Abstract
Background
Previous studies in occupational exposure and lung function have focused only on the main effect of occupational exposure or genetics on lung function. Some disease-susceptible genes may be missed due to their low marginal effects, despite potential involvement in the disease process through interactions with the environment. Through comprehensive genome-wide gene-environment interaction studies, we can uncover these susceptibility genes. Our objective in this study was to explore gene by occupational exposure interaction effects on lung function using both the individual SNPs approach and the genetic network approach.
Methods
The study population comprised the Offspring Cohort and the Third Generation from the Framingham Heart Study. We used forced expiratory volume in one second (FEV1) and ratio of FEV1 to forced vital capacity (FVC) as outcomes. Occupational exposures were classified using a population-specific job exposure matrix. We performed genome-wide gene-environment interaction analysis, using the Affymetrix 550 K mapping array for genotyping. A linear regression-based generalized estimating equation was applied to account for within-family relatedness. Network analysis was conducted using results from single-nucleotide polymorphism (SNP)-level analyses and from gene expression study results.
Results
There were 4,785 participants in total. SNP-level analysis and network analysis identified SNP rs9931086 (Pinteraction =1.16 × 10-7) in gene SLC38A8, which may significantly modify the effects of occupational exposure on FEV1. Genes identified from the network analysis included CTLA-4, HDAC, and PPAR-alpha.
Conclusions
Our study implies that SNP rs9931086 in SLC38A8 and genes CTLA-4, HDAC, and PPAR-alpha, which are related to inflammatory processes, may modify the effect of occupational exposure on lung function.
Background
Chronic obstructive pulmonary disease (COPD) is defined by decreased lung function, commonly measured by forced expiratory volume in one second (FEV1) and the ratio of FEV1 to forced vital capacity (FVC). Cigarette smoking is the most important environmental risk factor for COPD. However, COPD development in smoking populations is not universal. In a study conducted by the American Thoracic Society [1], approximately 15% of COPD may be attributable to workplace exposures; a similar estimate was reported in a more recent study [2]. Indeed, an association between COPD or poor lung function and occupational exposure to dust, gas, and fumes has been reported in both industry-based studies [3-6] and community-based [7-11] cross-sectional studies. In addition, mounting evidence indicates that genetic factors contribute to COPD. In fact, the disease aggregates in families [12], and genome-wide association studies (GWAS) [13-17] have identified single nucleotide polymorphisms (SNPs) that are associated with COPD or lung function.
To date, few study has comprehensively examined combined genetic and environmental effects on lung function. GWAS on lung function have considered genetic effects alone; a recent review [18] showed that most GWAS findings do not replicate consistently, which may imply a role played by environmental exposures and gene-environmental interactions. Studies of occupational exposure have only investigated the main effect of exposure on lung function. Therefore, some disease susceptibility genes may be missed due to low marginal effects, despite being involved in the disease process through interactions with environmental exposures [19]. Further, the majority of gene-environment interaction studies of lung function or lung function has focused on gene by smoking interaction [20-23]. However, no reports describe gene by occupational exposure interactions. In addition, most researchers investigated SNPs or genes through either in silico statistical methods or by gene expression experiments, but did not integrate these results. Thus, an integrated, genome-wide gene-environment interaction study can more readily identify disease susceptibility genes when environmental factors may also be contributing.
The purpose of this study was to investigate gene by occupational exposure interactions on FEV1 and FEV1/FVC thorough SNP-level analysis and network analysis. Specifically, we first performed a GWAS using data from the Framingham Heart Study (FHS) to identify SNPs that interact with occupational exposure using a population-specific job exposure matrix (JEM) to affect (FEV1) and ratio of FEV1 to FVC. We next combined our GWAS SNP results with gene expression results to build a network of biological processes that are driven by networks and not by individual genes. The findings from these analyses uncovered several SNPs/Genes that may offer avenues for future functional studies of genes contributing to lung function changes following occupational exposures.
Methods
Study population
Our study population derives from the FHS [24], which includes only Caucasians. This study has recruited participants since 1948; there have been three generations of participants: the Original Cohort, their Offspring, and the Third Generation. Spirometry measurements, detailed medical history, physical examinations, and laboratory tests were done approximately every two years. We used the 4,785 participants with complete spirometry phenotypes, occupational information, genotypes, and related covariates from the Offspring Cohort and the Third Generation Cohort.
Ethics statement
Written informed consents were provided by all participants. Protocols were approved by local institutional review boards.
Spirometry phenotypes and covariates
Spirometry from participant lung function Exam 8 and the Third Generation Exam were used in our study. We used the FEV1 and FEV1 ratio (FEV1/FVC) as continuous outcomes. Age, gender, height (inch), pack-years, and smoking status were used as covariates in our analysis. Smoking status (never, former, and current smokers) was coded as dummy variable.
Genotyping and quality control
Genotyping for 500,568 SNPs was conducted with approximately 550 K SNPs using the Affymetrix 500 K mapping array plus Affymetrix 50 K supplemental array in 9,237 subjects from the three generations of participants. We used 4,785 subjects from two generations in our study. A quality test was conducted using the PLINK software (version1.06, http://pngu.mgh.harvard.edu/~purcell/plink/). A total of 499 individuals with genotyping call-rate <95% were deleted, and the genotyping rate in the remaining individuals was 98.6%. We conducted the Hardy-Weinberg test for all SNPs, and found 19,546 SNPs had a p-value <1 × 10-6. These SNPs showed an obvious deviation from the 45-degree line of a QQ plot and were excluded from our analysis. A total of 34,110 SNPs had a per-SNP missing rate >5% among all subjects and were excluded. We also excluded 146,203 SNPs with minor allele frequency lower than 5% in our study subjects. After filtering, 300,709 SNPs remained for analysis.
Occupational exposure
We used a job exposure matrix (JEM) for occupational exposure assessment. Occupational exposure was classified as high versus low likelihood for dust exposure (coded as 1 and 0) according to their job categories based on a questionnaire in Offspring Exam 8 and the Third Generation Exam (Table 1). The questionnaire asked, “Using the occupation coding sheet choose the code that best describes your occupation”. There are 29 job categories (exclude retired) on the FHS occupation coding sheet, and we classified 4 of them as highly likely dust exposure including factory/assembly/mechanic, skilled labor, general labor, and heavy labor, as modified from UCSF COPD JEM (January, 2009 revision) [25].
Table 1.
Job categories for dust exposure classification
| Dust exposure group | Job category |
|---|---|
| Highly likely dust exposure |
Factory/assembly/mechanic |
| Skilled labor (e.g., plumber, carpenter, painter, hairdresser) | |
| General labor (e.g., custodian, delivery, mailman, truck driver) | |
| Heavy labor (e.g., construction, landscaping) | |
| Less likely dust exposure |
Nurse/medical personnel |
| Laboratory technician | |
| Physical/occupational/speech therapist | |
| Police/fire/security/military | |
| Restaurant/food worker | |
| Homemaker | |
| Self-employed business owner | |
| M.D./dentist/scientist/research | |
| Lawyer/judge | |
| Psychologist/social worker/mental health counselor | |
| Engineer/computer science | |
| Banker/accountant | |
| Manager/consultant (e.g., production manager) | |
| Administrative (e.g., personnel) | |
| Educator | |
| Secretary/clerk/data entry | |
| retail/cashier | |
| Sales/marketing/insurance | |
| Realtor | |
| Writer/editor/artist/graphic designer/craftsperson | |
| Musician | |
| Clergy (minister, priest, rabbi) | |
| Sports pro/coach/exercise instructor/other | |
| Statistician | |
| Student |
Statistical analysis
SNP-level analysis
The family-based cohort was analyzed using linear regression-based generalized estimating equations implemented in the GWAF package for R [26] to account for within-family relatedness. The within-pedigree correlation matrix was modeled using an exchangeable working correlation matrix. We analyzed each SNP separately using FEV1, and FEV1/FVC as outcomes. For each SNP, we included age, gender, height (inch), pack-years, smoking status, occupational exposure status, and tested for the main SNP effect. We next fit an interaction model by adding an SNP by occupational exposure status interaction to the model, and tested for the SNP-exposure interaction.
Network analysis
To identify important networks among the genes that interact with occupational exposure, we used Metacore software (GeneGo, St Joseph, MI, USA) tools to build networks using the gene lists. One gene list was obtained from a previous study [27], in which human airway epithelial cells obtained from 6 normal individuals were exposed to coarse, fine, or ultrafine particulate matter for 6 and 24 hours before gene expression was assessed. The authors reported 71 unique genes altered by the particulate matter (PM). To the best of our knowledge, no other studies have focused on the effects of dust exposure or PM on gene expression in human airway cells. Another list of genes was generated from our results. We chose SNPs whose interactions with occupational exposure had p-values smaller than 10-3 (around top 0.1% of SNPs), and mapped these SNPs to genes to generate a gene list. Gene annotation was performed using the gene prediction track “RefSeqGenes” in the UCSC browser (http://genome.ucsc.edu).
Results
This study comprised 4,785 participants, with 1,247 participants from the Offspring Cohort and 3,538 participants from the Third Generation. Among them, 4,238 participants were classified in the less likely dust exposure group and 547 participants in the highly likely dust exposure group.
Table 2 summarizes the characteristics for participants by both exposure group and cohort. Mean FEV1 was higher in the group with highly likely dust exposure, but the mean FEV1/FVC was similar in the two groups overall. However, the proportion of COPD cases (defined by FEV1/FVC less than 70%) was higher in the group with highly likely dust exposure. Lung function was higher and the proportion of COPD cases was lower in the Third Generation as compared to the Offspring Generation, most likely attributable to the younger age of participants in the Third Generation.
Table 2.
Characteristic of participants, stratified by cohort and dust exposure groups
| |
Offspring |
Third generation |
Total |
|||
|---|---|---|---|---|---|---|
|
(n = 1247) |
(n = 3538) |
(n = 4785) |
||||
| |
Less likely dust exposure |
Highly likely dust exposure |
Less likely dust exposure |
Highly likely dust exposure |
Less likely dust exposure |
Highly likely dust exposure |
| (n = 1126) | (n = 121) | (n = 3112) | (n = 426) | (n = 4238) | (n = 547) | |
| Male, n (%) |
444 (39.43) |
100 (82.64) |
1298 (41.71) |
378 (88.73) |
1742 (41.10) |
478 (87.39) |
| FEV1, L ± SD |
2.71 ± 0.78 |
3.09 ± 0.75 |
3.54 ± 0.78 |
4.02 ± 0.71 |
3.32 ± 0.86 |
3.81 ± 0.82 |
| FEV1/FVC, % ± SD |
0.73 ± 0.07 |
0.72 ± 0.08 |
0.78 ± 0.06 |
0.77 ± 0.06 |
0.77 ± 0.07 |
0.76 ± 0.07 |
| FEV1/FVC < 70%, n (%) |
337 (29.93) |
43 (35.54) |
318 (10.22) |
49 (11.50) |
655 (15.46) |
92 (16.82) |
| Age, years ± SD |
62.83 ± 8.02 |
60.48 ± 7.87 |
39.81 ± 8.58 |
41.03 ± 8.57 |
45.93 ± 13.21 |
45.33 ± 11.66 |
| Height, inches ± SD |
65.81 ± 3.76 |
67.99 ± 3.46 |
66.90 ± 3.65 |
69.29 ± 2.98 |
66.61 ± 3.71 |
69.01 ± 3.14 |
| Pack-years* ± SD |
21.40 ± 21.58 |
30.73 ± 23.32 |
12.56 ± 13.61 |
19.12 ± 15.90 |
15.50 ± 17.19 |
22.07 ± 18.74 |
| Smoking status, n (%) | ||||||
| Never smokers |
501 (44.49) |
36 (29.75) |
1854 (59.58) |
177 (41.55) |
2355 (55.57) |
213 (38.94) |
| Former smokers |
522 (46.36) |
58 (47.93) |
789 (25.35) |
115 (27.00) |
1311 (30.93) |
173 (31.63) |
| Current smokers | 103 (9.15) | 27 (22.31) | 469 (15.07) | 134 (31.46) | 572 (13.50) | 161 (29.43) |
*Pack-years mean and standard deviation calculated among current and former smokers.
SNP-level analysis
The genome-wide analysis for 300,709 SNPs (Bonferroni correctionα = 1.66 × 10-7) detected a significant interaction effect of SNP rs9931086 in gene SLC38A8 on chromosome 16q23.3 and occupational exposure on FEV1 (Pinteraction = 1.16 × 10-7). Further, gene-environment interaction effects of SNPs rs3889785 and rs4234966 in the MARCH1 gene and SNPs rs17508671, rs17430621, and rs17508706 in the ZNF804A gene and occupational exposure on FEV1 also had small P values (all Pinteraction < 3.04 × 10-5). The genomic inflation factor value of 1.045 calculated from the QQ plot suggested the p-values were appropriately distributed. The results for the top 30 SNPs with strongest occupational exposure interaction effects on FEV1 are shown in Table 3 and the Manhattan plot is shown in Figure S1 (see Additional file 1).
Table 3.
Top 30 strongest SNPs interacting with occupational exposure to affect FEV 1
| SNP | Chr | Position | Band | Gene | All1 | All2 | MAF | β SNP | P SNP | β interaction | P interaction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9931086 |
16 |
84046105 |
q23.3 |
SLC38A8 |
A |
C |
0.19 |
27.70 |
2.53E-02 |
-214 |
1.16E-07* |
| rs11252277 |
10 |
4065704 |
p15.1 |
|
G |
A |
0.09 |
2.90 |
1.05E-01 |
-263 |
6.32E-07 |
| rs3889785 |
4 |
165097529 |
q32.3 |
MARCH1 |
A |
C |
0.13 |
-2.02 |
1.69E-01 |
237 |
1.05E-06 |
| rs7098287 |
10 |
4050329 |
p15.1 |
|
G |
C |
0.09 |
3.60 |
4.88E-02 |
-247 |
2.47E-06 |
| rs32977 |
5 |
14119057 |
p15.2 |
|
A |
G |
0.06 |
5.50 |
7.38E-03 |
-310 |
3.98E-06 |
| rs17508671 |
2 |
185491847 |
q32.1 |
ZNF804A |
A |
G |
0.16 |
-0.27 |
8.32E-01 |
-173 |
4.45E-06 |
| rs2544785 |
19 |
49085710 |
q13.33 |
SULT2B1 |
C |
T |
0.16 |
0.03 |
0.983642 |
172 |
4.57E-06 |
| rs4234966 |
4 |
165078580 |
q32.3 |
MARCH1 |
C |
G |
0.12 |
-1.92 |
1.88E-01 |
217 |
6.35E-06 |
| rs32978 |
5 |
14119645 |
p15.2 |
|
C |
T |
0.05 |
5.05 |
1.75E-02 |
-302 |
6.63E-06 |
| rs32979 |
5 |
14119775 |
p15.2 |
|
T |
C |
0.05 |
5.05 |
1.75E-02 |
-302 |
6.63E-06 |
| rs17645582 |
6 |
82797317 |
q14.1 |
|
C |
T |
0.07 |
-1.07 |
5.75E-01 |
208 |
6.89E-06 |
| rs4932559 |
15 |
92133594 |
q26.1 |
|
A |
G |
0.36 |
-1.62 |
0.081898 |
132 |
7.10E-06 |
| rs745926 |
2 |
120482057 |
q14.2 |
|
T |
G |
0.41 |
-1.13 |
2.24E-01 |
136 |
9.90E-06 |
| rs1561577 |
15 |
89146968 |
q26.1 |
|
A |
G |
0.07 |
-3.17 |
0.104592 |
224 |
1.16E-05 |
| rs6445707 |
3 |
54881562 |
p14.3 |
CACNA2D3 |
G |
A |
0.18 |
0.40 |
7.47E-01 |
166 |
1.17E-05 |
| rs2544784 |
19 |
49085733 |
q13.33 |
SULT2B1 |
A |
T |
0.16 |
0.00 |
0.997879 |
161 |
1.25E-05 |
| rs17110400 |
14 |
26276467 |
q12 |
|
C |
T |
0.09 |
-0.31 |
0.855973 |
265 |
1.53E-05 |
| rs10231843 |
7 |
103884705 |
q22.2 |
|
C |
G |
0.27 |
-1.10 |
3.09E-01 |
160 |
1.64E-05 |
| rs17430621 |
2 |
185492469 |
q32.1 |
ZNF804A |
A |
T |
0.16 |
0.02 |
9.85E-01 |
-165 |
1.89E-05 |
| rs163814 |
5 |
14122497 |
p15.2 |
|
C |
T |
0.05 |
5.43 |
1.16E-02 |
-285 |
1.94E-05 |
| rs477920 |
1 |
74767844 |
p31.1 |
FPGT TNNI3K |
A |
G |
0.17 |
0.23 |
8.55E-01 |
-183 |
2.04E-05 |
| rs6972824 |
7 |
157473222 |
q36.3 |
PTPRN2 |
C |
A |
0.11 |
2.38 |
8.22E-02 |
-196 |
2.30E-05 |
| rs6124623 |
20 |
42746332 |
q13.12 |
JPH2 |
C |
G |
0.14 |
1.34 |
3.00E-01 |
-172 |
2.34E-05 |
| rs2832081 |
21 |
30152992 |
q21.3 |
|
C |
A |
0.31 |
-2.33 |
0.01353 |
137 |
2.36E-05 |
| rs7356986 |
6 |
12301462 |
p24.1 |
|
G |
A |
0.11 |
-1.68 |
3.20E-01 |
202 |
2.36E-05 |
| rs41328144 |
1 |
166319711 |
q24.1 |
|
C |
T |
0.13 |
-0.68 |
6.35E-01 |
155 |
2.54E-05 |
| rs2803543 |
10 |
88301331 |
q23.2 |
|
G |
A |
0.10 |
-2.35 |
1.49E-01 |
224 |
2.90E-05 |
| rs163813 |
5 |
14122154 |
p15.2 |
|
G |
A |
0.05 |
5.00 |
1.88E-02 |
-281 |
2.96E-05 |
| rs17508706 |
2 |
185524591 |
q32.1 |
ZNF804A |
G |
A |
0.16 |
-0.14 |
9.12E-01 |
-162 |
3.04E-05 |
| rs4130671 | 7 | 119277137 | q31.31 | A | C | 0.49 | -1.37 | 1.27E-01 | 122 | 3.71E-05 |
SNP-estimates are based on an additive model. Beta-estimates represent the increase or decrease FEV1 in mL.
*Significant after Bonferroni correction for testing 300,709 SNPs (α = 1.66E-07).
Chr: Chromosome; All1: allele1; All2: allele2.
For gene by occupational exposure interaction effects on FEV1/FVC, although no SNP reached the Bonferroni corrected significance level (α = 1.66 × 10-7), SNPs rs7297210, rs6486961, rs7297431, and rs1875467 in gene AEBP2 and SNPs rs10751811, rs2173524, and rs11595576 in ADARB2 had small P values (all Pinteraction < 4.32 × 10-5). In addition, SNP rs7314308 had small P values for both the main genetic effect (PSNP = 5.43 × 10-5) and the gene by occupational exposure effect (Pinteraction = 1.20 × 10-5). The genomic inflation factor value of 1.043 suggested the p-values were appropriately distributed. The results for the top 30 SNPs with strongest occupational exposure interaction effects on FEV1/FVC are shown in Table 4 and the Manhattan plot is shown in Figure S2 (see Additional file 1: Figure S2).
Table 4.
Top 30 strongest SNPs interacting with occupational exposure to affect FEV 1 /FVC
| SNP | Chr | Position | Band | Gene | All1 | All2 | MAF | β SNP | P SNP | β interaction | P interaction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17051547 |
4 |
53421172 |
q12 |
|
C |
A |
0.13 |
-0.44 |
3.31E-02 |
2.84 |
2.86E-06 |
| rs17051550 |
4 |
53425943 |
q12 |
|
C |
G |
0.11 |
-0.38 |
8.06E-02 |
2.91 |
6.01E-06 |
| rs11671079 |
19 |
29900846 |
q12 |
LOC284395 |
G |
A |
0.46 |
-0.10 |
0.397666 |
1.55 |
8.65E-06 |
| rs2804576 |
10 |
113775409 |
q25.2 |
|
A |
G |
0.13 |
0.34 |
9.56E-02 |
-2.83 |
1.01E-05 |
| rs7314308 |
12 |
59684714 |
q14.1 |
|
G |
A |
0.19 |
0.65 |
5.43E-05 |
-2.09 |
1.20E-05 |
| rs7297210 |
12 |
19630207 |
p12.3 |
AEBP2 |
T |
C |
0.40 |
0.18 |
2.22E-01 |
-1.62 |
1.53E-05 |
| rs2655364 |
18 |
26229339 |
q12.1 |
|
T |
A |
0.46 |
-0.20 |
1.43E-01 |
1.76 |
1.55E-05 |
| rs4322259 |
1 |
64339246 |
p31.3 |
ROR1 |
C |
G |
0.35 |
0.21 |
1.59E-01 |
-1.72 |
1.61E-05 |
| rs1504108 |
18 |
26224965 |
q12.1 |
|
A |
G |
0.45 |
-0.20 |
1.56E-01 |
1.76 |
1.82E-05 |
| rs2655360 |
18 |
26231390 |
q12.1 |
|
T |
C |
0.45 |
-0.20 |
1.54E-01 |
1.73 |
2.23E-05 |
| rs987124 |
18 |
26217351 |
q12.1 |
|
G |
A |
0.46 |
-0.20 |
1.38E-01 |
1.71 |
2.54E-05 |
| rs9878581 |
3 |
37969695 |
p22.2 |
CTDSPL |
C |
G |
0.13 |
-0.37 |
6.85E-02 |
2.23 |
2.92E-05 |
| rs10751811 |
10 |
1687171 |
p15.3 |
ADARB2 |
A |
G |
0.36 |
-0.17 |
2.46E-01 |
1.53 |
3.06E-05 |
| rs6486961 |
12 |
19626815 |
p12.3 |
AEBP2 |
A |
G |
0.40 |
0.13 |
3.60E-01 |
-1.59 |
3.13E-05 |
| rs6941466 |
6 |
168929950 |
q27 |
SMOC2 |
T |
C |
0.22 |
-0.21 |
2.54E-01 |
2.03 |
3.21E-05 |
| rs7297431 |
12 |
19600232 |
p12.3 |
AEBP2 |
T |
C |
0.45 |
0.15 |
2.75E-01 |
-1.59 |
3.30E-05 |
| rs10770501 |
12 |
19683476 |
p12.3 |
|
G |
A |
0.46 |
0.16 |
2.51E-01 |
-1.57 |
3.35E-05 |
| rs537693 |
6 |
159662136 |
q25.3 |
FNDC1 |
G |
A |
0.12 |
-0.11 |
6.18E-01 |
2.28 |
3.36E-05 |
| rs2173524 |
10 |
1687799 |
p15.3 |
ADARB2 |
T |
G |
0.35 |
-0.18 |
2.46E-01 |
1.51 |
3.41E-05 |
| rs1105791 |
9 |
81179317 |
q21.31 |
|
C |
A |
0.19 |
-0.36 |
0.026841 |
1.91 |
3.83E-05 |
| rs1875467 |
12 |
19622345 |
p12.3 |
AEBP2 |
C |
A |
0.46 |
0.13 |
3.43E-01 |
-1.54 |
4.06E-05 |
| rs9888671 |
15 |
26277939 |
q12 |
LOC100128714 |
C |
T |
0.13 |
0.17 |
3.79E-01 |
2.01 |
4.09E-05 |
| rs12141717 |
1 |
55388480 |
p32.3 |
|
G |
A |
0.29 |
-0.08 |
5.89E-01 |
1.80 |
4.24E-05 |
| rs11595576 |
10 |
1688497 |
p15.3 |
ADARB2 |
T |
C |
0.36 |
-0.16 |
2.96E-01 |
1.53 |
4.32E-05 |
| rs5996665 |
22 |
24469115 |
q11.23 |
CABIN1 |
G |
A |
0.08 |
0.35 |
8.54E-02 |
-2.65 |
4.41E-05 |
| rs7078873 |
10 |
61787587 |
q21.2 |
ANK3 |
T |
A |
0.38 |
0.30 |
3.34E-02 |
-1.62 |
4.49E-05 |
| rs4938871 |
11 |
57329402 |
q12.1 |
UBE2L6 |
C |
A |
0.26 |
-0.36 |
3.28E-02 |
1.90 |
4.63E-05 |
| rs1885166 |
9 |
17610211 |
p22.2 |
SH3GL2 |
T |
C |
0.39 |
0.10 |
0.487458 |
-1.70 |
4.77E-05 |
| rs10052266 |
5 |
6010711 |
p15.32 |
|
G |
A |
0.12 |
0.32 |
0.094656 |
-2.01 |
4.83E-05 |
| rs2763249 | 6 | 168926062 | q27 | SMOC2 | A | G | 0.22 | -0.21 | 2.38E-01 | 1.97 | 4.89E-05 |
SNP-estimates are based on an additive model. Beta-estimates represent the increase or decrease FEV1/FVC in percentage.
Chr: Chromosome; All1: allele1; All2: allele2.
Networks analysis
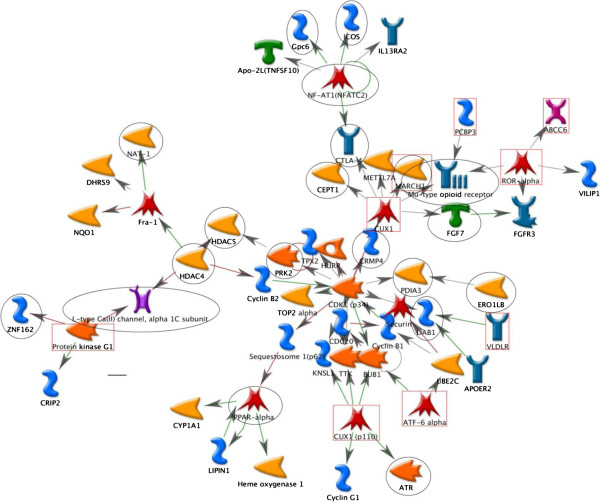
With a cut-off p-value of 10-3, we identified from our GWAS data 450 SNPs in 142 genes that interacted with occupational exposure to affect FEV1 and 360 SNPs in 116 genes that interacted with occupational exposure to affect FEV1/FVC. From our gene list, we found three network clusters consisting of 3, 6, and 13 genes. Integrating the gene list obtained from the gene expression study after exposure to PM, we found a network cluster consisting of 55 genes; the three clusters identified from our gene list are included in this big cluster (Figure 1). The genes obtained from the gene expression results linked all the network clusters together. Some key genes that interacted with occupational exposure in the network were HDAC, CTLA-4, PPAR-alpha, and OPRM1. These genes play important roles in connecting different network clusters into a big network cluster. In addition, we found that all network clusters were centered by the network cluster built from those genes identified from the expression study after exposure to PM.
Figure 1.
Network for genes interacting with occupational exposure to affect lung function. Network analysis revealed network clusters of genes with occupational exposure interaction effects on lung function (circled) that are connected by the network cluster built from genes affected by exposure to PM [27]. Genes modifying the occupational exposure effect on lung function are indicated by circles (FEV1) and rectangles (FEV1/FVC). All other symbols are explained in the Metacore legend (http://ftp.genego.com/files/MC_legend.pdf).
Discussion
To the best of our knowledge, this paper is the first study assessing gene by occupational exposure interactions on lung function using a community-based population. Our results from SNP-level analysis suggest that SNP rs9931086 in gene SLC38A8 may significantly modify the effect of occupational exposure on FEV1. The results of network analysis suggest that genes HDAC, PPAR-alpha, and CTLA-4 play important roles in the gene by occupational exposure interaction network.
The novel locus, SNP rs9931086, in gene SLC38A8 had a significant gene by occupational exposure interaction effect on FEV1, although its genetic marginal effect was not statistically significant. For each C allele increase, the FEV1 decreased by 214 mL. We did not identify this gene through the network analysis; this may result, in part, from missing some genes that connect SLC38A8 to the main gene network. For other top SNPs in our study, SNP rs6941466 in SMOC2 (p = 3.21 × 10-5) was associated with FEV1/FVC through gene by occupational exposure interaction. SMOC2 has been previously reported in both a gene main effect study [28] and a gene by smoking interaction study [23]. Another top SNP, rs1289714 (p = 5.25 × 10-4), in HHIP had a gene by occupational exposure effect modification on FEV1/FVC. The HHIP region was previously highlighted in the GWAS of FEV1/FVC focus on gene main effect using FHS data. The recent gene by smoking interaction on lung function study [23] discovered three gene regions, DNER, HLA-DQB1/HLA-DQA2, and KCNJ2/SOX9, with significant SNPs. However, the p-values for the SNPs in these genes were not statistically significant in our study.
The genes we identified through the network analysis are notable. Histone deacetylases (HDAC) are enzymes that regulate inflammatory gene expression. Several studies have shown the correlation between HDAC activity and COPD disease severity, measured by FEV1 and FEV1/FVC, and that HDAC activity is reduced in COPD patients [29-33]. Our results are consistent with a recent large-scale GWAS that identified an HDAC4 association with lung function [34], which was also found in the gene by smoking interaction study [23].
Another inflammation-related gene, PPAR-alpha, is an isotype of the peroxisome proliferator-activated receptors, which can regulate the induction of inflammatory response [35]. One study showed that COPD patients had higher percentages of PPAR-alpha-positive alveolar macrophages and cells in their alveolar wall in COPD patients [36]. In addition, PPAR-alpha agonists may enhance the inducible isoform of nitric oxide synthase (iNOS) [37], and the expression of iNOS was found to increase in patients with severe COPD [38].
CTLA-4, the cytotoxic T-lymphocyte antigen 4, plays a role in downregulation of T-cell activation [39]. Further, the amount of T-cells correlates with the severity of airway obstruction and alveolar damage [40-43]. Genome-wide linkage analysis of the Boston Early-Onset COPD Study also showed a significant peak for airflow limitation on chromosome 2q, in the region of genes CTLA-4 and ICOS[44,45]. CTLA-4 gene polymorphisms are also associated with chronic bronchitis or COPD [46,47].
In the network analysis, the cluster network built by results of the gene expression experiment after PM exposure was associated with cell cycle regulation. This network of cell cycle regulation connected genes HDAC, PPAR-alpha, CTLA-4 and may imply the possible underlying biological mechanism. Thus, the findings from SNP-level analyses and network analyses complement each other well. The genes found in our study are associated with inflammatory pathways. Genetic variation in the ability to mount an inflammatory response may explain varying individual responses to occupational exposure.
Importantly, we observed two genes through SNP-level analysis (ZNF804A) and network analysis (OPRM1) that interacted with occupational exposure to affect lung function. Although their functions remain to be uncovered, these two genes are significantly associated with many psychiatric diseases (e.g., schizophrenia, alcoholism, and tobacco use disorder) in several studies [48-53]. The comorbidity of abnormal pulmonary function or COPD and psychiatric diseases has been reported in many studies [54,55]. These psychiatric diseases might share some common pathway with COPD. However, we must exercise caution in interpreting these findings because smoking prevalence is higher among psychiatric patients [56]; nonetheless, another study controlling for smoking found schizophrenia to be an independent risk factor for COPD [57].
It should be noted that this study has several limitations. The first limitation is the lack of replication in population studies that use similar methods as the FHS, a community-based study with both occupational information and genome-wide genotyping. Therefore, our results are more exploratory in nature. Further, SNP-coverage was low for certain genes, which may result in missing several genes that might interact with occupational exposures. In addition, the mean FEV1 is higher in the group with highly likely dust exposure. This may be explained by gender distribution, since the proportion of males was higher in the group with high likelihood of dust exposure. Another potential explanation is the healthy worker effect confounding bias [58-60]. Participants in the highly likely dust exposure group were likely those in better health since most dusty jobs (e.g. heavy labor) require workers to be in better health. This bias may result in underestimation of the occupational exposure effect and reduce the effect size of our findings. Finally, our occupational exposure status was classified based on a JEM having less detailed occupational information than the UCSF COPD JEM; thus, our estimates may not reflect the true frequency. The occupational information in our study derives from a cross-sectional questionnaire, which may not reflect the longest-held job (e.g., participants may have worked in dusty job for 10 years but had switched to a non-dusty job by the time of the survey). These measurement errors might result in an underestimation of the gene by occupational exposure interaction association.
Our study indicates that integrating the results of a gene expression experiment into the GWAS of gene-environment interaction is informative for exploring novel genes. Future studies should increase the SNP coverage and measure occupational exposure more precisely by collecting more detail occupational information including the duration of each job and detail job titles. Moreover, future research could focus on examining the roles of CTLA-4, HDAC, and PPAR-alpha in the inflammatory airway process.
Conclusions
Our study found that SNP rs9931086 in the SLC38A8 gene and genes CTLA-4, HDAC, and PPAR-alpha, which are involved in inflammatory processes, may modify the effects of occupational exposure on lung function.
Abbreviations
COPD: Chronic obstructive pulmonary disease; FEV1: Forced expiratory volume in one second; FHS: Framingham heart study; FVC: Forced vital capacity; GWAS: Genome-wide association study; JEM: Job exposure matrix; SNP: Single nucleotide polymorphism.
Competing interests
The authors declare that they have no actual or potential competing financial interests.
Author contributions
SL: Conception, data cleaning, analysis and interpretation of data, drafting and revising manuscript, final approval of manuscript; XL: Conception, analysis and interpretation of data, revision and final approval of manuscript; DCC: Conception, analysis and interpretation of data, revision and final approval of manuscript. All authors read and approved the final manuscript.
Supplementary Material
Manhattan plot for interacting SNP with occupational exposure on FEV1. Figure S2. Manhattan plot for interacting SNP with occupational exposure on FEV1/FVC.
Contributor Information
Shu-Yi Liao, Email: shuyi.liao@mail.harvard.edu.
Xihong Lin, Email: xlin@hsph.harvard.edu.
David C Christiani, Email: dchris@hsph.harvard.edu.
Acknowledgements
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Funding support for the Framingham Social Network datasets was provided by NIA grant P01 AG 031093.
We would like to acknowledge the comments of Edwin K. Silverman, MD, PhD from Harvard Medical School and research assistance of Zhaoxi Wang from Harvard School of Public Health.
References
- Balmes J, Becklake M, Blanc P, Henneberger P, Kreiss K, Mapp C, Milton D, Schwartz D, Toren K, Viegi G. American Thoracic Society Statement: Occupational contribution to the burden of airway disease. Am J Respir Crit Care Med. 2003;12:787–797. doi: 10.1164/rccm.167.5.787. [DOI] [PubMed] [Google Scholar]
- Blanc PD, Toren K. Occupation in chronic obstructive pulmonary disease and chronic bronchitis: an update. Int J Tuberc Lung Dis. 2007;12:251–257. [PubMed] [Google Scholar]
- Higgins IT. The epidemiology of chronic respiratory disease. Prev Med. 1973;12:14–33. doi: 10.1016/0091-7435(73)90005-4. [DOI] [PubMed] [Google Scholar]
- Kauffmann F, Drouet D, Lellouch J, Brille D. Occupational exposure and 12-year spirometric changes among Paris area workers. Br J Ind Med. 1982;12:221–232. doi: 10.1136/oem.39.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxman AD, Muir DC, Shannon HS, Stock SR, Hnizdo E, Lange HJ. Occupational dust exposure and chronic obstructive pulmonary disease. A systematic overview of the evidence. Am Rev Respir Dis. 1993;12:38–48. doi: 10.1164/ajrccm/148.1.38. [DOI] [PubMed] [Google Scholar]
- Johnsen HL, Kongerud J, Hetland SM, Benth JS, Soyseth V. Decreased lung function among employees at Norwegian smelters. Am J Ind Med. 2008;12:296–306. doi: 10.1002/ajim.20557. [DOI] [PubMed] [Google Scholar]
- Bakke PS, Baste V, Hanoa R, Gulsvik A. Prevalence of obstructive lung disease in a general population: relation to occupational title and exposure to some airborne agents. Thorax. 1991;12:863–870. doi: 10.1136/thx.46.12.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heederik D, Kromhout H, Kromhout D, Burema J, Biersteker K. Relations between occupation, smoking, lung function, and incidence and mortality of chronic non-specific lung disease: the Zutphen Study. Br J Ind Med. 1992;12:299–308. doi: 10.1136/oem.49.5.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn RJ, Dockery DW, Speizer FE, Ware JH, Ferris BG Jr. Occupational exposures and chronic respiratory symptoms. A population-based study. Am Rev Respir Dis. 1987;12:298–304. doi: 10.1164/ajrccm/136.2.298. [DOI] [PubMed] [Google Scholar]
- Krzyzanowski M, Kauffmann F. The relation of respiratory symptoms and ventilatory function to moderate occupational exposure in a general population. Results from the French PAARC study of 16,000 adults. Int J Epidemiol. 1988;12:397–406. doi: 10.1093/ije/17.2.397. [DOI] [PubMed] [Google Scholar]
- Viegi G, Prediletto R, Paoletti P, Carrozzi L, Di Pede F, Vellutini M, Di Pede C, Giuntini C, Lebowitz MD. Respiratory effects of occupational exposure in a general population sample in north Italy. Am Rev Respir Dis. 1991;12:510–515. doi: 10.1164/ajrccm/143.3.510. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S. et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;12:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- Castaldi PJ, Cho MH, Cohn M, Langerman F, Moran S, Tarragona N, Moukhachen H, Venugopal R, Hasimja D, Kao E. et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum Mol Genet. 2010;12:526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG. et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;12:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A. et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;12:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V. et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;12:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, O’Connor GT. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;12:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolonska J, Wijmenga C, Postma DS, Boezen HM. Meta-analyses on suspected chronic obstructive pulmonary disease genes: a summary of 20 years’ research. Am J Respir Crit Care Med. 2009;12:618–631. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- Chapman J, Clayton D. Detecting association using epistatic information. Genet Epidemiol. 2007;12:894–909. doi: 10.1002/gepi.20250. [DOI] [PubMed] [Google Scholar]
- Curjuric I, Imboden M, Nadif R, Kumar A, Schindler C, Haun M, Kronenberg F, Kunzli N, Phuleria H, Postma DS. et al. Different genes interact with particulate matter and tobacco smoke exposure in affecting lung function decline in the general population. PLoS One. 2012;12:e40175. doi: 10.1371/journal.pone.0040175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg J, Iliadou A, Anderson M, de Verdier MG, Nihlen U, Dahlback M, Pedersen NL, Higenbottam T, Svartengren M. Genetic and environmental influence on lung function impairment in Swedish twins. Respir Res. 2010;12:92. doi: 10.1186/1465-9921-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Glutathione S-transferase variants and their interaction with smoking on lung function. Am J Respir Crit Care Med. 2004;12:388–394. doi: 10.1164/rccm.200312-1763OC. [DOI] [PubMed] [Google Scholar]
- Hancock DB, Artigas MS, Gharib SA, Henry A, Manichaikul A, Ramasamy A, Loth DW, Imboden M, Koch B, McArdle WL. et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;12:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB Sr, Fox CS, Larson MG, Murabito JM. et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;12:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- Blanc PD, Iribarren C, Trupin L, Earnest G, Katz PP, Balmes J, Sidney S, Eisner MD. Occupational exposures and the risk of COPD: dusty trades revisited. Thorax. 2009;12:6–12. doi: 10.1136/thx.2008.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;12:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Karoly ED, Dailey LA, Schmitt MT, Silbajoris R, Graff DW, Devlin RB. Comparison of gene expression profiles induced by coarse, fine, and ultrafine particulate matter. J Toxicol Environ Health A. 2011;12:296–312. doi: 10.1080/15287394.2010.516238. [DOI] [PubMed] [Google Scholar]
- Wilk JB, Herbert A, Shoemaker CM, Gottlieb DJ, Karamohamed S. Secreted modular calcium-binding protein 2 haplotypes are associated with pulmonary function. Am J Respir Crit Care Med. 2007;12:554–560. doi: 10.1164/rccm.200601-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adcock IM, Ito K, Barnes PJ. Histone deacetylation: an important mechanism in inflammatory lung diseases. COPD. 2005;12:445–455. doi: 10.1080/15412550500346683. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J. 2005;12:552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- Chen Y, Huang P, Ai W, Li X, Guo W, Zhang J, Yang J. Histone deacetylase activity is decreased in peripheral blood monocytes in patients with COPD. J Inflamm (Lond) 2012;12:10. doi: 10.1186/1476-9255-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, Barnes PJ. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;12:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- Lewis A, Riddoch-Contreras J, Natanek SA, Donaldson A, Man WD, Moxham J, Hopkinson NS, Polkey MI, Kemp PR. Downregulation of the serum response factor/miR-1 axis in the quadriceps of patients with COPD. Thorax. 2012;12:26–34. doi: 10.1136/thoraxjnl-2011-200309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;12:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol. 2002;12:748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- Marian E, Baraldo S, Visentin A, Papi A, Saetta M, Fabbri LM, Maestrelli P. Up-regulated membrane and nuclear leukotriene B4 receptors in COPD. Chest. 2006;12:1523–1530. doi: 10.1378/chest.129.6.1523. [DOI] [PubMed] [Google Scholar]
- Colville-Nash PR, Qureshi SS, Willis D, Willoughby DA. Inhibition of inducible nitric oxide synthase by peroxisome proliferator-activated receptor agonists: correlation with induction of heme oxygenase 1. J Immunol. 1998;12:978–984. [PubMed] [Google Scholar]
- Maestrelli P, Paska C, Saetta M, Turato G, Nowicki Y, Monti S, Formichi B, Miniati M, Fabbri LM. Decreased haem oxygenase-1 and increased inducible nitric oxide synthase in the lung of severe COPD patients. Eur Respir J. 2003;12:971–976. doi: 10.1183/09031936.03.00098203. [DOI] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G. et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;12:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J. 2003;12:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- Sun G, Stacey MA, Vittori E, Marini M, Bellini A, Kleimberg J, Mattoli S. Cellular and molecular characteristics of inflammation in chronic bronchitis. Eur J Clin Invest. 1998;12:364–372. doi: 10.1046/j.1365-2362.1998.00285.x. [DOI] [PubMed] [Google Scholar]
- Turato G, Zuin R, Saetta M. Pathogenesis and pathology of COPD. Respiration. 2001;12:117–128. doi: 10.1159/000050478. [DOI] [PubMed] [Google Scholar]
- Gamble E, Burns W, Zhu J, Ansari T, De Rose V, Kips J, Barnes NC, Jeffery PK. Variation of CD8+ T-lymphocytes around the bronchial internal perimeter in chronic bronchitis. Eur Respir J. 2003;12:992–995. doi: 10.1183/09031936.03.00115302. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ. et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet. 2002;12:1229–1239. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ. et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet. 2002;12:623–632. doi: 10.1093/hmg/11.6.623. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liang WB, Gao LB, Pan XM, Chen TY, Wang YY, Xue H, Zhang LS, Zhang L. CTLA4 and CD86 gene polymorphisms and susceptibility to chronic obstructive pulmonary disease. Hum Immunol. 2010;12:1141–1146. doi: 10.1016/j.humimm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhu G, Agusti A, Gulsvik A, Bakke P, Coxson H, Lomas DA, Silverman EK, Pillai SG. CTLA4 gene polymorphisms are associated with chronic bronchitis. Eur Respir J. 2009;12:598–604. doi: 10.1183/09031936.00141808. [DOI] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L. et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;12:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O’Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case–control Study of Schizophrenia sample. Mol Psychiatry. 2010;12:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, Mortensen PB, Andreassen OA, Sigurdsson E, Thorgeirsson TE, Bottcher Y. et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 2011;12:59–66. doi: 10.1038/mp.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EC, Chong SA, Mahendran R, Tan CH, Teo YY. Mu opioid receptor gene polymorphism and neuroleptic-induced tardive dyskinesia in patients with schizophrenia. Schizophr Res. 2003;12:61–63. doi: 10.1016/S0920-9964(02)00491-7. [DOI] [PubMed] [Google Scholar]
- Town T, Abdullah L, Crawford F, Schinka J, Ordorica PI, Francis E, Hughes P, Duara R, Mullan M. Association of a functional mu-opioid receptor allele (+118A) with alcohol dependency. Am J Med Genet. 1999;12:458–461. doi: 10.1002/(SICI)1096-8628(19991015)88:5<458::AID-AJMG3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kendler KS, Chen X. The mu-opioid receptor gene and smoking initiation and nicotine dependence. Behav Brain Funct. 2006;12:28. doi: 10.1186/1744-9081-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Lolak S. Psychiatric aspects of chronic lung disease. Curr Psychiatry Rep. 2009;12:219–225. doi: 10.1007/s11920-009-0034-9. [DOI] [PubMed] [Google Scholar]
- Leucht S, Burkard T, Henderson J, Maj M, Sartorius N. Physical illness and schizophrenia: a review of the literature. Acta Psychiatr Scand. 2007;12:317–333. doi: 10.1111/j.1600-0447.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- Chaves L, Shirakawa I. Nicotine use in patients with schizophrenia evaluated by the Fagerstrom Tolerance Questionnaire: a descriptive analysis from a Brazilian sample. Rev Bras Psiquiatr. 2008;12:350–352. doi: 10.1590/S1516-44462008005000014. [DOI] [PubMed] [Google Scholar]
- Copeland LA, Mortensen EM, Zeber JE, Pugh MJ, Restrepo MI, Dalack GW. Pulmonary disease among inpatient decedents: Impact of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2007;12:720–726. doi: 10.1016/j.pnpbp.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Eisen EA, Wegman DH, Louis TA, Smith TJ, Peters JM. Healthy worker effect in a longitudinal study of one-second forced expiratory volume (FEV1) and chronic exposure to granite dust. Int J Epidemiol. 1995;12:1154–1161. doi: 10.1093/ije/24.6.1154. [DOI] [PubMed] [Google Scholar]
- Li CY, Sung FC. A review of the healthy worker effect in occupational epidemiology. Occup Med (Lond) 1999;12:225–229. doi: 10.1093/occmed/49.4.225. [DOI] [PubMed] [Google Scholar]
- Monson R, editor. Occupational Epidemiology. Boca Raton: CRC Press; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Manhattan plot for interacting SNP with occupational exposure on FEV1. Figure S2. Manhattan plot for interacting SNP with occupational exposure on FEV1/FVC.