Abstract
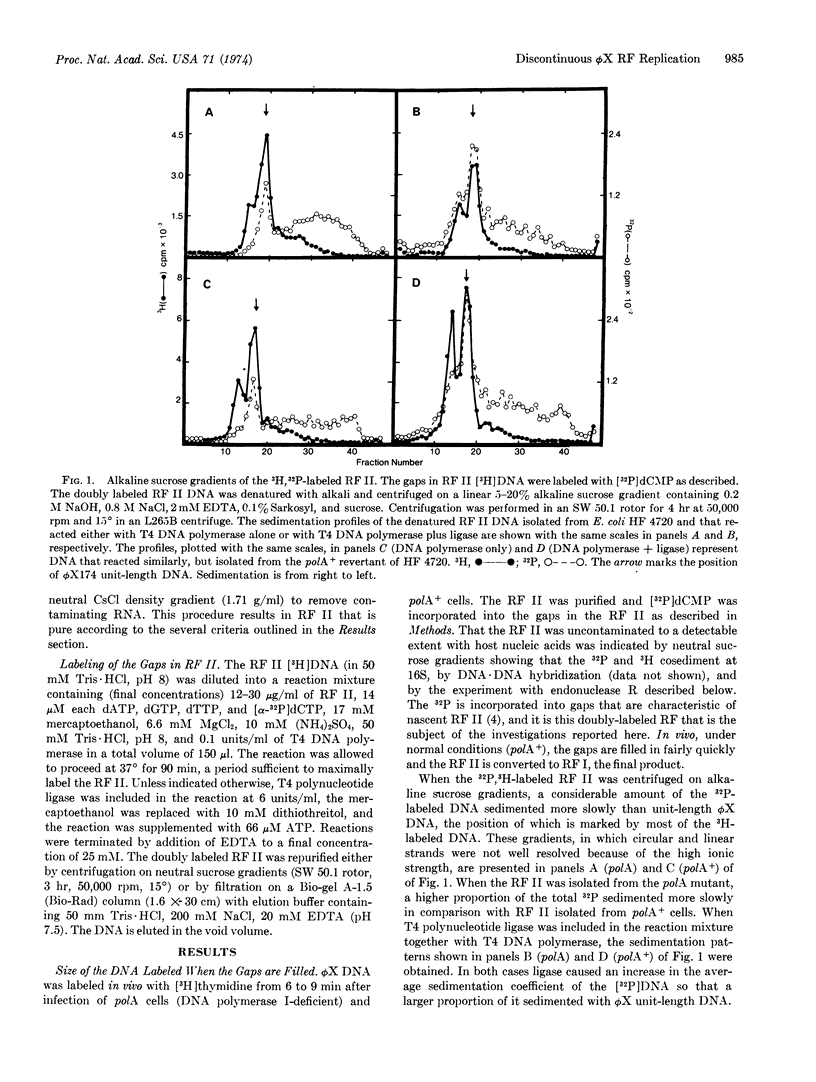
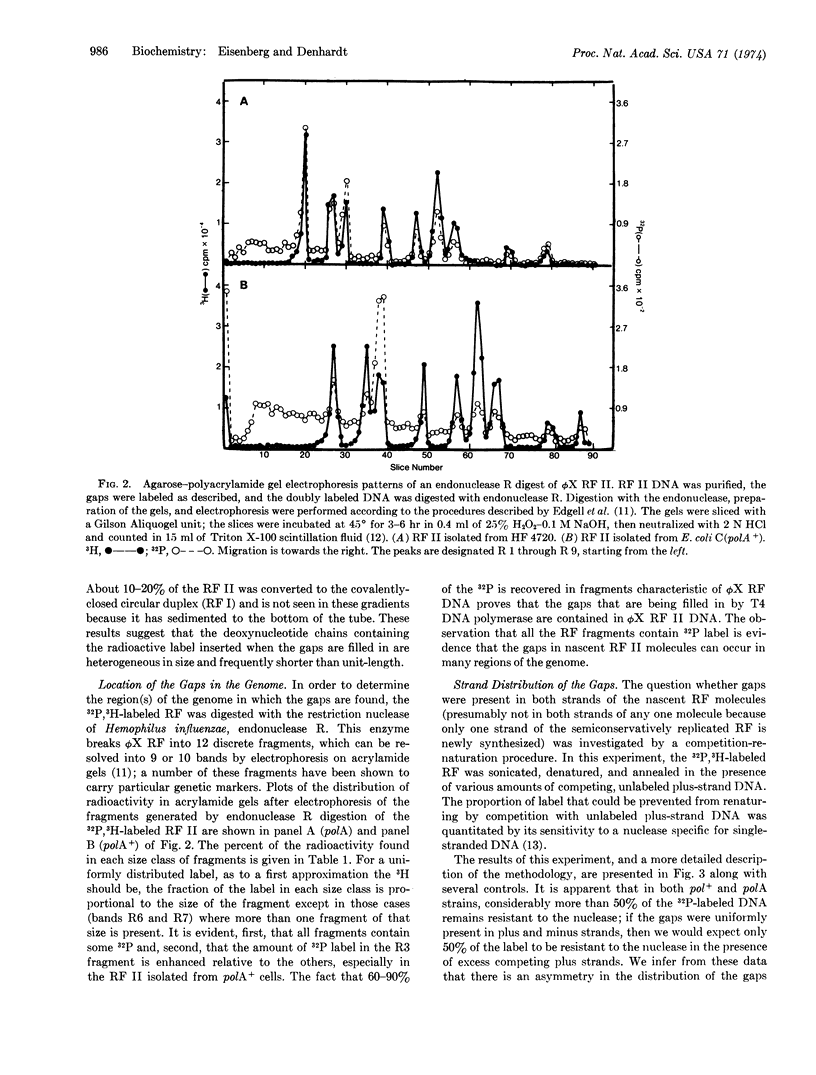
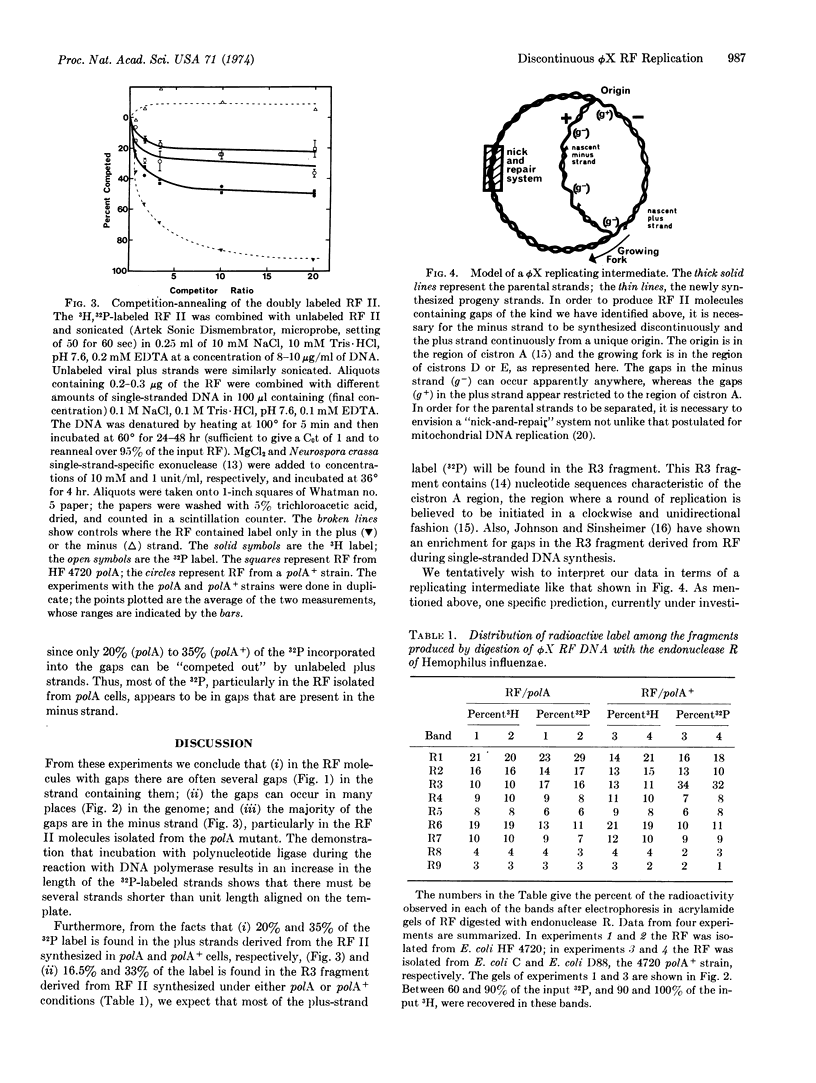
ϕX174 RF (replicative form) II DNA, labeled in vivo with [methyl-3H]thymidine, was isolated from Escherichia coli polA (DNA polymerase I-deficient) and polA+ cells during RF replication. [32P]dCMP was incorporated into the gaps present in the RF II DNA with [α-32P]dCTP and T4 DNA polymerase. Sedimentation in alkaline sucrose gradients revealed that much of the incorporated 32P was present in a heterogeneous collection of fragments shorter than unit length. Inclusion of polynucleotide ligase in the gap-filling reaction increased the average size of the 32P-labeled fragments. Gel electrophoresis of the products formed by digestion of the 32P-labeled RF II molecules with the restriction nuclease, endonuclease R, indicated that in the population of RF II molecules gaps could occur anywhere in the genome. Competition-annealing experiments provided evidence that the majority of the label incorporated into gaps was present in the minus strand. RF II molecules isolated from polA+ cells were enriched for gaps in a unique region of the genome in comparison with RF II molecules isolated from polA cells. The presence of multiple gaps in the minus strand implies that it is synthesized by a discontinuous mechanism during ϕX RF replication.
Keywords: DNA polymerase, restriction nuclease, competition hybridization, gap-filling, polA
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. PhiX174 replicative form DNA replication, origin and direction. J Mol Biol. 1972 Feb 14;63(3):569–576. doi: 10.1016/0022-2836(72)90448-2. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Hutchison C. A., 3rd, Edgell M. H. Isolation and genetic localization of three phi-X174 promoter regions. Nat New Biol. 1973 Jun 20;243(129):233–236. doi: 10.1038/newbio243233a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A theory of DNA replication. J Theor Biol. 1972 Mar;34(3):487–508. doi: 10.1016/0022-5193(72)90137-3. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. Formation of ribosylthymine in Escherichia coli. Studies on pulse labeling with thymine and thymidine. J Biol Chem. 1969 May 25;244(10):2710–2715. [PubMed] [Google Scholar]
- Edgell M. H., Hutchison C. A., 3rd, Sclair M. Specific endonuclease R fragments of bacteriophage phiX174 deoxyribonucleic acid. J Virol. 1972 Apr;9(4):574–582. doi: 10.1128/jvi.9.4.574-582.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Iwaya M., Eisenberg S., Bartok K., Denhardt D. T. Mechanism of replication of single-stranded PhiX174 DNA. VII. Circularization of the progeny viral strand. J Virol. 1973 Oct;12(4):808–818. doi: 10.1128/jvi.12.4.808-818.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H., Deutscher M. P., Littauer U. Z. Rates of DNA chain growth in Escherichia coli. J Mol Biol. 1971 Nov 14;61(3):503–524. doi: 10.1016/0022-2836(71)90062-3. [DOI] [PubMed] [Google Scholar]
- Rabin E. Z., Tenenhouse H., Fraser M. J. An exonuclease of Neurospora crassa specific for single-stranded nucleic acids. Biochim Biophys Acta. 1972 Jan 18;259(1):50–68. doi: 10.1016/0005-2787(72)90473-x. [DOI] [PubMed] [Google Scholar]
- Robberson D. L., Kasamatsu H., Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A. 1972 Mar;69(3):737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R. W., Iwaya M., Bromstrup K., Denhardt D. T. The mechanism of replication of phi X174 single-stranded DNA. 3. An enzymic study of the structure of the replicative form II DNA. J Mol Biol. 1971 Apr 28;57(2):177–199. doi: 10.1016/0022-2836(71)90340-8. [DOI] [PubMed] [Google Scholar]
- Sebring E. D., Kelly T. J., Jr, Thoren M. M., Salzman N. P. Structure of replicating simian virus 40 deoxyribonucleic acid molecules. J Virol. 1971 Oct;8(4):478–490. doi: 10.1128/jvi.8.4.478-490.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsheimer R. L. Bacteriophage phi-X174 and related viruses. Prog Nucleic Acid Res Mol Biol. 1968;8:115–169. [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Sugino A., Hirose S., Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Preparation of [alpha-32P]nucleoside and deoxynucleoside 5'-triphosphates from 32Pi and protected and unprotected nucleosides. Biochim Biophys Acta. 1969 Oct 22;190(2):548–550. doi: 10.1016/0005-2787(69)90105-1. [DOI] [PubMed] [Google Scholar]
- Wickner W., Brutlag D., Schekman R., Kornberg A. RNA synthesis initiates in vitro conversion of M13 DNA to its replicative form. Proc Natl Acad Sci U S A. 1972 Apr;69(4):965–969. doi: 10.1073/pnas.69.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]


