Abstract
It has been repeatedly shown that in case-control association studies, analysis of a secondary trait which ignores the original sampling scheme can produce highly biased risk estimates. Although a number of approaches have been proposed to properly analyze secondary traits, most approaches fail to reproduce the marginal logistic model assumed for the original case-control trait and/or do not allow for interaction between secondary trait and genotype marker on primary disease risk. In addition, the flexible handling of covariates remains challenging. We present a general retrospective likelihood framework to perform association testing for both binary and continuous secondary traits which respects marginal models and incorporates the interaction term. We provide a computational algorithm, based on a reparameterized approximate profile likelihood, for obtaining the maximum likelihood (ML) estimate and its standard error for the genetic effect on secondary trait, in presence of covariates. For completeness we also present an alternative pseudo-likelihood method for handling covariates. We describe extensive simulations to evaluate the performance of the ML estimator in comparison with the pseudo-likelihood and other competing methods.
2 Introduction
The retrospective case-control design is one of the most important tools for epidemiology, and for rare diseases/traits may offer tremendous savings in time and expense compared to a prospective design. Even so, case-control designs remain costly, and efficiency is further maximized by gathering additional clinical phenotypes/traits for the sampled individuals. We refer to the dichotomous case/control variable as the primary trait (or phenotype), and other traits, which may be discrete or continuous, as secondary. Methods for analyzing such data have received considerable recent attention due to the availability of genome-wide association (GWA) datasets, which often follow a case-control design and include numerous secondary traits, which may be correlated with the primary trait (Frayling et al., 2007; Weedon et al., 2008; Spitz et al., 2008). In such studies the risk variables of main interest are genotypes of single nucleotide polymorphisms (SNPs), with possible additional covariate effects. The approaches described here apply generally to secondary analysis of case-control data, but the notation and examples are applicable to genetic association studies.
It is widely understood that, for case-control designs, a (prospective) analysis which ignores the sampling scheme yields consistent estimates of the odds ratio for disease risk (e.g., using logistic regression models (Prentice and Pyke, 1979)). For the analysis of secondary traits within a case-control design, a number of approaches have been described (Jiang et al., 2006; Lin and Zeng, 2009; He et al., 2012; Wang and Shete, 2011, 2012). The existing methods can be broadly divided as follows: a) the naïve method of analyzing the combined sample of cases and controls, without accounting for the case-control ascertainment; b) performing analysis within case and control groups; c) combining estimates from the case and control groups either by meta-analysis with inverse variances as weights or by using case/control status as a covariate; d) correcting bias using weighting schemes originally developed for sample surveys; and d) explicitly accounting for the case/control sampling scheme via a retrospective likelihood.
The naïve method can lead to severely biased estimates of the risk effects for the secondary traits, except under restrictive conditions (Nagelkerke et al., 1995; Jiang et al., 2006; Lin and Zeng, 2009; Monsees et al., 2009). Simple methods which account for the biased sampling include adjusting for the disease status in a regression model or restricting the analysis to cases or controls only. For rare diseases, the controls-only analysis is approximately unbiased, but may be highly inefficient. If the association between the primary and the secondary traits does not depend on the SNP genotype, then the cases also give a valid risk effect estimate and can be combined with the controls-only estimate via a inverse-variance meta-analytic procedure to provide a weighted estimate with improved efficiency. Li et al. (2010) describe an adaptively weighted method for rare diseases with binary secondary trait and genotype, that further combines the controls-only and the weighted estimates. Recently, they proposed another adaptive procedure to analyze secondary phenotypes for data from a case-control study of a primary disease that is not rare (Li and Gail, 2012).
The survey approach uses weights inversely proportional to the sampling fractions (Jiang et al., 2006; Scott and Wild, 2002). Richardson et al. (2007) and Monsees et al. (2009) use this standard survey-weighted approach, applied to the analysis of binary secondary traits, termed the inverse-probability-of-sampling-weighted (IPW) regression by Monsees et al. (2009). Technically this approach requires knowledge of the case-control sampling fractions, but the disease prevalence is often available externally and may be used as in Wang and Shete (2011).
In general, most of the approaches described above result in inconsistent risk estimates, although the bias may be low under some specific assumptions. The efficiency of the procedures also varies widely (Jiang et al., 2006). An alternative approach to account for the sampling mechanism is to use the retrospective likelihood, explicitly conditioning on the sampling scheme (Jiang et al., 2006; Scott and Wild, 1997b, 2001, 1991; Lee et al., 1998; Lin and Zeng, 2009; He et al., 2012). The retrospective likelihood models the joint distribution for the primary and secondary traits as a function of the genotype and other covariates. There are a number of attractive features to this approach. If the model is correct, then it will provide large-sample optimality for the maximum likelihood (ML) estimates. Provided the model is sufficiently rich, it enables partitioning of the correlations between primary and secondary phenotypes into the portion due to the risk variable (genotype), as well as a residual portion that may be due to other genes or environment. In addition, standard semi-parametric maximum likelihood (SPML) approaches are available to handle covariates in a flexible manner, with minimal assumptions for the potentially complex interplay of covariates with other data. For this reason the retrospective approaches are collectively termed “SPML” in the taxonomy of Jiang et al. (2006).
Although the motivation for the retrospective likelihood is clear, specific implementations vary. One approach is to factor the joint distribution into the marginal for the secondary trait and the conditional for the primary trait given secondary (SPML2 in Jiang et al., 2006). For example, Lin and Zeng (2009) handle both binary and continuous secondary traits by modeling the disease status given the secondary trait as a logistic regression. Wang and Shete (2011) use this joint model and apply a method of moments approach to produce bias-corrected odds ratio estimates for binary secondary traits using prevalence estimates for the primary and secondary traits from the literature. Recently they have shown that their method is robust even when there is an interactive effect of the SNP and secondary phenotype on the primary disease risk (Wang and Shete, 2012). He et al. (2012) have proposed a Gaussian copula-based approach that models the joint distribution in terms of the marginals for the primary and secondary phenotypes (SPML3 in Jiang et al., 2006) and uses the multivarite normal distribution to build in correlation between the phenotypes. Their method can handle multiple correlated secondary phenotypes.
Despite all of these efforts, a number of deficiencies remain. Most retrospective likelihood approaches do not incorporate interaction between the genetic variant and the secondary trait on primary disease risk. Other than the Gaussian copula approach (He et al., 2012), retrospective methods do not generally preserve the marginal logistic model for the disease trait, creating a contradiction between the primary trait risk estimates obtained from the marginal model and from the joint primary-secondary analysis. Additionally, the nonparametric handling of continuous covariates remains challenging; when there are multiple covariates, including one or more continuous ones, direct maximization of the likelihood is infeasible.
We propose an approach that specifies the joint distribution of primary and secondary traits in terms of the marginals for the two traits, with terms to govern their association (SPML3 in Jiang et al., 2006). Our framework enables association testing for both binary and continuous secondary traits, while respecting the desired logistic model for the primary trait and standard marginal models for the secondary trait. We demonstrate how this approach can incorporate covariates, and easily allow for interaction between the genetic variant and the secondary trait on primary disease risk. To handle the computational complexity introduced by the covariates, we reparameterize the profile likelihood, and derive a closed form expression which provides ML estimates of risk effects. For completeness, we briey describe a pseudo-likelihood approach that bypasses the need to involve the potentially high-dimensional covariate distribution. We perform extensive simulations to evaluate the performance of our profile likelihood method and compare it with the pseudo-likelihood and other competing methods.
The remainder of the paper is organized as follows. In Section 3 we lay down the details for our proposed profile likelihood-based method. In particular, in Subsection 3.1 we describe our joint model for the primary and secondary traits and in the following subsection we provide details of the estimation procedure. In Section 4 we demonstrate the performance of the proposed method as compared to other competing methods via a real data example and simulations. Section 5 concludes with some comments on related work and future directions. In Section 7 we expand on the theoretical details.
3 Methods
Let D denote the disease status or the primary trait (0=control, 1=case), Y the secondary trait, G the genotype at a biallelic locus, and Z the vector of covariates. Under additive model, G represents the number of minor alleles (0, 1, or 2) at the locus; under dominant (recessive) model, G denotes whether the individual carries at least one minor allele (two minor alleles). We randomly sample n0 and n1 individuals from the controls (D = 0) and the cases (D = 1) in the population respectively, and observe their Y-, G-, and Z-values. The log-likelihood for our case-control sampled data (du, yu, gu, zu), u = 1, 2, …, n, takes the retrospective form where
We model P(d, y|g, z) parametrically, the regression P(y|g, z) being of primary interest. Note that P(g, z), the joint distribution for G and Z in the population, cannot be ignored, since
We deal with P(g, z) nonparametrically, thereby taking a semi-parametric approach to modeling the likelihood. In the following subsections we describe the joint model for the bivariate response (D, Y) and the estimation procedure.
3.1 Joint modeling of the primary and secondary traits
In modeling the joint distribution for the traits we try to preserve the marginal logistic model typically assumed for the original case-control trait D. We specify the bivariate distribution, P(D, Y|g, z), by parametrically modeling the marginals for the primary and secondary traits and also building a parametric model for their association given the genotype and the covariates. The natural choice for the marginal distribution for disease status is logistic, and that for the secondary trait is logistic or normal depending on whether the trait is binary or continuous respectively. For binary Y, the Palmgren model (Palmgren, 1989), the Bahadur model (Bahadur, 1959), and models based on copula theory (Meester and MacKay, 1994) have been used previously to specify joint distributions for bivariate binary responses. But for continuous Y, there is no standard bivariate distribution that yields logistic and normal marginals for D and Y respectively.
3.1.1 Binary secondary trait
The bivariate logistic model, considered by Palmgren (1989), has been used previously to model the joint distribution for correlated binary data (Jiang et al., 2006; Lee et al., 1998) and is conceptually very simple. It is based on the fact that the joint distribution of two binary variables can be specified in terms of their marginal probabilities and their odds ratio. Thus, for a randomly sampled individual in the population we specify the joint distribution of D and Y given g and z as
and .
We are interested in inferring about β2, the risk effect for the secondary trait.
3.1.2 Continuous secondary trait
For continuous Y, we consider joint models for (D, Y |g, z) such that the marginal distributions for D and Y given g and z correspond to the standard logistic and linear regressions respectively, that is,
To come up with a joint model that complies with the above marginals, we start with the following bivariate normal distribution,
where , and . We then introduce another latent variable to produce the logistic marginal. To transform the normal variate V, we define . It follows that the density of U given g and z is logistic with location parameter μ1 and scale parameter 1,
We then threshold U at 0 to derive D. That is, D, defined as
follows
As in the binary case, β2 is our parameter of interest.
3.2 Estimation of β2
3.2.1 ML estimate in absence of covariates
Let us first consider the situation without any covariates. The retrospective log-likelihood is
| (1) |
We assume Hardy-Weinberg equilibrium and parameterize the genotype distribution in terms of the minor allele frequency, δ. We use θ to denote the set of parameters describing the joint distribution for D and Y given g. We write θ as (θ1, θ2)′, where θ1 = (α1, β1, γ1)′ parameterizes the disease model. Then the log-likelihood in (1) can be written as
| (2) |
We maximize (2) with respect to η = (θ, δ)′ to derive η̂, the ML estimate for η. We can use the standard Newton-Raphson method to obtain the ML estimate iteratively, or other optimization tools such as quasi-Newton methods, Nelder-Mead simplex algorithm for derivative-free maximization, simulated annealing. Specifically, using Newton-Raphson method, we update the parameter in the (k + 1)th iteration by
The inverse of the observed information matrix, , gives standard error estimate for η̂.
3.2.2 ML estimate via profiling in presence of covariates
Let us now consider the situation where the joint model for the disease and the secondary trait involves covariates. The retrospective log-likelihood is
| (3) |
We can reasonably assume that G and Z are independently distributed in the underlying population, but must somehow account for the distribution of Z. Since the covariate structure is usually far too complicated to model parametrically (considering that it may be correlated with the primary trait), we want to make no assumptions about the form of the covariate distribution.. For a single binary covariate parameterized by ψ, we can easily derive the ML estimate by maximizing the retrospective likelihood with respect to (η, ψ). However, this approach is infeasible for a continuous covariate as the ML estimate involves maximization with respect to a high-dimensional nuisance parameter. In the following section we describe a computational technique to derive a closed-form expression, which may be thought of as an approximate profile likelihood up to one nuisance parameter, that upon maximization provides the ML estimate for the parameters of interest.
Let {z1, z2, …, zL} represent unique values of Z in the case-control sample. We parameterize the distribution function for Z in terms of the probability masses {ψ1, ψ2, …, ψL} that we assign to {z1, z2, …, zL}. The retrospective log-likelihood in (3) can now be written as
| (4) |
When ψ is high-dimensional, maximizing the log-likelihood with respect to (η, ψ) can be a daunting task. We take the approximate profile likelihood approach described by Chatterjee and Carroll (2005) to handle the nuisance parameter ψ (hereafter dropping the “approximate” qualifier). To obtain the overall ML estimate for η, we maximize the profile log-likelihood, l(η) = supψl(η, ψ) with respect to η. Following Scott and Wild (1997a, 2001) and Chatterjee and Carroll (2005), we show in the Appendix (Section 7.1) that the profile log-likelihood l(η) can be equivalently expressed as
where
The parameter κ represents the ratio of the sampling fractions,
and satisfies the equation . We thus have effectively reduced the number of parameters from (p + L − 1) to (p + 1), with p used to denote the dimension of η. We provide a closed form expression, l*(ϕ), that on maximization with respect to ϕ = (η, κ)′ gives η̂, thereby bypassing the need to numerically maximize the likelihood with respect to a high-dimensional nuisance parameter.
Although l*(ϕ) is not a true log-likelihood the relevant asymptotic theory for ϕ̂ can be obtained by working with , referred to as the “pseudo” score-equations by Scott and Wild (2001). Under the assumption that n goes to infinity with n1/n and n2/n remaining fixed, we show in the Appendix (Section 7.2) that converges in distribution to a normal random vector with mean zero and covariance matrix
where
and
with and . In practice, we can use
the inverse of the observed information matrix based on l*, to estimate Cov(ϕ̂). Since Γ(ϕ) ≥ 0, J*(ϕ̂)−1 provides a conservative estimate for Cov(ϕ̂). Note that if the disease prevalence is known or well-estimated, we fix κ at its true value and work with l*(η) to obtain η̂ and its standard error.
3.2.3 Pseudo-likelihood estimate
Motivated by the fact that β2, the parameter of interest, appears only in the first term of the expression for the log-likelihood in (3), one might attempt to handle the covariates via a previously described pseudo-likelihood approach (Gong and Samaniego, 1981). For the sake of completeness, we lay down the framework for this approach to estimate β2 and its standard error, and evaluate its performance via simulations. The retrospective log-likelihood in (4) can be viewed as
| (5) |
where
| (6) |
and
As β2 is of primary interest, we treat θ1 as nuisance parameter. Gong and Samaniego proposed first obtaining an estimate for θ1. When disease prevalence is known, we derive an estimate for θ1, say θ̃1, from l2(θ1, δ, ψ) (in general different from the ML estimate θ̂1 derived from l). We then plug this estimate into l1(θ1, θ2) to obtain the pseudo log-likelihood
| (7) |
The pseudo-likelihood estimate, θ̃2, is obtained by maximizing l̃1 with respect to θ2. We discuss the asymptotic properties of the pseudo-likelihood estimate and provide formulae for its variance estimation in the Appendix (Section 7.3).
4 Results
We present a data example and simulation results to demonstrate the performance of the profile likelihood-based ML estimate in comparison with the pseudo-likelihood estimate and other competing methods: the naïve method ignoring the sampling mechanism, the cases-only and controls-only methods, the weighted method combining the cases-only and controls-only estimators via inverse-variance, the adaptively weighted method, proposed by Li et al. (2010), combining the controls-only and weighted estimators, the adjusted method including the disease status as a covariate in the regression for the secondary trait, and finally the survey approach using weights inversely proportional to the sampling fractions. All analyses were performed in R v.2.15.0. In the table and figures we use ‘Wtd’, ‘Awtd’, and ‘Adj’ to denote the weighted, the adaptively weighted, and the adjusted estimates respectively.
4.1 Data example
We reanalyze the data on colorectal cancer, smoking, and N-acetyltransferase 2 (NAT2) presented in Li et al. (2010). The authors compare different analysis methods for a binary secondary trait using case-control data for colorectal adenoma described in Moslehi et al. (2006). Moslehi et al. explore how variants in NAT1 and NAT2 genes affect the smoking-colorectal cancer relationship using cases with colorectal adenoma and controls selected from the screening arm of the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial. Of 42,037 participants who provided a blood sample, 4,834 were excluded. 772 cases were randomly selected from 1234 participants who had at least one advanced colorectal adenoma detected at baseline screen and 777 gender and age-matched controls were sampled from 26,651 participants with a negative baseline sigmoidoscopy screening. Li et al. use this case-control data for colorectal adenoma to analyze the effect of NAT2 gene on smoking (secondary trait). In Table 1 we display the data analyzed in Li et al. (2010) and in Table 2 we illustrate our re-analysis of the data.
Table 1.
Data for studying effect of NAT2 on smoking from a case-control study for colorectal adenoma
| Cases of colorectal adenoma | Matched controls | ||||||
|---|---|---|---|---|---|---|---|
| Smoking status | Total | Smoking status | Total | ||||
| Never/Former | Current | Never/Former | Current | ||||
| NAT2=0 | 199 | 380 | 579 | NAT2=0 | 255 | 317 | 572 |
| NAT2=1 | 18 | 13 | 31 | NAT2=1 | 10 | 23 | 33 |
| Total | 217 | 393 | 610 | Total | 265 | 340 | 605 |
Table 2.
Estimates of effect of NAT2 on smoking from a case-control study for colorectal adenoma
| Method | log(OR) | SE |
|---|---|---|
| Naïve | −0.18 | 0.26 |
| Cases | −0.97 | 0.37 |
| Controls | 0.62 | 0.39 |
| Adj | −0.17 | 0.26 |
| Wtd | −0.21 | 0.27 |
| Awtd | 0.57 | 0.40 |
| ML/Survey | 0.54 | 0.37 |
There are no covariates here, so the ML estimate based on the Palmgren model is easily derivable from the retrospective likelihood, without requiring additional profiling or pseudo-likelihood. We used a prevalence of 0.04 (=1234/(1234+26651)) based on Moslehi et al. (2006). Under known prevalence, the ML based on the Palmgren model and the survey approach are the same. Li et al., in their paper, treat the robust controls-only estimate as the gold standard and show that the adaptively weighted estimate is similar to the controls-only estimate. The authors point out that the non-zero interaction between NAT2 and smoking on colorectal adenoma risk results in high bias for the commonly used methods, apparent in the table for the naïve, the cases-only, the weighted, and the adjusted estimates. In contrast, our results show that the ML/survey estimate is similar to the adaptively weighted and controls-only estimates, with the ML/survey estimate enjoying smaller standard error.
4.2 Simulations
We compare performances of the different methods for both binary and continuous secondary traits, for biologically plausible scenarios, by generating data from the corresponding bivariate models (see Methods). We consider a biallelic SNP with an additive mode of inheritance and a minor allele frequency of 0.25. For demonstration purposes we include two covariates, one following a Bernoulli distribution with 0.45 success probability and the other a standard normal variate. For our simulations we consider the SNP genotype and the covariates as independent in the population. We fix the disease prevalence at 5% and the mean or prevalence of the secondary trait at 20%. Rather than specifying α1 and α2 directly, it is more interpretable to solve for them for specified values of the prevalences and other parameters. We fix β1 at 0.25 (odds ratio = 1.28). We consider β2 = 0 to reect the null and compare Type I error for different methods. To simulate data under alternative hypotheses and examine power, we fix β2 at either −0.25 or 0.25 for the binary secondary trait; for the continuous setup we use −0.15 and 0.15. We assign the values (−0.94, −0.37)′ and (−1.82, 0.30)′, each generated independently from the standard normal, to the parameters γ1 and γ2, respectively. We fix α3 at 1.0 and examine β3 ranging from −2.0 to 2.0. We use σ2 = 1.0 for the continuous scenario. For fixed prevalences, α1 and α2 are calculated according to the following equations:
and
For each combination of parameter values, we ran 10,000 simulations, each consisting of 1500 cases and 1500 controls. For each simulated dataset, we derive the ML, pseudo-likelihood, naïve, cases-only, controls-only, weighted, adaptively weighted, adjusted and survey-weighted estimates of the risk effect for the secondary trait and the corresponding standard error estimates. We used the “nmk” function within the “dfoptim” package in R to implement the robust Nelder-Mead algorithm for derivative-free optimization of the likelihood and the “hessian” function from the “numDeriv” package to derive Hessian matrices at parameter estimates. We judge the different estimators based on the amount of bias they incur in estimating the risk effect and their mean squared error (MSE). Using the standard error estimates we construct 95% Wald-type confidence intervals for the risk effect and study their coverage probabilities. Also, we examine Type I error and power for corresponding Wald tests conducted at an intended 5% significance level.
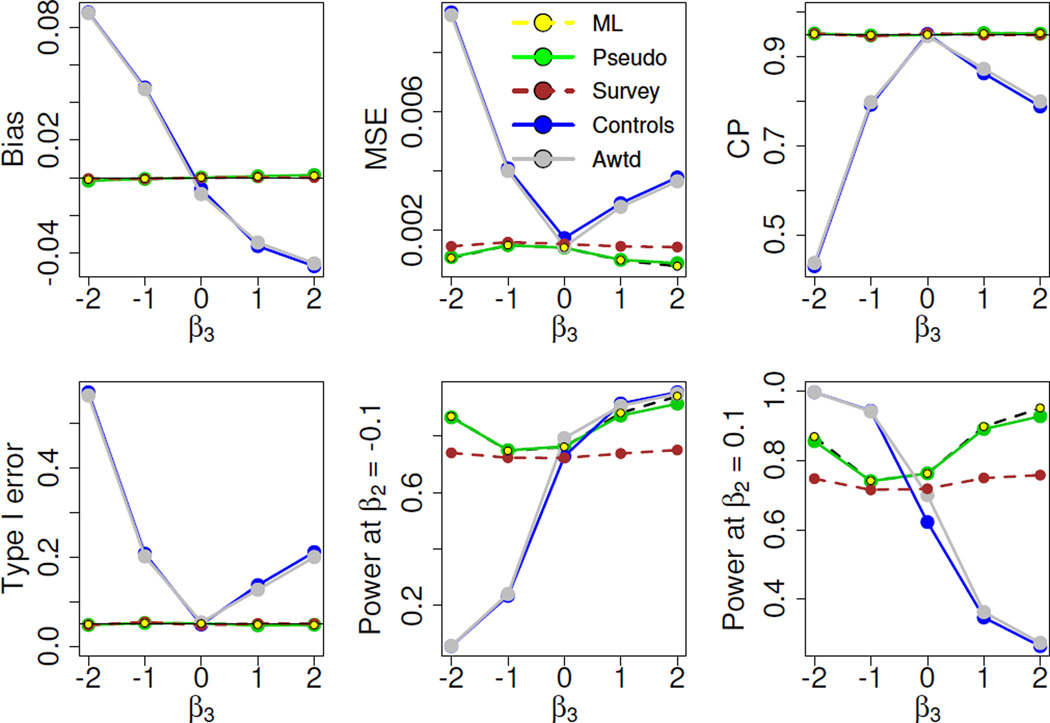
Figures 1 and 2 present the key simulation results for the binary and continuous secondary traits, respectively. The upper row plots the bias, MSE, and coverage probability estimates for the different estimators. In the lower row the estimated Type I error and power are displayed for the corresponding tests under the null and the alternative scenarios. In Figures 1 and 2 we demonstrate the performance of the likelihood-based methods in comparison with only three other methods: the controls-only, adaptively weighted, and survey approaches. The remaining methods (naïve, cases-only, weighted and adjusted), are hugely biased, exhibit extremely large MSEs, and suffer from poor coverage (Figure 4 in Appendix).
Figure 1.
Binary secondary trait
Figure 2.
Continuous secondary trait
We observe the same performance patterns for binary (Figure 1) and continuous (Figure 2) secondary traits. The leftmost plot in the upper row displays the bias incurred by the different estimators. The controls-only and the adaptively weighted methods are considerably biased for non-zero values of β3. The likelihood-based and survey-weighted estimators are essentially unbiased. The corresponding MSEs for the estimators are shown in the middle plot of the upper row. Both the ML and pseudo-likelihood estimators, as well as the survey-weighted estimator, have smaller mean squared errors than the controls-only and the adaptively weighted estimators for most β3 values examined. In particular, the two likelihood-based estimators have practically the same MSEs and perform slightly better than the survey-weighted estimator, especially for the continuous trait. The rightmost plot in the upper row presents the estimated coverage probabilities. The coverage for the controls-only and the adaptively weighted methods drops considerably for large β3. For the confidence intervals corresponding to the likelihood-based and survey methods, the coverage is maintained at the nominal level.
Only the likelihood and the survey approaches maintain the correct Type I error throughout. The power for the Wald tests corresponding to the likelihood and survey methods ranges between 62% to 75% for the binary setup, and varies roughly from 71% to 95% for the continuous setup, whereas the controls-only and the adaptive methods have power as low as 6% (21%) to as high as 100% (95%) for the continuous (binary) trait. But their apparently superior power for large values of β3 is largely illusory, as they suffer from substantially inflated Type I error for those β3 values. The key message from the simulations is that the relative advantage of the likelihood and the survey methods over the controls-only and the adaptively weighted methods is preserved across the simulation setups. Furthermore, the likelihood methods can have substantial power advantage over the survey method, evident in the plots for the continuous secondary trait. The mean squared error estimates for the continuous trait suggest that the likelihood estimators can be almost one and a half times more efficient than the survey-weighted estimator. The performances of the ML and pseudo-likelihood estimators are comparable.
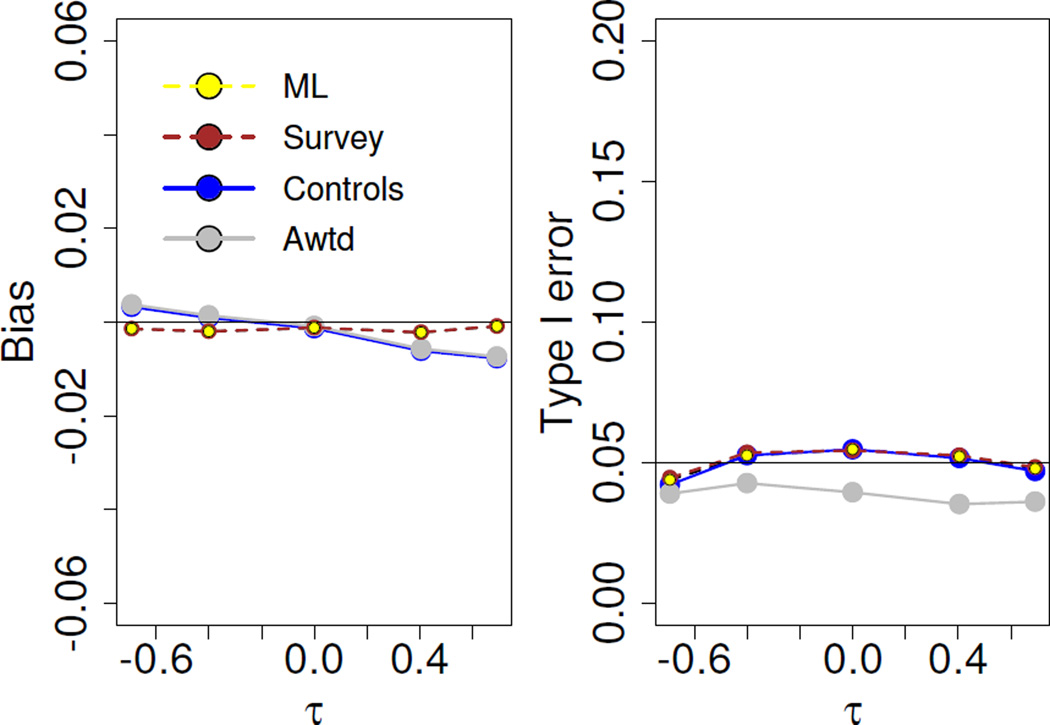
4.2.1 Evaluation of robustness
We illustrate the robustness of the proposed ML method by generating data for the binary secondary trait under an alternative model. Earlier, we had simulated data from the Palmgren model. Here we generate data from the following bivariate model, proposed by Lin and Zeng (2009),
We note that the disease model above does not preserve the standard logistic marginal distribution for primary disease risk and does not allow for interaction between the genetic variant and the secondary trait on disease risk. The simulation setup is as before. We vary the correlation between the primary and secondary phenotypes, denoted by τ in the disease model, between −0.7 and 0.7. We note that estimates from all the methods are practically unbiased and that Type I error is maintained; the adaptively weighted method is slightly conservative.
5 Discussion
The recent attention to the analysis of secondary traits in case/control genome scans has spurred a variety of methodological efforts. However, it is not clear that the genetics literature has utilized the full range of longstanding methods, including survey-based weighted estimators (Scott and Wild, 2002; Binder, 1983), the Palmgren model (Palmgren, 1989) and the work of Lee et al. (1998). The paper by Jiang et al. (2006) provided a relatively complete taxonomy of various approaches to the problem, but key choices in the modeling of primary and secondary effects had remained open. The more recent work of Lin and Zeng (2009); Li et al. (2010); Li and Gail (2012); He et al. (2012); Wang and Shete (2011, 2012) have provided specific solutions, relevant for the genomic scan context, but with constraints such that it is not clear that a comprehensive approach has been available.
Our likelihood approach is designed to encompass the data types typically encountered for genome scans. The proposed framework can handle continuous secondary traits, as well as the binary traits which have received more attention. In addition, our models allow for interaction between the genetic variant and the secondary trait on primary disease risk. We do not require the rare-disease assumption, i.e., our method can be used to to analyze secondary phenotypes for data from case-control studies of both rare and common primary diseases. We have developed our approach in the context of genome scans, but it is relevant to any standard case/control design. However, our joint model may be particularly useful in a genetic context, for which the parameters are interpretable quantities elucidating genetic effects on risk for both traits, as well as additional trait-trait correlation unexplained by the SNP.
Although demonstrated for a single SNP in this paper, our method can, in principle, be applied to genome-wide association data. However, it will be computationally challenging, as will also be any other retrospective method handling covariates nonparametrically. Derivative-based numerical optimization techniques, as well as C routines for numerical optimization, are likely to provide a faster solution. To reduce computational burden of a genome scan, we can employ a two-stage approach whereby we first screen SNPs using one of the faster but reasonably accurate methods, such as the survey approach, to identify SNPs that might be potentially significant and then follow-up on the interesting SNPs using the ML method.
Our formulation of the joint model for the two traits arises from viewing the pair as a bivariate response, and differs from the usual models based on conditional factorization of the joint distribution. In particular, specifying a logistic model for the conditional distribution of disease status given the secondary trait distorts the natural logistic marginal distribution for primary disease risk, resulting in incompatible models for the primary trait when performing primary vs. secondary analysis. Our parameterization of the joint distribution respects the conventional logistic choice for the marginal distribution of the disease trait, whether the secondary trait is binary or continuous. For binary secondary traits, this property is achieved using a well-known bivariate (Palmgren) logistic model. For continuous secondary traits we achieve the intended marginal model via a two-stage latent variable approach. It turns out that our model for the continuous trait is a special case of the Gaussian copula model (He et al., 2012) for a single normally distributed secondary phenotype.
We emphasize that we provide algorithms that allow us to deal with covariates nonparametrically. In our approximate profile likelihood approach, we reparameterize the profile likelihood to provide a closed form expression eliminating the need for explicit maximization over high-dimensional nuisance parameters, thus moving a step forward from theoretically discussing profile likelihood approaches to handling covariates nonparametrically. We present the pseudo-likelihood approach as another attractive way of dealing with the covariate distribution. We note that both the likelihood approaches yield almost identical results. The closed form expression for the reparameterized profile likelihood can be treated just like a likelihood to obtain the ML estimate and its standard error. The pseudo-likelihood approach, on the other hand, is intuitively very appealing and easily provides estimates; but variance estimation adds to the computational complexity. Yet another option would be to use Bayesian techniques. As one reviewer pointed out, we can use a Dirichlet prior on the probability masses ψ = {ψ1, ψ2, …, ψL} for Z and then integrate out ψ as in Zhang and Liu (2007). However, in absence of prior knowledge on covariate distribution, the likelihood and Bayesian approaches will provide very similar results.
We have assumed the genetic and environmental exposures to be independently distributed in the underlying population. We emphasize that the independence assumption applies to the general population, so that after selection by case-control status, they may be dependent (due to their common dependence on D). Thus the assumption is not as restrictive as it may seem. Moreover, for specific genetic applications, the assumption may be reasonable. For random environmental exposures, for instance, it may be entirely reasonable to assume that the genotype does not cause the exposure. The reciprocal assumption is also very standard, i.e. the exposure does not ”cause” genotype, which is fixed from conception. However, if gene-environment independence assumption seems biologically implausible, we can easily relax this assumption. Earlier, we used Z to denote covariates and G the SNP genotype. Instead, Z will now contain all genetic and environmental factors together, including gene-environment interactions, and we model P(z) nonparametrically as before. On the other hand, if it is reasonable to assume independence of genetic and environmental exposures in the population, our framework allows us to exploit this and still leave the distribution of the environmental exposures to be nonparametric.
Our setup lends itself to performing corrections for secondary trait risk estimates which are subject to significance bias, commonly known as the “winner’s curse”. Such bias correction is necessary if the SNPs have been initially selected based on association with the primary trait, and approaches based on likelihood models (Ghosh et al., 2008) can be extended to encompass our joint likelihood.
Several proposed methods for secondary analysis require knowledge of the disease prevalence. The survey approach uses the ratio of sampling fractions, a function of disease prevalence, for appropriate weighting of cases and the controls. The method of moments approach proposed by Wang and Shete (2011) involves knowing the prevalence for the secondary trait in addition to the disease prevalence. In principle, likelihood-based methods do not require specification of the disease prevalence. However, when disease prevalence is assumed unknown, the estimation of intercept parameter α1 can lead to numerically unstable solutions, as has been noted by others (Lin and Zeng, 2009; Chatterjee and Carroll, 2005). In practice, a sensitivity analysis is warranted to determine the degree to which inference is affected by the assumed prevalence, and robust approaches to estimate the prevalence in the context of our models is an important area for further exploration.
Supplementary Material
Figure 3.
Performance under alternative model
Acknowledgements
The authors appreciate the insightful and constructive comments made by the two anonymous reviewers. The authors thank Dr. Ravi Varadhan for helpful suggestions regarding numerical optimization in R. Part of Dr. Ghosh’s research was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute.
Appendix
7.1 The profile log-likelihood
Let n+++l denote the number of observations in the sample with Z = zl, l = 1, …, L. The retrospective log-likelihood in (4) can then be expressed as
| (8) |
Using a Lagrange multiplier λ to account for the constraint and then maximizing the retrospective log-likelihood with respect to ψl, we obtain
| (9) |
Multiplying (9) by ψl and summing over l gives us λ = 0 which on substitution results in
| (10) |
where . We note that . Substituting ψl in the expression for μi we have
| (11) |
We substitute (10) in (8) to obtain the following expression that is equivalent to the profile log-likelihood up to two additional nuisance parameters, μi, i = 0, 1,
where μi’s satisfy (11) and is defined as
We note that
| (12) |
Comparing (12) with (11), we realize that (11) can be replaced by . Thus, we can treat l*(η, μ) as a log-likelihood, although it is not an actual log-likelihood, and maximize it with respect to (η, μ) to obtain η̂. We further note that l*(η, μ) can be expressed in terms of η and as follows:
| (13) |
with
7.2 Asymptotic theory for ML estimate
We first show that E(S*(ϕ)) = 0. The proof is presented in the following subsection. We then assume that n goes to infinity with n1/n and n2/n remaining fixed. Under this assumption it can be shown, by expanding S*(ϕ̂) about the true value ϕ and then applying standard procedures, that converges in distribution to a normal random variable with mean zero and covariance matrix
We then show in Subsection 7.2.2 that Cov(S*(ϕ)) = J*(ϕ) − Γ(ϕ). This leads to
7.2.1 E(S*(ϕ)) = 0
7.2.2 Cov(S*(ϕ)) = J*(ϕ) − Γ(ϕ)
Using the same argument as before it can be shown that
which implies that
We have also shown before that
where . Then
∴
7.3 Asymptotic theory for pseudo-likelihood estimate
Gong and Samaniego (1981), under some regularity conditions, showed that θ̃2 is consistent when θ̃1 is consistent. Also, suppose that
where is the true value of (θ1, θ2)′ and
Then, under some regularity conditions, discussed in Gong and Samaniego,
with
| (14) |
where
I22 is equal to the variance-covariance matrix of the score function for θ2 evaluated at and is the limiting variance-covariance matrix of .
7.3.1 Variance Estimation
We use
and
We note that
If θ̃1 is the ML estimate obtained by maximizing g(θ1), by Taylor expansion, is asymptotically equivalent to
which leads to
We can easily estimate the asymptotic variance of θ̃1 by
Now we have only to estimate . If we can write
and
then we can use the following to estimate the covariance,
Thus,
Also,
Plugging in all the corresponding estimates in (14) gives
| (15) |
References
- Bahadur R. A representation of the joint distribution of responses to n dichotomous items. In: Solomon H, editor. In Studies in Item Analysis and Prediction. Stanford, California: Stanford University Press; 1959. Stanford Mathematical Studies in the Social Sciences VI. [Google Scholar]
- Binder D. On the variances of asymptotically normal estimators from complex surveys. International Statistical Review/Revue Internationale de Statistique. 1983:279–292. [Google Scholar]
- Chatterjee N, Carroll R. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92:399–418. [Google Scholar]
- Frayling T, Timpson N, Weedon M, Zeggini E, Freathy R, Lindgren C, Perry J, Elliott K, Lango H, Rayner N, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Zou F, Wright F. Estimating odds ratios in genome scans: an approximate conditional likelihood approach. The American Journal of Human Genetics. 2008;82:1064–1074. doi: 10.1016/j.ajhg.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Samaniego F. Pseudo maximum likelihood estimation: theory and applications. The Annals of Statistics. 1981:861–869. [Google Scholar]
- He J, Li H, Edmondson A, Rader D, Li M. A gaussian copula approach for the analysis of secondary phenotypes in case–control genetic association studies. Biostatistics. 2012;13:497–508. doi: 10.1093/biostatistics/kxr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Scott AJ, Wild CJ. Secondary analysis of case-control data. Stat Med. 2006;25:1323–1339. doi: 10.1002/sim.2283. [DOI] [PubMed] [Google Scholar]
- Lee A, McMURCHY L, Scott A. Re-using data from case-control studies. Statistics in medicine. 1998;16:1377–1389. doi: 10.1002/(sici)1097-0258(19970630)16:12<1377::aid-sim557>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Li H, Gail M. Efficient adaptively weighted analysis of secondary phenotypes in case-control genome-wide association studies. Human Heredity. 2012;73:159–173. doi: 10.1159/000338943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gail M, Berndt S, Chatterjee N. Using cases to strengthen inference on the association between single nucleotide polymorphisms and a secondary phenotype in genome-wide association studies. Genetic epidemiology. 2010;34:427–433. doi: 10.1002/gepi.20495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Zeng D. Proper analysis of secondary phenotype data in case-control association studies. Genetic epidemiology. 2009;33:256–265. doi: 10.1002/gepi.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meester S, MacKay J. A parametric model for cluster correlated categorical data. Biometrics. 1994;50:954–963. [PubMed] [Google Scholar]
- Monsees G, Tamimi R, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genetic epidemiology. 2009;33:717–728. doi: 10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi R, Chatterjee N, Church T, Chen J, Yeager M, Weissfeld J, Hein D, Hayes R. Cigarette smoking, N-acetyltransferase genes and the risk of advanced colorectal adenoma. Pharmacogenomics. 2006;7:819–829. doi: 10.2217/14622416.7.6.819. [DOI] [PubMed] [Google Scholar]
- Nagelkerke N, Moses S, Plummer F, Brunham R, Fish D. Logistic regression in case-control studies: The effect of using independent as dependent variables. Statistics in medicine. 1995;14:769–775. doi: 10.1002/sim.4780140806. [DOI] [PubMed] [Google Scholar]
- Palmgren J. Regression models for bivariate binary responses; UW Biostatistics Working Paper Series; 1989. p. 101. [Google Scholar]
- Prentice R, Pyke R. Logistic disease incidence models and case-control studies. Biometrika. 1979;66:403–411. [Google Scholar]
- Richardson DB, Rzehak P, Klenk J, Weiland SK. Analyses of case-control data for additional outcomes. Epidemiology. 2007;18:441–445. doi: 10.1097/EDE.0b013e318060d25c. [DOI] [PubMed] [Google Scholar]
- Scott A, Wild C. Fitting logistic regression models in stratified case-control studies. Biometrics. 1991:497–510. [Google Scholar]
- Scott A, Wild C. Fitting regression models to case-control data by maximum likelihood. Biometrika. 1997a;84:57–71. [Google Scholar]
- Scott A, Wild C. Maximum likelihood for generalised case-control studies. Journal of Statistical Planning and Inference. 2001;96:3–27. [Google Scholar]
- Scott A, Wild C. On the robustness of weighted methods for fitting models to case-control data. Journal of the Royal Statistical Society. Series B, Statistical Methodology. 2002:207–219. [Google Scholar]
- Scott AJ, Wild CJ. Fitting regression models to case-control data by maximum likelihood. Biometrika. 1997b;84:57–71. [Google Scholar]
- Spitz M, Amos C, Dong Q, Lin J, Wu X. The chrna5-a3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. Journal of the National Cancer Institute. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shete S. Estimation of odds ratios of genetic variants for the secondary phenotypes associated with primary diseases. Genetic epidemiology. 2011;35:190–200. doi: 10.1002/gepi.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shete S. Analysis of secondary phenotype involving the interactive effect of the secondary phenotype and genetic variants on the primary disease. Annals of Human Genetics. 2012;76:484–499. doi: 10.1111/j.1469-1809.2012.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon M, Lango H, Lindgren C, Wallace C, Evans D, Mangino M, Freathy R, Perry J, Stevens S, Hall A, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nature Genetics. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu J. Bayesian inference of epistatic interactions in case-control studies. Nature genetics. 2007;39:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.