed to zinc oxide fumes. Toxicol Sci. 2001;60:144–51. doi: 10.1093/toxsci/60.1.144. [DOI] [PubMed] [Google Scholar]
- 26.Lichtlen P, Wang Y, Belser T, Georgiev O, Certa U, Sack R, Schaffner W. Target gene search for the metal-responsive transcription factor MTF-1. Nucleic Acids Res. 2001;29:1514–23. doi: 10.1093/nar/29.7.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusakabe T, Nakajima K, Nakazato K, Suzuki K, Takada H, Satoh T, Oikawa M, Arakawa K, Nagamine T. Changes of heavy metal, metallothionein and heat shock proteins in Sertoli cells induced by cadmium exposure. Toxicol In Vitro. 2008;22:1469–75. doi: 10.1016/j.tiv.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Vallee BL. Zinc: biochemistry, physiology, toxicology and clinical pathology. Biofactors. 1988;1:31–6. [PubMed] [Google Scholar]
- 29.Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3:625–34. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- 30.Graff DW, Cascio WE, Brackhan JA, Devlin RB. Metal particulate matter components affect gene expression and beat frequency of neonatal rat ventricular myocytes. Environ Health Perspect. 2004;112:792–8. doi: 10.1289/ehp.112-1241994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauman JW, Liu J, Klaassen CD. Production of metallothionein and heat-shock proteins in response to metals. Fundam Appl Toxicol. 1993;21:15–22. doi: 10.1006/faat.1993.1066. [DOI] [PubMed] [Google Scholar]
- 32.Arya R, Mallik M, Lakhotia SC. Heat shock genes - integrating cell survival and death. J Biosci. 2007;32:595–610. doi: 10.1007/s12038-007-0059-3. [DOI] [PubMed] [Google Scholar]
- 33.Ryan MT, Pfanner N. Hsp70 proteins in protein translocation. Adv Protein Chem. 2001;59:223–42. doi: 10.1016/s0065-3233(01)59007-5. [DOI] [PubMed] [Google Scholar]; Takayama S, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;22:9041–7. doi: 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]; van der Laan M, Hutu DP, Rehling P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta. 2010;1803:732–9. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Sharma V, Shukla RK, Saxena N, Parmar D, Das M, Dhawan A. DNA damaging potential of zinc oxide nanoparticles in human epidermal cells. Toxicol Lett. 2009;185:211–8. doi: 10.1016/j.toxlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Frampton MW, Ghio AJ, Samet JM, Carson JL, Carter JD, Devlin RB. Effects of aqueous extracts of PM(10) filters from the Utah valley on human airway epithelial cells. Am J Physiol. 1999;277:L960–7. doi: 10.1152/ajplung.1999.277.5.L960. [DOI] [PubMed] [Google Scholar]
- 36.Gilmour PS, Schladweiler MC, Nyska A, McGee JK, Thomas R, Jaskot RH, Schmid J, Kodavanti UP. Systemic imbalance of essential metals and cardiac gene expression in rats following acute pulmonary zinc exposure. J Toxicol Environ Health A. 2006;69:2011–32. doi: 10.1080/15287390600746173. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem. 2003;278:49246–53. doi: 10.1074/jbc.M306764200. [DOI] [PubMed] [Google Scholar]
- 38.MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, Hayes JD. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–80. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CH, Lee CT, Tsai FC, Hsu SJ, Yang PM. Gastroduodenal corrosive injury after oral zinc oxide. Ann Emerg Med. 2006;47:296. doi: 10.1016/j.annemergmed.2005.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.