Abstract
Bacteria use conjugation systems, a subfamily of the type IV secretion systems, to transfer DNA to recipient cells. Despite 50 years of research, the architecture and mechanism of action of the channel mediating DNA transfer across the bacterial cell envelope remains obscure. By use of a sensitive, quantifiable assay termed transfer DNA immunoprecipitation (TrIP), we identify contacts between a DNA substrate (T-DNA) and 6 of 12 components of the VirB/D4 conjugation system of the phytopathogen Agrobacterium tumefaciens. Our results define the translocation pathway for a DNA substrate through a bacterial conjugation machine, specifying the contributions of each subunit of the secretory apparatus to substrate passage.
The translocation of nucleic acids across membrane barriers is central to many cellular processes. Bacterial conjugation systems are a subfamily of the type IV secretion systems (T4SS), which collectively mobilize the transfer of macromolecules such as monomeric proteins, multimeric toxins, and DNA-protein complexes across the cell envelope (1, 2). Conjugation systems mediate horizontal gene transfer, thus contributing to genome plasticity, evolution of infectious pathogens, and dissemination of antibiotic resistance and other virulence traits (3). Since the early discovery of the Escherichia coli F plasmid transfer system, many regulatory and mechanistic features of this and other T4SS have been described (1, 2, 4 ). Surprisingly, however, we still lack a fundamental understanding of the channel through which DNA substrates are delivered across the donor cell envelope.
In nature, Agrobacterium tumefaciens uses the VirB/D4 conjugation system (fig. S1) to deliver oncogenic transfer DNA (T-DNA) and effector proteins to susceptible plant cells, often inciting crown gall disease, which can devastate agriculturally important crop species. In the laboratory, the capacity of this bacterium to transfer DNA between kingdom boundaries has been exploited to genetically engineer a large number of plant, fungal, and other eukaryotic species (5). Here, we sought to define the translocation route for the T-DNA through this archetypal T4SS (1).
We developed a sensitive assay termed transfer DNA immunoprecipitation (TrIP) to identify close contacts between the T-DNA substrate as it exits the cell and subunits of the VirB/D4 T4SS (6) (fig. S2). This assay was adapted from the chromatin immunoprecipitation (ChIP) assay commonly used to study chromatin and transcription complexes in eukaryotic cells (7 ). In this three-stage assay, we treat vir gene–induced A. tumefaciens cells with formaldehyde to cross-link proteins to DNA in vivo, and then we precipitate a Vir protein of interest from detergent-solubilized cell extracts. Finally, we assay for coprecipitation of DNA by the polymerase chain reaction (PCR). We amplify DNA with two sets of primers, one specific for the left end of the transmissible TL-DNA carried on the tumor-inducing (Ti) plasmid pTiA6NC of strain A348. The second set is specific for the nontransferred octopine catabolism region (ophDC) positioned ~25 kb from the T-DNA on the Ti plasmid. We further developed a quantitative version of TrIP to compare levels of DNA substrate recovered in the immunoprecipitates (fig. S3).
Initially, we defined the genetic requirements for two early reactions associated with type IV translocation: substrate processing and recruitment to the secretory apparatus. Reminiscent of the processing of conjugative plasmids (8), the A. tumefaciens VirD2 relaxase binds origin of transfer–like T-DNA border sequences and cleaves the strand destined for transfer (T-strand). The relaxase is thought to remain covalently bound to the 5′ end of the T-strand, resulting in a VirD2–T-strand nucleoprotein particle. We isolated this presumptive transfer intermediate by immunoprecipitation with antibodies to VirD2 (Fig. 1A). The antibodies precipitated VirD2 as well as the T-strand—but not the ophDC Ti plasmid control fragment—from extracts of wild-type cells as well as mutants (table S1) defective for synthesis of the secretory apparatus or one of its protein substrates, the VirE2 single-stranded DNA binding protein (SSB) (5). With the quantitative TrIP assay, we confirmed that immunoprecipitates from wild-type and mutant cell extracts contained similar levels of T-strand (Fig. 1B). Thus, the presence of the secretory apparatus or the VirE2 substrate does not stimulate the T-DNA processing reaction.
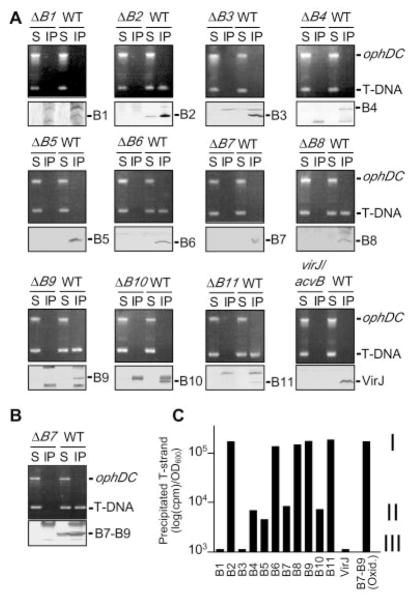
Fig. 1. Isolation of the VirD2–T-strand transfer intermediate.
(A) Antibodies to VirD2 precipitated VirD2 detected by SDS–polyacrylamide gel electrophoresis and immunostaining (lower panels) and the T-DNA substrate (T-DNA) but not the pTi control (ophDC) fragment detected by PCR amplification and gel electrophoresis (upper panels). Strain A348 (wild-type, WT) and the isogenic mutants (table S1) were untreated (−) or treated (+) with formaldehyde (FA) before lysis. S, supernatant after immunoprecipitation; IP, immunoprecipitate. Upper bands in the immunoblots are immunoreactive immunoglobulin G (IgG) heavy chain. Molecular mass markers (left lane) are in kilobases (upper) or kilodaltons (lower). (B) T-strand levels in material precipitated with antibodies to VirD2 (αVirD2) from the strains indicated, as determined by quantitative TrIP. Data are presented as counts per minute (CPM) of incorporated radionucleotide during one cycle in the logarithmic phase of PCR amplification. Values for the mutant strains are presented as a fraction of WT, normalized to 1.0.
The absence of effect of VirE2 on T-DNA processing is of special interest because this SSB is known to interact with the VirD2–T-strand at some point during translocation for targeting to the plant nuclear pore (5). We also failed to detect an interaction between VirE2 and the T-strand when A. tumefaciens cells were treated with formaldehyde before lysis (fig. S4A). These and earlier results (9) indicate that A. tumefaciens separately exports the VirD2–T-strand and VirE2 substrates and that the VirD2–T-strand–VirE2 complex (T-complex) assembles in the plant cytosol.
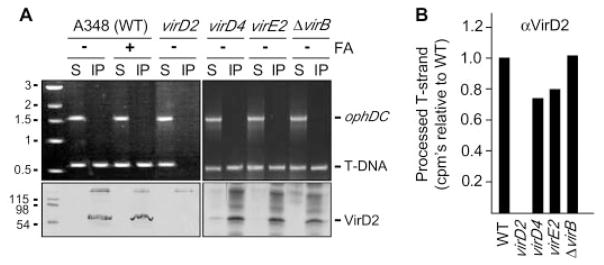
One T4SS subunit, the coupling protein or T4CP, functions in substrate selection and thus is postulated to recruit substrates to the secretory apparatus (10–12). We tested this model by assaying for a VirD4 T4CP interaction with T-strand in wild-type cells and various mutant strains. Antibodies to VirD4 precipitated a presumptive VirD4–T-strand complex from extracts of formaldehyde-treated wild-type cells, but not from untreated wild-type cells or mutant strains lacking VirD4 or the VirD2 relaxase (Fig. 2A). Moreover, the antibodies coprecipitated the VirD4–T-strand complex from mutants defective for synthesis of the T4SS or the VirE2 SSB. As above, the quantitative TrIP assay confirmed that the immunoprecipitates from these mutants contained wild-type levels of T-strand (Fig. 2B). The VirD4 T4CP therefore recruits the transfer intermediate independently of contributions from the VirB components of the T4SS or the VirE2 protein substrate. These and additional findings below demonstrate that the T4CP is the first T4SS subunit of an ordered substrate export pathway.
Fig. 2. VirD4 binds the VirD2–T-strand.
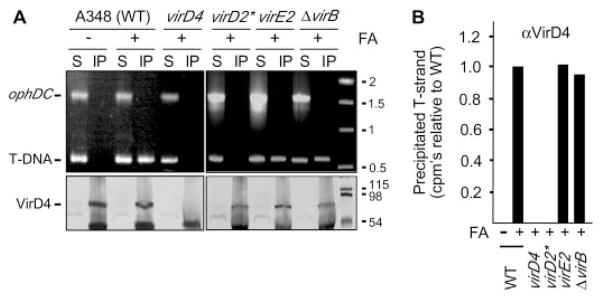
(A) Antibodies to VirD4 coprecipitated VirD4 (lower panels) and the T-DNA substrate (upper panels) from extracts of formaldehyde-treated (+) strain A348 (WT) and mutants defective for synthesis of the T4SS or the VirE2 SSB. virD2* is a virD2 mutant expressing virD4 from an IncP replicon. Lower bands in the immunoblots are IgG heavy chain. (B) T-strand levels in material precipitated with antibodies to VirD4 from the strains indicated, as determined by quantitative TrIP. Data are presented as CPM of incorporated radionucleotide during one cycle in the logarithmic phase of PCR amplification. Values for the mutant strains are presented as a fraction of WT, normalized to 1.0.
Upon substrate recruitment, the VirD4 T4CP functionally interacts with the VirB proteins to drive substrate translocation across the cell envelope (1, 9). How this or other conjugation machines are structurally arranged is unknown, but a general architecture can be inferred (fig. S1) (1). For example, structural studies have shown that T4CPs and VirB11 adenosine triphosphatases (ATPases) from different T4SS assemble as structurally dynamic, homohexameric rings with central channels large enough to accommodate DNA or unfolded polypeptides (13–16 ). In A. tumefaciens, VirD4 and VirB11 coordinate their activities with the VirB4 ATPase, the polytopic protein VirB6, and two bitopic proteins, VirB8 and VirB10, to drive substrate transport across the inner membrane (IM). One or more of these IM proteins interact with the VirB2 pilin, VirB3, VirB5, VirB7 lipoprotein, and the VirB9 secretin-like protein to convey substrates across the periplasm and outer membrane (OM) (1). VirB2 pilin polymerizes as an extracellular pilus to establish contact with target cells, but its physical or functional relationship to the secretion channel is not known. VirB1 is a putative transglycosylase whose activity is nonessential for type IV secretion (17).
We asked whether specific VirB subunits form close contacts with the T-DNA during translocation. Remarkably, antibodies to five of the 11 VirB proteins—VirB2, VirB6, VirB8, VirB9, and VirB11—coprecipitated the cognate VirB protein and T-strand at abundant levels from extracts of formaldehyde-treated wild-type cells, as assessed by gel electrophoresis (Fig. 3A) and quantitative TrIP (Fig. 3C). We propose that these class I proteins correspond to channel subunits of the secretory apparatus. In striking contrast, antibodies to VirB1, VirB3, VirB4, VirB5, VirB7, and VirB10, and to another presumptive secretory component, VirJ (18), precipitated the cognate protein, but the T-strand was not detectable by gel electrophoresis (Fig. 3A). Upon further examination, however, the quantitative TrIP assay showed that these proteins in fact were divisible into two classes. Antibodies to the class III proteins ( VirB1, VirB3, and VirJ) did not precipitate detectable levels of T-strand, which suggests that these proteins do not form close contacts with the transfer intermediate. By contrast, antibodies to the class II proteins ( VirB4, VirB5, VirB7, and VirB10) reproducibly precipitated the T-strand at ~2 to 4% of levels precipitated by antibodies to the class I proteins (Fig. 3C).
Fig. 3. VirB/D4 T4SS subunits that associate with the T-DNA transfer intermediate.
(A) Antibodies to VirB2, VirB6, VirB8, VirB9, and VirB11 coprecipitated the cognate protein (lower panels) and the T-DNA substrate (upper panels) from extracts of formaldehyde-treated A348 (WT) but not from extracts of the corresponding virB null mutant (ΔBx). ophDC, pTi control fragment; S, supernatant; IP, immunoprecipitate. (B) Under oxidizing conditions, antibodies to VirB7 coprecipitated a disulfide–cross-linked VirB7-VirB9 dimer (lower panel; blot development with antibodies to VirB7) and the T-DNA substrate (upper panel). (C) T-strand levels in material precipitated from the WT strain with antibodies to the Vir protein listed at the bottom, as determined by quantitative TrIP. Data are presented as CPM of incorporated radionucleotide during one cycle of PCR amplification on a log scale. Vir proteins were classified according to the relative level of coprecipitated T-strand: class I, abundant; class II, low; class III, undetectable.
Possibly, the class II subunits interact with the transfer intermediate in some way; however, results obtained for the VirB7 lipoprotein favor an alternative explanation. VirB7 forms a disulfide bridge with VirB9 in vivo (19–22). When cell extracts were treated with reducing agent before the TrIP assay, this cross-link was broken and the antibodies to VirB7 precipitated low (class II) levels of T-strand (Fig. 3, A and C). However, in the absence of reducing agent, the antibodies precipitated the disulfide–cross-linked VirB7-VirB9 dimer and abundant levels of T-strand (Fig. 3, B and C). VirB7 thus does interact with the T-strand, but only indirectly through its covalent association with VirB9. In vivo formaldehyde treatment can induce cross-linking, albeit inefficiently, between two closely interacting partner proteins. Hence, we suggest that the antibodies to VirB7 and the other class II subunits coprecipitated low levels of T-strand indirectly through in vivo formaldehyde cross-linking of a small fraction of class I and class II subunit contacts.
The class I subunits distribute across the cell envelope (fig. S1). To test whether the TrIP findings describe a pathway through which the T-strand exits the cell, we assayed strains defective for production of each class I subunit for stage-specific blocks in substrate transfer (Fig. 4A). Initial quantitative TrIP assays confirmed that each of the mutant strains used in this study produced similar levels of the T-strand substrate (fig. S4B). As expected (Fig. 2), deletion of the VirD4 T4CP blocked substrate transfer to the class I VirB proteins. Very strikingly, however, deletions of the remaining class I subunits conferred transfer defects consistent with the subcellular location of the missing subunit (fig. S1). A deletion of VirB11 did not affect substrate docking with the VirD4 T4CP but arrested transfer to the VirB6 and VirB8 integral IM and the VirB2 and VirB9 OM subunits. Deletions of VirB6 or VirB8 did not affect transfer to VirB11 but arrested transfer to the VirB8 or VirB6 IM subunits, respectively, as well as the OM subunits. Finally, deletions of VirB2 or VirB9 did not affect transfer to VirB6 and VirB8 but arrested transfer to VirB9 or VirB2, respectively. At present, we are unable to assign a temporal order to substrate contacts with VirB6 and VirB8 or with VirB2 and VirB9, possibly because these subunit pairs act in concert for translocation across the IM and OM, respectively. In summary, results of these TrIP studies strongly support the T-strand translocation pathway depicted in Fig. 4C.
Fig. 4. Nonpolar virB and virD4 null mutations block T-strand translocation.
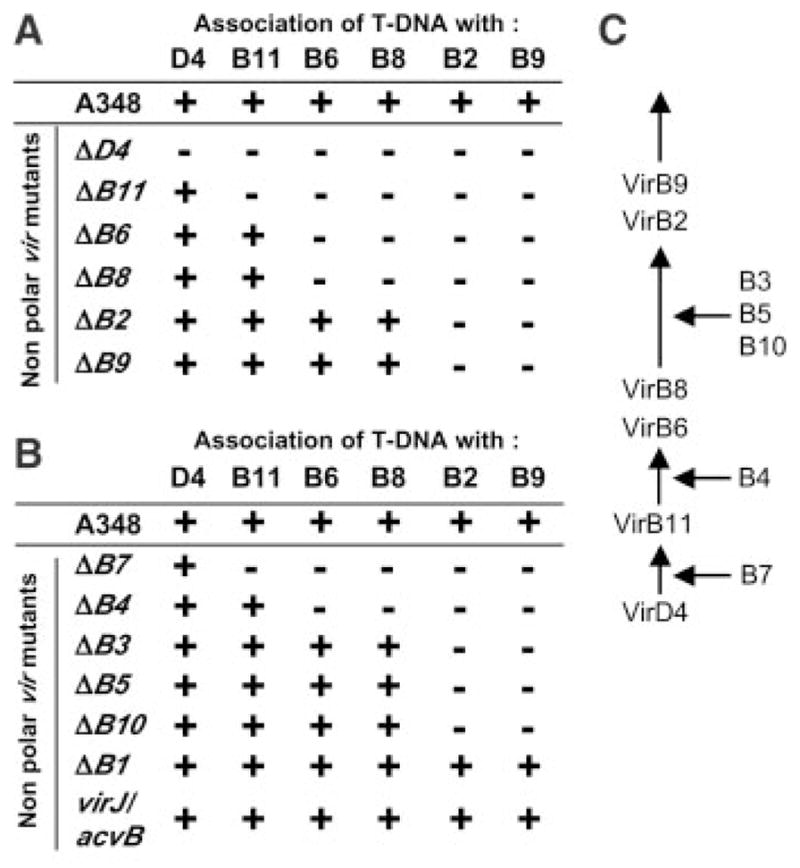
(A) TrIP summary for mutants lacking one of the class I channel subunits; (+) denotes a T-strand interaction with the protein listed at the top; (−), no interaction. (B) TrIP summary for mutants lacking one of the class II or class III subunits. (C) A postulated contact pathway for the T-strand during translocation through the A. tumefaciens VirB/D4 T4SS. Proteins listed at right are required for passage of the T-strand at the step indicated.
Deletions of remaining VirB proteins also arrested substrate transfer at specific steps in the translocation pathway (Fig. 4, B and C). For example, deletion of the VirB7 lipoprotein blocked transfer very early in the pathway, from VirD4 to VirB11, in agreement with results showing that synthesis of this lipoprotein is essential for assembly of a stable secretory apparatus (23, 24). Deletion of the VirB4 ATPase arrested transfer at the next step, from VirB11 to VirB6/VirB8, consistent with evidence that the VirB4 and VirB11 ATPases functionally interact to drive substrate export (21, 25). Deletions of VirB10, VirB5, or VirB3 arrested transfer from the integral IM to the OM class I subunits (Fig. 4, B and C). Interestingly, VirB10 interacts with the T4CP at the IM (26, 27) and the VirB7-VirB9 dimer at the OM (24); apparently, this coupling of IM and OM VirB subcomplexes is necessary for substrate passage across the cell envelope. Finally, mutations of two class III subunits, VirB1 and VirJ, did not affect substrate transfer from VirD4 to VirB2/VirB9. The dispensability of VirB1 is consistent with previous findings (17). The lack of effect of VirJ (and its chromosomal homolog AcvB) on substrate transfer, as well as our inability to detect a VirJ–T-strand interaction (Fig. 3A), together contradict a proposal that VirJ or AcvB function as subunits of this T4SS (18).
We have described the translocation route for a DNA substrate through a bacterial conjugation machine (Fig. 4C). The steps of the pathway and the contributions specified for each of the VirB/D4 T4SS subunits are compatible with a large amount of structure-function data about this and related conjugation systems of Gram-negative bacteria (1, 4, 13, 14). We anticipate that further applications of the TrIP assay will ultimately present a detailed mechanistic picture of how type IV machines translocate their substrates across the bacterial cell envelope. TrIP should also prove highly useful for studies of many other fundamental processes of bacteria and eukaryotes that involve the movement of DNA across biological membranes (e.g., bacterial transformation and transduction, chromosomal partitioning and movement, and viral infection cycles). Finally, there is increasing evidence that conjugation systems are actually protein secretion systems that have evolved the capacity to recognize and translocate relaxases as well as, coincidentally, the “hitchhiker” T-strand (1). In this context, our studies have shown that DNA serves as a useful marker for monitoring the translocation of a protein substrate through its cognate secretory apparatus.
Supplementary Material
Footnotes
References and Notes
- 1.Cascales E, Christie PJ. Nature Rev Microbiol. 2003;1:137. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An updated list of type IV secretion systems is available at http://mmg.uth.tmc.edu/webpages/faculty/supplements/pchristie/T4SS-updated.pdf.
- 3.Hacker J, Hentschel U, Dobrindt U. Science. 2003;301:790. doi: 10.1126/science.1086802. [DOI] [PubMed] [Google Scholar]
- 4.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. FEMS Microbiol Lett. 2003;224:1. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 5.Gelvin SB. Microbiol Mol Biol Rev. 2003;67:16. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.See supporting data on Science Online.
- 7.Kuo MH, Allis CD. Methods. 1999;19:425. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 8.Lessl M, Lanka E. Cell. 1994;77:321. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 9.Vergunst AC, et al. Science. 2000;290:979. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 10.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Mol Microbiol. 2002;45:1. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding Z, Atmakuri K, Christie PJ. Trends Microbiol. 2003;11:527. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atmakuri K, Ding Z, Christie PJ. Mol Microbiol. 2003;49:1699. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomis-Ruth FX, et al. Nature. 2001;409:637. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 14.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Mol Cell. 2000;6:1461. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 15.Hormaeche I, et al. J Biol Chem. 2002;277:46456. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 16.Savvides SN, et al. EMBO J. 2003;22:1969. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger BR, Christie PJ. J Bacteriol. 1994;176:3646. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pantoja M, Chen L, Chen Y, Nester EW. Mol Microbiol. 2002;45:1325. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 19.Spudich GM, Fernandez D, Zhou XR, Christie PJ. Proc Natl Acad Sci USA. 1996;93:7512. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson LB, Hertzel AV, Das A. Proc Natl Acad Sci USA. 1996;93:8889. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward DV, Draper O, Zupan JR, Zambryski PC. Proc Natl Acad Sci USA. 2002;99:11493. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakubowski SJ, Krishnamoorthy V, Christie PJ. J Bacteriol. 2003;185:2867. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez D, Spudich GM, Zhou XR, Christie PJ. J Bacteriol. 1996;178:3168. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaupré CE, Bohne J, Dale EM, Binns AN. J Bacteriol. 1997;179:78. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang TAT, Zhou XR, Graf B, Christie PJ. Mol Microbiol. 1999;32:1239. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Mol Microbiol. 2003;49:105. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- 27.Llosa M, Zunzunegui S, Alfonso C, Rivas G, de la Cruz F. Proc Natl Acad Sci USA. 2003;100:10465. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supported by NIH grant GM48746. We thank V. Schramke for advice toward the development of the TrIP assay, members of our laboratory for helpful discussions, and W. Margolin, H. Kaplan, G. Dunny, and anonymous referees for critiques of this manuscript.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.