Summary
The maintenance of protein solubility is a fundamental aspect of protein homeostasis, as aggregation is associated with cytotoxicity and a variety of human diseases. Numerous proteins unrelated in sequence and structure, however, can misfold and aggregate, and widespread aggregation can occur in living systems under stress or ageing. A crucial question in this context is why only certain proteins aggregate in vivo while others do not. We identify here the proteins most vulnerable to aggregation as those whose cellular concentrations are high relative to their solubilities. These supersaturated proteins represent a metastable sub-proteome involved in pathological aggregation during stress and ageing, and are overrepresented in biochemical processes associated with neurodegenerative disorders. Consequently, such cellular processes become dysfunctional when the ability to keep intrinsically supersaturated proteins soluble is compromised. Thus, the simultaneous analysis of abundance and solubility can rationalize the diverse cellular pathologies linked to neurodegenerative diseases and aging.
Introduction
Neurodegenerative disorders are increasingly prevalent in our society and represent a very significant challenge to healthcare systems (Balch et al., 2008; Dobson, 2003). A number of explanations of the fundamental origins of these diseases have been proposed, including mitochondrial dysfunction, disruptions of the endoplasmic reticulum and membrane trafficking, effects on protein folding and clearance, and the activation of inflammatory responses (Balch et al., 2008; Dobson, 2003; Querfurth and LaFerla, 2010; Selkoe, 2011). One common feature associated with these conditions, however, is the aggregation of certain peptides and proteins, which generates a cascade of pathological events, including the secondary aggregation of various other proteins and the consequent failure of protein homeostasis to preserve normal biological function (Balch et al., 2008; Dobson, 2003; Gidalevitz et al., 2006; Selkoe, 2011).
Given the evidence that protein aggregation is a widespread phenomenon (Chapman et al., 2006; David et al., 2010; Gidalevitz et al., 2006; Koga et al., 2011; Koplin et al., 2010; Liao et al., 2004; Narayanaswamy et al., 2009; Olzscha et al., 2011; Reis-Rodrigues et al., 2012; Wang et al., 2005; Xia et al., 2008), two key questions are why some proteins, but not others, aggregate in vivo and generate pathological states, and whether the identities of these proteins differ substantially between diseases. If particular proteins aggregate in response to specific stresses, different sets of aggregated proteins will appear under each condition. Alternatively, the various sets of aggregating proteins may correspond to a fraction of the proteome with distinctive characteristics that increase the risk of aggregation under many kinds of stress. The latter possibility is consistent with observations that aggregation-prone proteins share general physicochemical features (Chiti et al., 2003; Fernandez-Escamilla et al., 2004; Olzscha et al., 2011; Tartaglia et al., 2008).
Our aim in this work has been to answer a fundamental question about widespread protein aggregation – why certain proteins aggregate in stress, ageing, or disease, while others do not. To address this problem, we have sought to establish a proteome-wide method of identifying the proteins that are vulnerable to aggregation in vivo. Using this method, we have identified a number of proteins that are expressed at levels that are high relative to their solubilities. These proteins are supersaturated, as their concentrations exceed their critical solubility levels. Early evidence that supersaturation predisposes proteins to aggregate was provided by the finding that the rate and extent of aggregation of hemoglobin S, which is associated with sickle cell anemia, is strongly concentration-dependent (Hofrichter et al., 1976). More recently, this idea has been used to compare the aggregation and crystallisation behaviour of proteins (Yoshimura et al., 2012). Here, we have extended the concept of supersaturation to the proteome level by considering both the unfolded and folded states that can be populated by individual proteins, as well as their association into complexes. Thus, for example, an intrinsically aggregation-prone protein is not necessarily dangerous unless it is expressed at a relatively high concentration. Similarly, a highly concentrated protein may not be at risk of losing its solubility unless its intrinsic propensity to aggregate is relatively high.
By predicting supersaturation from estimated protein concentration and aggregation propensity at a proteome scale, we are able to rationalize a variety of phenomena associated with aggregation and misfolding diseases. We find through our analysis of the human and C. elegans proteomes that those proteins known to interact with aggregates or to aggregate upon aging are highly supersaturated, and that the cellular processes known to be associated with neurodegenerative diseases are at risk of disruption because they involve an exceptionally large number of supersaturated proteins. These results show how the initial appearance of protein aggregates in the presence of other vulnerable proteins can precipitate a series of uncontrolled aggregation events with severe pathological consequences, and that proteins in a supersaturated state compose the sub-proteome most at risk of misfolding and aggregation under conditions of stress. These proteins and the biochemical pathways to which they belong may be the first to suffer from an impairment of protein homeostasis, and therefore represent the underlying basis for the cellular damage caused by diseases of misfolding, including neurodegenerative conditions such as Alzheimer’s and Parkinson’s diseases.
Results
Prediction of protein supersaturation from concentration and aggregation propensity
In order to identify those proteins most at risk of misfolding and aggregating in vivo, we calculated their level of supersaturation using a score that we define in terms of the concentrations of proteins relative to their aggregation propensities (see Methods and Supplementary Information). The cellular concentrations of proteins with high supersaturation scores are more likely to exceed their critical values under varying conditions, leading these proteins to become insoluble. We here used the aggregation propensities of proteins as estimates of their solubility, as experimental measurements of critical concentrations of proteins in vivo are extremely difficult to carry out at the proteome level. To evaluate the risk of proteins to aggregate from their unfolded or native states, we define the parameters σu and σf as the supersaturation scores, respectively (Fig. 1). The risk of aggregation is different in these two states since in the folded state the most aggregation-prone regions tend to be buried in the core of the structure, and thus they are prevented from forming intermolecular interactions (Tartaglia et al., 2008). The critical concentrations of proteins in their unfolded states thus are generally expected to be lower than in their folded states, hence the necessity of introducing the σu and σf scores separately. Since the largest pool of unfolded proteins corresponds to newly synthesized proteins, whose concentrations can be estimated from the corresponding mRNA concentrations, we used the logarithmic average of scores derived from microarray analysis of over 70 types of human tissue or of the nematode C. elegans at a range of ages to represent levels of newly synthesized proteins (Golden et al., 2008; Su et al., 2004). For folded proteins, in order to define σf the score, we used the logarithm of the normalized spectral abundance factors (NSAFs) derived from mass spectrometry (Schrimpf et al., 2012).
Figure 1. Protein aggregation in vivo can occur through different routes.
Proteins may misfold and aggregate as they emerge from the ribosome or when they unfold at least transiently from the native state. The proteins most at risk of aggregation are those whose concentration is high with respect to their solubility. We quantify this risk by defining the supersaturation σu and σf scores. The supersaturation score σu, which measures the tendency of proteins to aggregate from the unfolded state, is based on the Zyggregator score (Tartaglia et al., 2008) (Zagg) and mRNA expression levels. The supersaturation score σf, which measures the tendency of proteins to aggregate from the folded state, is based on the structurally-corrected Zyggregator score (Tartaglia et al., 2008) and protein concentration.
We estimated the propensity of proteins to aggregate from the unfolded state using the Zagg score calculated with the Zyggregator method (Tartaglia et al., 2008), which is based on the analysis of the physicochemical properties of amino acid sequences (Chiti et al., 2003). The Zyggregator method employs algorithms that have been parameterized to reproduce the aggregation behavior of a set of known amyloidogenic proteins, and has been validated in a series of studies in which it has been shown to lead to accurate predictions of aggregation rates both in vitro and in vivo (Belli et al., 2011; Luheshi et al., 2007; Roodveldt et al., 2012; Tartaglia et al., 2008). For proteins that aggregate from the native state, we used an aggregation propensity score that accounts for the protective effects of the folded structure, which is defined by assigning corrections to the aggregation propensities of individual residues on the basis of the extent of the structural fluctuations that they experience in the folded state and that lead to their temporary exposure to the solvent (Tartaglia et al., 2008). This structurally-corrected aggregation propensity score has been shown to correlate well with protein solubility determined from an in vitro reconstituted bacterial ribosome system (Agostini et al., 2012). We then summed the logarithms of the concentration and aggregation propensity values (see Methods and Supplementary Information) to construct a human database of σu scores for 16,263 proteins and σf scores for 6,155 proteins and C. elegans database of σu scores for 16,623 proteinsand σf scores for 10,149 proteins (Table S2). The sizes of our databases were limited primarily by the availability of expression and abundance data that could be unambiguously mapped to specific protein identifiers.
Supersaturation scores rationalize widespread protein aggregation under stress
As a wide range of proteins are known to form fibrillar assemblies within the cell (Chapman et al., 2006; Chiti and Dobson, 2006; David et al., 2010; Fowler et al., 2007; Gidalevitz et al., 2006; Haass and Selkoe, 2007; Hartl et al., 2011; Koplin et al., 2010; Liao et al., 2004; Olzscha et al., 2011; Reis-Rodrigues et al., 2012; Wang et al., 2005; Xia et al., 2008) we investigated the relationship between the phenomena of supersaturation and aggregation. For instance, the function of actin, a highly abundant protein also identified as being highly supersaturated in our calculations (Table S2), relies on its ability to assemble reversibly into filaments, and it has been suggested that given the typical cytosolic concentration of actin, it would always remain in a polymerized form were it not for the presence of specific regulatory mechanisms (Pollard et al., 2000).
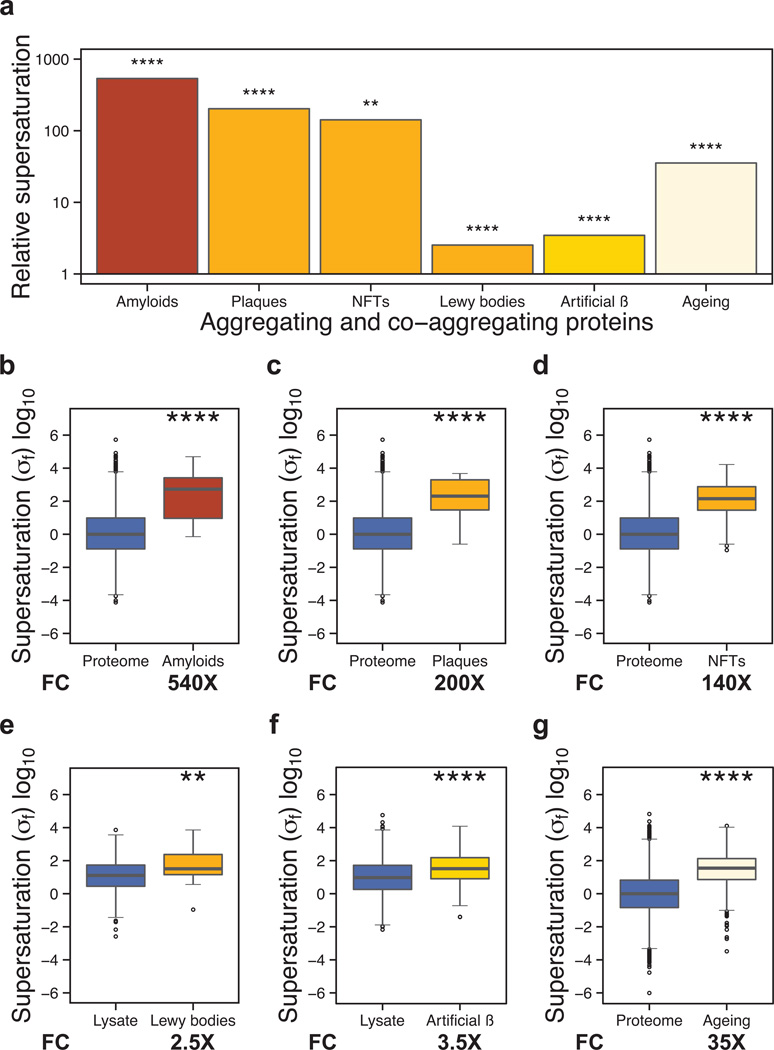
More generally, we find that the ‘amyloid proteins,’ as annotated in the UniProt database (UniProt Consortium, 2012), have elevated supersaturation scores (Figs. 2a,b, S1a,b and S1h,i). For these proteins, the score is more than 500-fold greater than the median value over the proteome (540×, p=1.9•10−5), indicating that these proteins are on average at greater risk of aggregation upon accumulation in the cell (Fig. 2a,b) than when they are in the process of being synthesized (Fig. S1a,b). This conclusion is consistent with the appearance of such proteins as the predominant constituents of either intracellular inclusions or extracellular deposits in a variety of diseases (Balch et al., 2008; Dobson, 2003).
Figure 2. Widespread protein aggregation is associated with high supersaturation scores.
(a) Summary of the results for the different classes of aggregating proteins analyzed in this work. Results are given in terms of the increases in the supersaturation scores over the average value for the whole proteome or for an experimental lysate (“fold change”). We compared the supersaturation scores σf for: (b) the whole proteome and the human “amyloid proteins” in UniProt (UniProt Consortium, 2012), (c) the whole proteome and proteins that co-precipitate in amyloid plaques (Liao et al., 2004), (d) the whole proteome and proteins that co-precipitate in neurofibrillary tangles (Wang et al., 2005), (e) the whole lysate and proteins that co-precipitate in Lewy bodies (Xia et al., 2008), (f) the whole lysate and proteins that co-precipitate in artificial β-sheet protein aggregates (Olzscha et al.), (g) the whole proteome and proteins found to aggregate in C. elegans during aging (David et al., 2010; Reis-Rodrigues et al., 2012). Boxplots extend from the lower to the upper quartiles, with the internal lines refer to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. The statistical significance was assessed by Wilcoxon/Mann-Whitney U test with the Bonferroni-corrected p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Given that high supersaturation scores correspond to an increased risk of proteins becoming insoluble, we investigated whether such scores can rationalize additional aspects of protein aggregation, including co-aggregation with large, insoluble deposits associated with disease (Liao et al., 2004; Wang et al., 2005; Xia et al., 2008) and artificial β-sheet protein (Olzscha et al., 2011) aggregates. Our results indicate that the σf score can identify proteins found to incorporate into amyloid plaques (Liao et al., 2004) (200×, p=1.7•10−7, Fig. 2a,c), neurofibrillary tangles (Wang et al., 2005) (140×, p=1.2•10−18, Fig. 2a,d), and Lewy bodies (Xia et al., 2008) (2.5×, p=2.9•10−3, Fig. 2a,e) isolated from autopsy samples of neurodegenerative disease patients. We find that proteins that co-aggregate with artificial β-sheet proteins in human cell cultures (Olzscha et al., 2011) are characterized by increased values of the σf score (3.5×, p=1.7•10−6, Fig. 2a,f), as well. Such proteins also have elevated σu scores (1.4×, p=1.4•10−2, Fig. S1a,f), consistent with observations from pulse-chase experiments that both newly-synthesized and preexisting proteins interact with aggregates (Olzscha et al., 2011). The σf (Fig. 2) and σu (Fig. S1) scores are broadly consistent for all of these sets, with proteins that co-aggregate with large inclusions tending to be relatively highly supersaturated in both the folded and unfolded states.
We then turned our attention to the analysis of proteins that are likely to become susceptible to aggregation when the control of protein homeostasis declines, as in ageing (David et al., 2010; Reis-Rodrigues et al., 2012). We observe that the σu scores (0.61×, p=2.0•10−85, Fig. S1a,g) of proteins that aggregate upon ageing in C. elegans (David et al., 2010; Reis-Rodrigues et al., 2012) are lower than those of the proteome as a whole; by contrast, the σf scores are much higher for these proteins (35×, p=1.9•10−158, Fig. 2a,g). The low values of the σu scores can be attributed to the fact that global gene expression in C. elegans declines with age (Golden et al., 2008), thus reducing the levels of newly synthesized proteins that may aggregate from their unfolded states. Instead, proteins that aggregate during ageing have low expression levels but relatively high concentrations, and tend to be metastable by virtue of their high σf scores.
Since both the estimates of concentration and the predictions of aggregation propensity are subject to considerable potential errors, to test the robustness of our results against these errors, we introduced Gaussian noise into the calculations of the σf and σu scores, finding that the results are indeed stable against high levels of noise (in many cases, more than 50-fold error, Figs. S2–S3).
Biochemical pathways associated with neurodegenerative diseases are enriched in supersaturated proteins
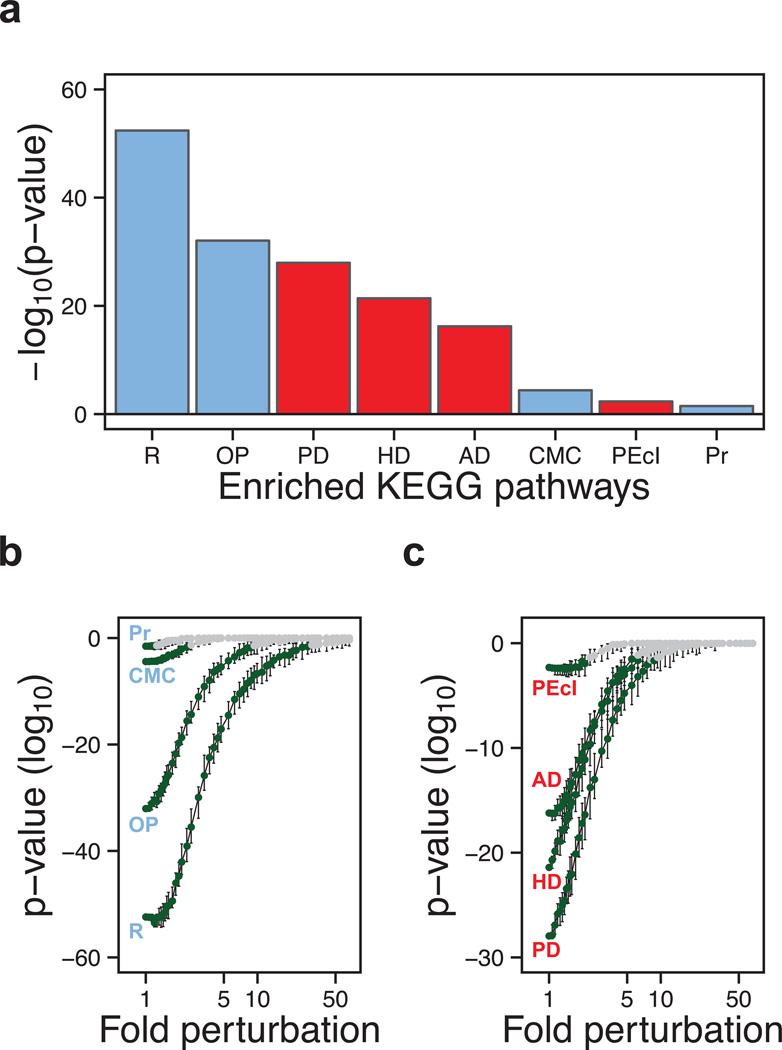
Since supersaturation scores could explain diverse phenomena related directly to aggregation, we wondered whether they might help to identify cellular processes that were particularly sensitive to stress. Therefore, we asked whether particular biochemical pathways are at high risk of disruption by virtue of the supersaturation levels of their constituent proteins. For our analysis we considered the pathways that are listed in the Kyoto Encyclopedia of Genes and Genomes (KEGG), which are based on manually-curated sets of proteins involved in cellular processes or proposed to be involved in disease on the basis of reports in the literature. We used the DAVID software (Huang et al., 2009) to determine if any of the KEGG pathways (Kanehisa et al.) have a large number of human proteins with high σu or σf scores. We carried out an unbiased search of the roughly 200 KEGG pathways and found only eight pathways significantly enriched in the proteins with σu scores at or above the 95th percentile. Strikingly, all these pathways involve proteins that either form well-defined functional complexes or are related to diseases that involve pathological protein complexes. The three disease pathways that are most dramatically enriched in supersaturated proteins are those of Alzheimer’s (p=6.0•10−17), Parkinson’s (p=1.1•10−28), and Huntington’s (p=4.0•10−22) diseases (Fig. 3a). A fourth pathway, involved in pathogenic E. coli infection, which is closely associated with the cytoskeleton, is also enriched, but to a much less significant level (p=4.6•10−3). KEGG pathways are broadly constructed, with those related to neurodegenerative diseases including not only proteins known to aggregate, but also the proteins that process these aggregates and the cellular systems that become impaired as a result of aggregation (Kanehisa et al.). Despite this disparate collection of proteins, a staggering two-thirds of proteins in the KEGG Alzheimer’s disease pathway have supersaturation scores above the median value for the human proteome (Fig. 6).
Figure 3. Biochemical processes associated with neurodegenerative diseases are highly enriched in supersaturated proteins.
(a) List of the KEGG pathways (Kanehisa et al., 2010) identified here as significantly enriched (Bonferroni-corrected p-values) in proteins at or above the 95th percentile of supersaturation (σu): (R) ribosome, (OP) oxidative phosphorylation, (PD) Parkinson’s disease, (HD) Huntington’s disease, (AD) Alzheimer’s disease, (CMC) cardiac muscle contraction, (PEcI) pathogenic E. coli infection, (Pr) proteasome; physiological and pathological pathways are shown in blue and red, respectively. (b,c) Test of the robustness of the significance of the enrichment of the KEGG pathways according to their supersaturation scores. Gaussian noise was introduced 100 independent times into the proteome scores at 50 different levels and plotted (1× = no noise) for: (b) physiological pathways, which are robust up to 28× (R), 8.8× (OP), 2.3× (CMC) and 1.2× (Pr), and (c) pathological pathways, which are robust up to 8.2× (PD), 6.2× (HD), 5.5× (AD) and 2.1× (PEcI). Error bars indicate interquartile ranges, green points indicate error levels below the p=0.05 significance (red dashed line) by the Wilcoxon/Mann-Whitney U test.
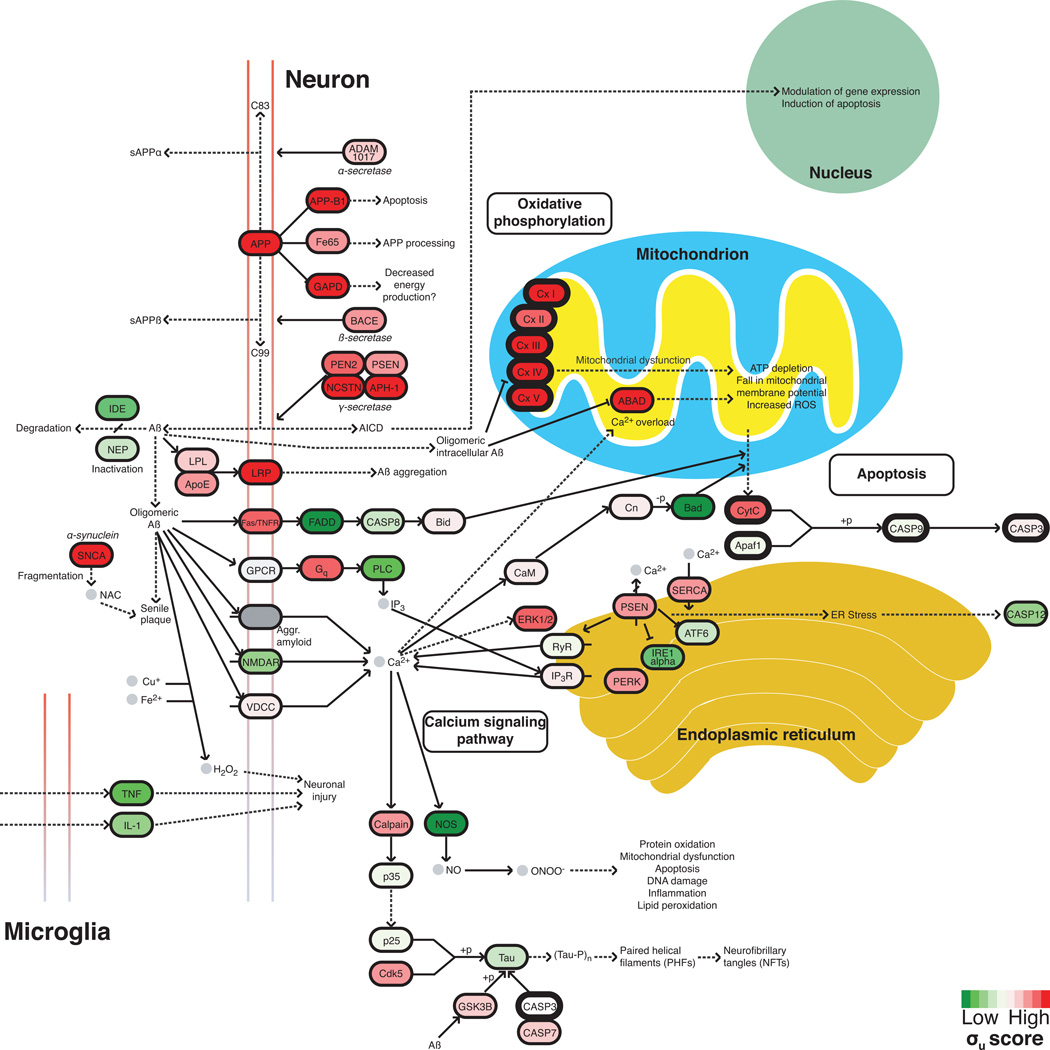
Figure 6. Supersaturated proteins are over-represented in the KEGG Alzheimer’s disease pathway.
The KEGG pathway for Alzheimer’s disease (Kanehisa et al.) is curated from the literature and includes many proteins not directly associated with the disruption of the homeostasis of the Aβ peptide. Those proteins that are part of the overlap (see Fig. 4a) of the Alzheimer’s, Parkinson’s, and Huntington’s disease pathways are shown in bold. Approximately 65% of proteins in the Alzheimer’s pathway have high σu scores. Colors are assigned based on the division of the σu database into deciles from low (green) to high (red).
In addition to these disease pathways, we also find that the KEGG pathways associated with the assembly of the ribosome (p=4.0•10−53) and the proteasome (p=3.2•10−2), and with the processes of oxidative phosphorylation (p=8.8•10−33) and cardiac muscle contraction (p=3.9•10−5), are enriched in supersaturated proteins (Fig. 3a). These results, particularly those associated with the ribosome and oxidative phosphorylation, are highly robust against errors in the calculation of σu scores (Fig. 3b). All of these pathways involve the assembly of major cellular macromolecular complexes, the components of which must contain interactive surfaces that tend to be aggregation-prone (Pechmann et al., 2009). In agreement with this finding, proteins involved in such assemblies, especially the ribosome, have been observed consistently in widespread aggregation studies (David et al., 2010; Koplin et al., 2010; Reis-Rodrigues et al., 2012). Significantly, none of the pathways identified by using supersaturation scores is identifiable from aggregation propensities alone.
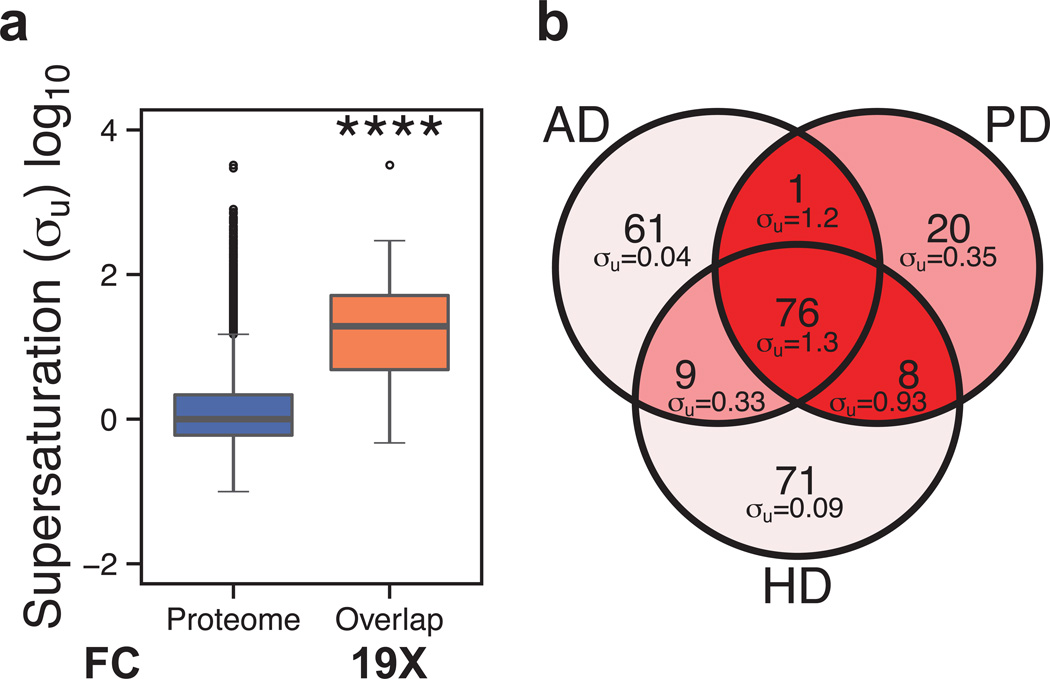
These results suggest that widespread aggregation under conditions of stress is defined not only by the specific nature of the stress itself, but also by the level of supersaturation of the proteins that aggregate. If this is the case, some proteins should have characteristics that render them susceptible to aggregation under a variety of conditions. To investigate this possibility, we determined the overlap between the Alzheimer’s, Parkinson’s, and Huntington’s disease KEGG pathways. The 76 proteins common to the pathways represented by these diseases have on average a much larger σu score (19×, p=4.2•10−31) than that of the proteome as a whole (Fig. 4a), or indeed, that of the remaining proteins in the three individual pathways (Fig. 4b).
Figure 4. Supersaturated proteins are common to different neurodegenerative pathways.
(a) Comparison of the σu scores for the proteome and the set of 76 proteins common among the Alzheimer’s, Parkinson’s, and Huntington’s KEGG pathways (Kanehisa et al., 2010); this set of proteins is denoted as ‘overlap’. (b) Comparison of the scores for the proteins in the Alzheimer’s, Parkinson’s, and Huntington’s pathways. Colors are assigned based on the division of the σu scores into deciles from low (green) to high (red).
Of the eight pathways enriched in proteins with high σu scores, five — including those for Alzheimer’s, Parkinson’s, and Huntington’s diseases — are also enriched in proteins with high σf scores, although in the latter case the data are less statistically significant (Fig. S4). The fact that the σu scores so strongly single out pathways involved in neurodegeneration suggests that the proteins that misfold and aggregate during the course of these diseases aggregate primarily from unstructured or partially unstructured states. In particular, since many proteins in the KEGG neurodegenerative disease pathways are membrane proteins, they are unlikely to aggregate from their folded state in the membrane environment. They are, however, likely to be at risk during folding or upon removal from the membrane for degradation (MacGurn et al., 2012; Notterpek et al., 1999; Skach, 2009), especially if protein homeostasis dysfunction impairs membrane trafficking, as has been reported (Cooper et al., 2006). Consistent with the view that membrane proteins may have a somewhat elevated risk in the unfolded state is that the median σu score score is modestly elevated for proteins with ‘membrane’ Gene Ontology (Ashburner et al., 2000) tag (1.2×, p=1.7•10−20), while the σf score is low for such proteins (0.47×, 5.9•10−52). This risk, therefore, is small compared to that which we show for most sets of aggregating proteins in Fig. 2. In order to test whether the observation that the Alzheimer’s, Parkinson’s, and Huntington’s disease pathways have elevated numbers of supersaturated proteins is simply a consequence of their richness in membrane proteins, we determined whether or not the sets of proteins at or above the 95th percentile of σu or σf scores are significantly enriched in membrane proteins compared to the proteome at large. Our results indicate that the most supersaturated proteins are not enriched in this way (σu: p=0.16, σf: p=1.0). However, we do find that the proteins most supersaturated in the unfolded state are enriched in those associated with the Gene Ontology tag ‘organelle membrane’ (σu: p=6.9•10−19, σf: p=1.0). This result is consistent with recent evidence that membrane proteins in the mitochondrial respiratory chain can misfold when the Parkinson’s-associated gene PINK1 is mutated (Pimenta de Castro et al., 2012). In fact, chief among the overlapping proteins in the Alzheimer’s, Parkinson’s, and Huntington’s disease pathways are the proteins that are members of the respiratory chain.
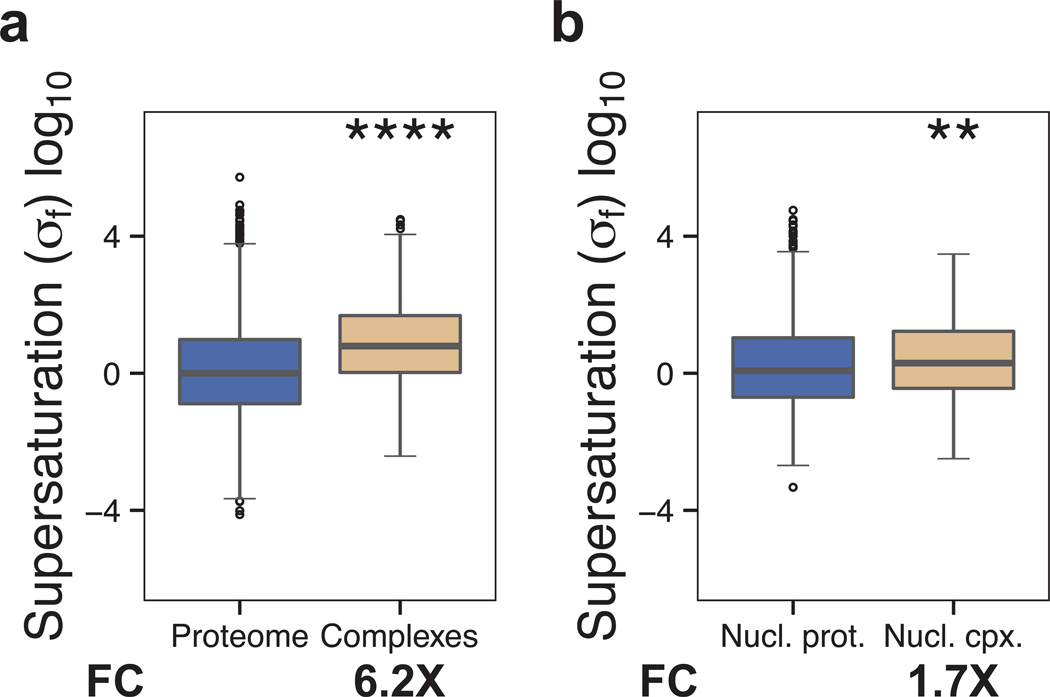
Since co-aggregating proteins are more highly supersaturated in the folded state than in the unfolded state, the ability of the σu score, in addition to the σf score, to identify disease pathways suggests at least three possibilities. First, it may be that many proteins involved with disease aggregate independently instead of associating with inclusions, as suggested by the tendency of proteins toward homomeric aggregation (Matsumoto et al., 2006; Rajan et al., 2001; Wright et al., 2005). Second, proteins involved with disease may be degraded if they fail to fold into their native conformation (Goldberg, 2003). Third, mass spectrometry experiments designed to identify aggregating or co-aggregating proteins tend to ignore membrane proteins because of the difficulty of distinguishing membrane-associated proteins from truly insoluble ones, and therefore may be missing an important component of the disease process. Despite the need for such details to be resolved in the future, our results suggest that supersaturation in the folded state is reporting on significant functional properties of proteins in the cell, as well as potentially pathological processes. We find, for example, that in addition to proteins that aggregate (Fig. 2), those proteins that form functional complexes tend to have high σf scores (complexes (Licata et al., 2012): 6.2×, p=2.3•10−58; nuclear complexes (Luc and Tempst, 2004): 1.7×, p=1.2•10−3, Fig. 5). These results are consistent with evidence that the features that mediate normal protein interactions are similar to those that promote aggregation (Pechmann et al., 2009).
Figure 5. Proteins that form complexes tend to be more supersaturated than the proteome as a whole.
We compared the σf scores for: (a) the proteome and proteins involved in complexes, and (b) the nuclear proteome and proteins involved in nuclear complexes. Boxplots extend from the lower to the upper quartiles, with the internal lines refer to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. The statistical significance was assessed by Wilcoxon/Mann-Whitney U test with the Bonferroni-corrected p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
To test our results for the σu scores, we used another predictor of aggregation from the unfolded state, the TANGO algorithm (Fernandez-Escamilla et al., 2004). A supersaturation score based on this algorithm (σuT) (Table S2) produces similar results to those obtained with the σu score (Fig. S1). Furthermore, to test the possibility that the σu and σf scores may give too much weight to the expression levels relative to the aggregation propensities, we increased the exponential weight of aggregation propensities in the σu andσf score. At the highest reweighting that we tested (Fig. S5–S6), supersaturation scores are more strongly correlated with aggregation propensity scores (Human Zagg, σu: 0.88, Human ,σf: 0.88, Worm Zagg, σu: 0.99, Worm , σf: 0.96) than concentration score (Human mRNA expression, σu: 0.35, Human protein abundance, σf: 0.53, Worm mRNA expression, σu: 0.10, Worm protein abundance, σf: −0.015). After reweighting, we find that most of our results are robust over a wide range of aggregation propensity values (Figs. S5–S6).
Still, we observed that concentration is a strong predictor of the variety of aggregation-related phenomena that we have analyzed in this work, as an important component of the predictive power of σf is attributable to this property. Indeed, the widespread aggregation data sets that we considered tend to exhibit protein abundance levels more elevated than corresponding mRNA levels, and while they also tend to be more elevated in than Zagg, the difference is much smaller (Fig. S1). In addition, the ratio of relative σf to σu values is positively correlated with the ratio of relative protein abundance to mRNA levels, but negatively correlated with the ratio of relative to Zagg levels (Fig. S1). The relevance of the concentration levels in rationalizing widespread aggregation measurements is a further indication that the concepts of solubility and supersaturation are key in understanding these data.
Moreover, since both the procedure adopted for pathway construction in the KEGG database (Kanehisa et al., 2010) and the identification of aggregating proteins using mass spectrometry (Olzscha et al., 2011) typically consider only the more abundant proteins, the use of the supersaturation score is likely to help to identify additional proteins that aggregate in disease, particularly those with low concentrations and high aggregation propensities. For example, pathways related to cell surface receptors, including olfactory transduction (p=4.0•10−90) and neuroactive ligand-receptor interaction (p=4.9•10−5), are enriched among those supersaturated proteins that are present at low concentrations.
Discussion
Previous studies that have investigated the causes of proteome-wide aggregation have considered the intrinsic aggregation propensities of proteins (Goldschmidt et al., 2010; Monsellier et al., 2008; Tartaglia et al., 2005; Tartaglia and Vendruscolo, 2010). It has thus been suggested that a diverse collection of proteins can form aggregates, although the presence of molecular chaperones and clearance processes largely prevents proteome-wide aggregation under stress-free conditions (Dobson, 2003; Goldschmidt et al., 2010; Lindquist and Kelly, 2011; Monsellier et al., 2008; Olzscha et al., 2011). It has also been shown that destabilizing mutations may drive soluble proteins towards aggregation, and that a genetic background predisposed to such defects can exacerbate this problem (Gidalevitz et al., 2006; Luheshi et al., 2007). More generally, since protein aggregation in the cellular environment has potentially devastating effects, the expression of aggregation-prone proteins is generally maintained at low levels (Tartaglia et al., 2007) and tightly regulated (Gsponer and Babu, 2012). It has been observed, however, that proteins are only just soluble at the levels at which they are expressed in the cell (Tartaglia et al., 2007) and as these trends are conserved in the yeast, mouse, and human proteomes, it has been suggested that monomeric and aggregate forms of proteins are in an effective homeostatic state (Gsponer and Babu, 2012). Similarly, it was recently shown that highly abundant proteins have fewer aggregation-prone surfaces, an observation consistent with their low aggregation propensities (Levy et al., 2012; Tartaglia et al., 2007). Given these evolutionary constraints, it may be surprising that in vivo aggregation is such a common phenomenon under stress (Chapman et al., 2006; Gidalevitz et al., 2006; Koplin et al., 2010; Liao et al., 2004; Narayanaswamy et al., 2009; Olzscha et al., 2011; Wang et al., 2005; Xia et al., 2008) or aging (David et al., 2010; Koga et al., 2011; Reis-Rodrigues et al., 2012). Our results on protein supersaturation provide an explanation for these observations, as they indicate that not only the intrinsic propensities of proteins to aggregate, but also their cellular concentrations are key factors that distinguish aggregation-prone proteins from those whose homeostasis is more robust (Tables 1, S1). While it has been observed that widespread aggregation occurs upon overexpression, which raises the supersaturation levels (Gsponer and Babu, 2012; Narayanaswamy et al., 2009; Sopko et al., 2006), our results indicate that a substantial fraction of the proteome is intrinsically supersaturated and therefore requires the constant aid of quality control mechanisms such as molecular chaperones to remain soluble.
Table 1. Summary of widespread aggregation data sets analyzed in this work.
Dataset: description of the data used. References: references for the data. Species: species to which the data refers. Original #: number of data points listed in the original published set. # UniProt IDs: number of data points listed after conversion to UniProt ID; for human proteins, only the reviewed UniProt IDs are counted. # σu, # σf, # σuT: of those proteins in the # UniProt IDs column, the number for which the given score is available.
| Dataset | References | Species | Original # |
# UniProt IDs |
# σu | # σf | # σuT |
|---|---|---|---|---|---|---|---|
| Amyloids | (UniProt Consortium, 2012) | Human | 27 | 27 | 20 | 13 | 20 |
| Plaques (Co-aggregators) | (Liao et al., 2004) | Human | 26 | 26 | 26 | 18 | 26 |
| NFTs (Co-aggregators) | (Wang et al., 2005) | Human | 72 | 88 | 75 | 52 | 75 |
| Lewy bodies (Co-aggregators) | (Xia et al., 2008) | Human | 36 | 34 | 33 | 22 | 33 |
| Lewy bodies (Detected) | (Xia et al., 2008) | Human | 707 | 557 | 538 | 380 | 538 |
| Artificial β (Co-aggregators) | (Olzscha et al., 2011) | Human | 133 | 151 | 141 | 97 | 140 |
| Artificial β (Lysate) | (Olzscha et al., 2011) | Human | 3,055 | 3,032 | 2,778 | 1,858 | 2,758 |
| Ageing aggregators | (David et al., 2010) | Worm | 720 | 719 | 517 | 577 | 512 |
| Ageing aggregators | (Reis-Rodrigues et al., 2012) | Worm | 203 | 203 | 160 | 178 | 159 |
| Complexes | (Licata et al., 2012) | Human | 1,729 | 1,637 | – | 941 | – |
| Nuclear complexes | (Luc and Tempst, 2004) | Human | 604 | 630 | – | 319 | – |
| Nuclear proteome | (Ashburner et al., 2000; Huang et al., 2009) | Human | – | 1,942 | – | 1,942 | – |
| Membrane proteins | (Ashburner et al., 2000; Huang et al., 2009) | Human | – | – | 6,167 | 2,285 | 6,140 |
The example of serum albumin illustrates some of the strategies adopted by supersaturated proteins to avoid aggregation, as well as their limitations. Albumin, which is exceptionally abundant, is classified in our analysis as supersaturated (Table S2). This protein, which is ubiquitous in the serum, is usually considered to be very soluble, and yet it has been observed to aggregate in vitro (Costantino et al., 1995; Maruyama et al., 2001) and to form toxic aggregates in the synovial fluid of rheumatoid arthritis patients (Oates et al., 2006). These findings reflect two important aspects that regulate the behaviour of supersaturated proteins – the volume that they occupy and the interactions that they form. Although albumin is highly abundant in the serum, its concentration is still relatively low owing to the large volume of the serum itself. By contrast, when confined in the synovial fluid, which has a smaller volume, the concentration of albumin may become substantially higher. In addition, albumin forms numerous complexes with proteins and other molecules, which may protect it from aggregation, as has been observed for the ribosomal proteins (David et al., 2010; Koplin et al., 2010; Reis-Rodrigues et al., 2012). Future studies should account for the volume occupied by proteins and the complexes they form in order to increase the accuracy of supersaturation predictions.
Although abundant proteins have evolved to be more soluble than those that are of low abundance (Tartaglia et al., 2007), some proteins are expressed at such high concentrations that it may be impossible for them to achieve the necessarily solubility with the constraints of functionality and the need for a stable hydrophobic core. We have found that abundance itself is also a good predictor of widespread aggregation in vivo. This result indicates that highly abundant proteins are intrinsically more at risk of aggregation than low abundant proteins (Table S3), which in turns suggests that highly abundant proteins must be maintained at high solubility levels by the protein homeostasis system. These proteins are therefore more susceptible to aggregation in processes that impair protein homeostasis, such as stress, ageing, or disease. The strong predictive power of abundance underscores the importance of solubility in this phenomenon.
These supersaturated proteins are kinetically, but not thermodynamically, stable in their soluble states (Baldwin et al., 2011; Gazit, 2002; Yoshimura et al., 2012), and are thus likely to be highly dependent on the systems that control protein homeostasis in order to remain folded. The instability of supersaturated proteins is thus expected to arise from the failure of cellular systems that contribute to keeping them soluble. The disruption of this machinery under stress and in disease conditions leads to the aggregation of such proteins by shifting the protein homeostasis boundary for their solubility (Hutt et al., 2009; Taylor and Dillin, 2011). The present study suggests that a widespread failure to maintain proteins in their soluble functional states underlies the diverse and complex pathophysiology of neurodegenerative diseases. The sensitivity of neurons to protein aggregation, therefore, may arise from their high dependence on classes of proteins, such as those identified here, that are inherently and unavoidably at risk. Thus, the initial aggregation of the primary amyloid proteins, such as A β and α-synuclein, may trigger an aggregation cascade that disrupts cellular pathways involving these supersaturated proteins.
In conclusion, we have shown that the presence of a large number of supersaturated proteins in the human proteome rationalizes a wide variety of aggregation phenomena associated with aging and disease. The finding that such proteins are overrepresented in a broad range of biochemical processes linked to neurodegenerative diseases reveals why such processes are particularly vulnerable to the appearance of aggregated species or other factors that compromise proteins homeostasis. We anticipate that the type of analysis that we have described can provide a general and widely applicable basis for tracking the instability of proteomes under specific circumstances. By exploiting recent advances in techniques for proteomic analysis, it may soon become possible to use supersaturation measures to assess quantitatively the vulnerability of the human proteome to aggregation and the risk of neurodegenerative disease in individuals over the course of their lives.
Methods
The concentration and aggregation propensity of a given protein were combined to produce the supersaturation score:σ = C + Z, where C is the logarithm of the concentration derived from the mRNA expression or protein abundance levels (see Tables S1 and S2), and Z is the Zyggregator score. The σ scores were then re-centered such that the median of each database was zero and used to analyze proteins associated with widespread aggregation (Chapman et al., 2006; David et al., 2010; Gidalevitz et al., 2006; Koplin et al., 2010; Liao et al., 2004; Olzscha et al., 2011; Reis-Rodrigues et al., 2012; Wang et al., 2005; Xia et al., 2008), by comparing them to a control lysate when available, or to the full proteome database otherwise. In other cases, comparisons were made against the whole proteome data set. Similar procedures were followed for protein complexes (Licata et al., 2012; Luc and Tempst, 2004). A summary of data sets used in this study is provided in Tables 1 and S1. DAVID (Gidalevitz et al., 2006) was used to find KEGG (Kanehisa et al., 2010) pathways enriched in proteins at or above the 95th percentile of each σ score, with the full database set as background. Error tests were performed by introducing Gaussian noise into the full σu and σf databases in 100 independent trials, or by changing the weighting of aggregation propensity in the un-centered scores, and then re-computing Mann-Whitney U p-values and median fold changes each time.
Supplementary Material
Protein supersaturation is an intrinsic aspect of protein homeostasis
Supersaturated proteins form a metastable sub-proteome
Supersaturated proteins are overrepresented in neurodegenerative pathways
Supersaturated proteins undergo aggregation upon cellular stress or ageing
Acknowledgements
P.C. was supported by grants from the US-UK Fulbright Commission and St John’s College, University of Cambridge. R.I.M. was supported by grants from the National Institutes of Health (NIGMS, NIA, NINDS), the Ellison Medical Foundation, and the Daniel F. and Ada L. Rice Foundation. C.M.D. and M.V. were supported by grants from the Wellcome Trust and the UK Biotechnology and Biological Sciences Research Council. P.C. thanks Francis J. DiTraglia for valuable discussions regarding robustness testing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes 6 Figures, 3 Tables, Supplemental Experimental Procedures, and Supplemental References, which can be found with this article online at (to be filled)
References
- Agostini F, Vendruscolo M, Tartaglia GG. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012;421:237–241. doi: 10.1016/j.jmb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Knowles TPJ, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- Belli M, Ramazzotti M, Chiti F. Prediction of amyloid aggregation in vivo. EMBO Rep. 2011;12:657–663. doi: 10.1038/embor.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc. Natl. Acad. Sci. USA. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Bioch. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu KN, Xu KX, Strathearn KE, Liu F, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino HR, Langer R, Klibanov AM. Aggregation of a lyophilized pharmaceutical protein, recombinant human albumin - effect of moisture and stabilization by excipients. Nat. Biotech. 1995;13:493–496. doi: 10.1038/nbt0595-493. [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotech. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid - from bacteria to humans. Trends Bioch. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Gazit E. The"correctly folded" state of proteins: Is it a metastable state. Angewandte Chemie-International Edition. 2002;41:257-+. doi: 10.1002/1521-3773(20020118)41:2<257::aid-anie257>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Golden TR, Hubbard A, Dando C, Herren MA, Melov S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging Cell. 2008;7:850–865. doi: 10.1111/j.1474-9726.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Babu MM. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Rep. 2012;2:1425–1437. doi: 10.1016/j.celrep.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hofrichter J, Ross PD, Eaton WA. Supersaturation in sickle cell hemoglobin solutions. Proc. Natl. Acad. Sci. USA. 1976;73:3035–3039. doi: 10.1073/pnas.73.9.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hutt DM, Powers ET, Balch WE. The proteostasis boundary in misfolding diseases of membrane traffic. FEBS Lett. 2009;583:2639–2646. doi: 10.1016/j.febslet.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucl. Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res. Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, De S, Teichmann SA. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc. Natl. Acad. Sci. USA. 2012;109:20461–20466. doi: 10.1073/pnas.1209312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Cheng D, Wang J, Duong DM, Losik TG, Gearing M, Rees HD, Lah JJ, Levey AI, Peng J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J. Biol. Chem. 2004;279:37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucl. Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases-progress and prognosis. Cold Spring Harb. Perspect. Biol. 2011;3:a004507. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luc PV, Tempst P. PINdb: A database of nuclear protein complexes from human and yeast. Bioinformatics. 2004;20:1413–1415. doi: 10.1093/bioinformatics/bth114. [DOI] [PubMed] [Google Scholar]
- Luheshi LM, Tartaglia GG, Brorsson A-C, Pawar AP, Watson IE, Chiti F, Vendruscolo M, Lomas DA, Dobson CM, Crowther DC. Systematic in vivo analysis of the intrinsic determinants of amyloid beta pathogenicity. PLoS Biol. 2007;5:2493–2500. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: From cradle to grave. Annu. Rev. Bioch. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Katoh S, Nakajima M, Nabetani H. Mechanism of bovine serum albumin aggregation during ultrafiltration. Biotechnol. Bioeng. 2001;75:233–238. doi: 10.1002/bit.10001. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Kim S, Morimoto RI. Huntingtin and mutant sod1 form aggregate structures with distinct molecular properties in human cells. J. Biol. Chem. 2006;281:4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- Monsellier E, Ramazzotti M, Taddei N, Chiti F. Aggregation propensity of the human proteome. PLoS Comp. Biol. 2008;4:e1000199. doi: 10.1371/journal.pcbi.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notterpek L, Ryan MC, Tobler AR, Shooter EM. PMP22 accumulation in aggresomes: Implications for cmt1a pathology. Neurobiol. Dis. 1999;6:450–460. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- Oates KMN, Krause WE, Jones RL, Colby RH. Rheopexy of synovial fluid and protein aggregation. J. R. Soc. Interface. 2006;3:167–174. doi: 10.1098/rsif.2005.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Levy ED, Tartaglia GG, Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc. Natl. Acad. Sci. USA. 2009;106:10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SHY, Martins LM. Genetic analysis of mitochondrial protein misfolding in drosophila melanogaster. Cell Death Differ. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM. Alzheimer's disease. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- Rajan RS, Illing ME, Bence NF, Kopito RR. Specificity in intracellular protein aggregation and inclusion body formation. Proc. Natl. Acad. Sci. USA. 2001;98:13060–13065. doi: 10.1073/pnas.181479798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, Gaman EA, Vantipalli M, Mooney SD, Gibson BW, Lithgow GJ, et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11:120–127. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodveldt C, Andersson A, De Genst EJ, Labrador-Garrido A, Buell AK, Dobson CM, Tartaglia GG, Vendruscolo M. A rationally designed six-residue swap generates comparability in the aggregation behavior of alpha-synuclein and beta-synuclein. Biochemistry. 2012;51:8771–8778. doi: 10.1021/bi300558q. [DOI] [PubMed] [Google Scholar]
- Schrimpf SP, von Mering C, Bendixen E, Heazlewood JL, Bumann D, Omenn G, Hengartner MO. The initiative on model organism proteomes (iMOP) Proteomics. 2012;12:346–350. doi: 10.1002/pmic.201290015. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease. Cold Spring Harb. Perspect. Biol. 2011;3:a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang DQ, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Bioch. Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pellarin R, Cavalli A, Caflisch A. Organism complexity anti-correlates with proteomic beta-aggregation propensity. Protein Science. 2005;14:2735–2740. doi: 10.1110/ps.051473805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, Vendruscolo M. Proteome-level interplay between folding and aggregation propensities of proteins. J. Mol. Biol. 2010;402:919–928. doi: 10.1016/j.jmb.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. Reorganizing the protein space at the universal protein resource (UniProt) Nucl. Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- Xia Q, Liao L, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Proteomic identification of novel proteins associated with Lewy bodies. Front. Biosci. 2008;13:3850–3856. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Lin YX, Yagi H, Lee YH, Kitayama H, Sakurai K, So M, Ogi H, Naiki H, Goto Y. Distinguishing crystal-like amyloid fibrils and glass-like amorphous aggregates from their kinetics of formation. Proc. Natl. Acad. Sci. USA. 2012;109:14446–14451. doi: 10.1073/pnas.1208228109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.