Variation in Rubisco activase gene promoters (RCAβ) could affect diversity of expression level, which provided a new approach for enhancing soybean productivity by altering the expression level of RCAβ
Key words: Allele mining, eQTL, photosynthesis, promoter, Rubisco activase, seed yield, soybean [Glycine max (L.) Merr.]
Abstract
Understanding the genetic basis of Rubisco activase (RCA) gene regulation and altering its expression levels to optimize Rubisco activation may provide an approach to enhance plant productivity. However, the genetic mechanisms and the effect of RCA expression on phenotype are still unknown in soybean. This work analysed the expression of RCA genes and demonstrated that two RCA isoforms presented different expression patterns. Compared with GmRCAα, GmRCAβ was expressed at higher mRNA and protein levels. In addition, GmRCAα and GmRCAβ were positively correlated with chlorophyll fluorescence parameters and seed yield, suggesting that changes in expression of RCA has a potential applicability in breeding for enhanced soybean productivity. To identify the genetic factors that cause expression level variation of GmRCAβ, expression quantitative trait loci (eQTL) mapping was combined with allele mining in a natural population including 219 landraces. The eQTL mapping showed that a combination of both cis- and trans-acting eQTLs might control GmRCAβ expression. As promoters can affect both cis- and trans-acting eQTLs by altering cis-acting regulatory elements or transcription factor binding sites, this work subsequently focused on the promoter region of GmRCAβ. Single-nucleotide polymorphisms in the GmRCAβ promoter were identified and shown to correlate with expression level diversity. These SNPs were classified into two groups, A and B. Further transient expression showed that GUS expression driven by the group A promoter was stronger than that by the group B promoter, suggesting that promoter sequence types could influence gene expression levels. These results would improve understanding how variation within promoters affects gene expression and, ultimately, phenotypic diversity in natural populations.
Introduction
Soybean [Glycine max (L.) Merr.] is the world’s most widely grown legume, and it provides high levels of protein, oil, and other nutrients for humans. The future demand for soybean will increase not only as the world population increases but also as incomes improve and diets become more meat intensive (Ainsworth et al., 2012). As crop productivity gains through traditional breeding begin to lag, novel strategies for improving crop yield potential have begun to examine supercharging photosynthesis to drive a new ‘green revolution’ (Raines, 2011).
Rubisco activase (RCA), which catalyses the activation of the key photosynthesis enzyme Rubisco (Portis, 2003), has been identified as one of several new potential targets for improving plant productivity and seed yield (Yin et al., 2010b). In several plant species, there are two closely related forms of RCA proteins, with molecular masses of 41kDa (β-isoform) and 47kDa (α-isoform), although tobacco only has the β-isoform (Salvucci et al., 1987). These two isoforms are capable of activating Rubisco (Salvucci et al., 2003), but they also have physiological significance in thermal sensitivity under heat stress conditions (Sanchez de Jimenez et al., 1995; Crafts-Brandner and Salvucci, 1997, 2004; Law and Crafts-Brandner, 2001; Portis, 2003). Studies on rice (Wang et al., 2010) and spinach (Crafts-Brandner et al., 1997) demonstrated that the α-isoform was more thermostable than the β-isoform, suggesting that the α-RCA isoform may play an important role in photosynthetic acclimation to moderate heat stress in vivo, whereas the β-RCA isoform was shown to play a major role in maintaining Rubisco’s initial activity under normal conditions (Wang et al., 2010). Understanding the genetic basis of RCA gene regulation and the differential expression of the two isoforms may provide a mechanism for optimizing Rubisco activation under prevailing environmental conditions (Crafts-Brandner et al., 1997).
In contrast to the RCA gene and its encoded protein, which have been well studied in a number of species, the molecular mechanism of transcriptional regulation about RCA gene has thus far been only studied in spinach (Orozco and Ogren, 1993), Arabidopsis (Liu et al., 1996), potato (Qu et al., 2011), and rice (Yang et al., 2012). Gene transcription is a primary intermediate between the information encoded in the genome and the final phenotype. In this context, variation in expression level may be an important cause of phenotypic variation (Alonso-Blanco et al., 2009; Bullard et al., 2010). A previous study demonstrated that many phenotypic differences between species may be due to alteration in gene expression regulation rather than protein structure (King and Wilson, 1975). Recent studies on soybean RCA genes have shown that the expression of the two isoforms was positively correlated with Rubisco activity, photosynthetic rate, and seed yield (Yin et al., 2010b). Thus, an analysis of the determinants of RCA expression not only helps in understanding the genetic basis of RCA regulation but also provides an approach to improve soybean photosynthesis and productivity by altering RCA expression levels to optimize Rubisco activation under the prevailing environmental conditions.
Expression quantitative trait locus (eQTL) mapping has been used to identify loci that might affect gene expression in a cis- or trans-acting manner (Delker and Quint, 2011). However, the nature of the sequence variation in the eQTLs that cause expression changes associated with phenotypic variation is a key and usually unknown feature for researchers and breeders (Vuylsteke and van Eeuwijk, 2008). Currently, mining for sequence variation is gaining more importance in the context of gene expression (Kumar et al., 2010). When a eQTL is cis-acting, cis-regulatory polymorphisms may affect the gene expression level in an allele-specific manner by altering cis-acting regulatory elements in the promoter or by modifying target sites for mRNA processing and stability in the 3′-untranscribed regions (3′-UTR), whereas trans-acting polymorphisms modify the sequence polymorphisms of the transcription factor or transcription factor-binding sites at cis-regulatory sequences in an allele-specific manner for both alleles, causing expression changes in trans-acting factors (Vuylsteke and van Eeuwijk, 2008). Interestingly, both types of polymorphisms are associated with regulatory sequence variations of the promoter, which affect cis-regulatory polymorphisms and trans-acting polymorphisms by altering the 5′-upstream cis-acting regulatory elements or transcription factor-binding sites. This work attempted to use a new strategy that combines eQTL mapping and allele mining in a natural population to understand the genetic basis of RCA gene regulation in soybean.
Both of the genes encoding the larger isoform (GmRCAα) and smaller isoform (GmRCAβ) in soybean have been cloned and characterized (Yin et al., 2010b). However, the expression pattern, genetic basis of the genes’ regulation, and the correlation between RCA expression and phenotype under natural conditions remained unknown. In this study, further analysis found that different patterns of RCA isoform expression were present in soybean, which may be necessary to control enzyme activity and increase overall soybean photosynthesis and productivity. In addition, both genes had a positive correlation with soybean photosynthesis and productivity under natural conditions. To understand the genetic basis of GmRCAβ gene regulation, a new strategy was used to identify the single-nucleotide polymorphisms (SNPs) that determine the expression level by combining eQTL mapping and allele mining in a natural population. All SNPs associated with GmRCAβ gene expression existed in a linked manner with the same P-value, indicating that these SNPs might affect gene expression together, and any of the SNPs might be used as an allele-specific molecular marker (Kumar et al., 2010) to optimize gene expression. These allele-specific molecular markers would contribute to understanding the molecular basis of the genetic regulation of gene expression and facilitate the introgression of the novel alleles through marker-assisted breeding.
Materials and methods
Plant materials and plant growth conditions
Soybean Kefeng No. 1 was grown under natural conditions in the field at Nanjing Agricultural University. Extracts prepared from the plants were used for immunoblot and expression pattern analysis of the RCA gene. Sowing was done on 17 June 2011. Once the upper-third leaves had fully expanded, they were collected and frozen immediately in liquid nitrogen for Western blot analysis. Leaves, stems, roots, and flowers collected at the flowering stage (R2 stage) and seeds harvested at the full seed stage (R6 stage) were used for the gene expression analysis. At the R2 stage, the mature upper-third leaves were collected individually from three plants at different time points during a 48-h cycle for analysis of the diurnal pattern of RCA expression.
A panel of 219 soybean landraces collected at 53–24 ° N 134–97 ° E in China was used to evaluate RCA gene expression, empirical chlorophyll transient parameters, seed weight, and seed yield. Of the 219 landraces, 191 landraces that had been genotyped for 1142 SNPs with minor allele frequencies of higher than 10% were used for the association analysis. These landraces originated from 24 provinces in China and covered four ecological regions: northeast region, north region, Huanghuai region, and south region (Supplementary Fig. S1, available at JXB online). These four regions represented the four major planting areas of soybean in China (Li et al., 2008). Seeds of all landraces were obtained from germplasm storage in the National Center for Soybean Improvement (Nanjing, China). The trials were performed under natural conditions at Jiangpu Experimental Station, Nanjing Agricultural University. All landraces were grown using previously described procedures (Yin et al., 2010b).
To control the environmental effects on phenotypic evaluation, the natural population was divided into four groups according to their time to maturity as observed in previous years (data not shown). Each group was sown at different times so that when trait data were collected, all landraces were at a similar growth stage. Sowing was done on 7, 17, and 27 June and 7 July 2011. Nutrition and water were supplied sufficiently throughout the experiment to avoid potential nutrient and drought stresses. At the R6 stage of development, the mature upper-third leaves were collected individually from three plants of each landrace in the morning (09:00–11:30) on a sunny day, frozen immediately in liquid nitrogen, and stored at –80 °C. In order to reduce systematic errors caused by the time of sampling, time segment was taken as the block, which represented one repetition of each landrace, and sampling of each block by three groups of people was accomplished in a short period of time (less than 50 minutes) to reduce the effect of time of sampling on each material within block. The variance analysis of GmRCAα and GmRCAβ expression is shown in Supplementary Table S1.
RNA isolation and synthesis of cDNA
Total RNA was isolated from leaves using a RNA Plant Extraction Kit (Tiangen, China) and treated with RNase-free DNase I (Takara, Japan) to remove contamination by genomic DNA. Approximately 2 µg of purified total RNA was reverse transcribed using AMV reverse transcriptase (Takara) with oligo-dT-18 as the primer (Takara), according to the manufacturer’s instructions.
DNA sequencing and genotyping
Genomic DNA was extracted from the young leaves of soybean plants using a DNA plant extraction kit (Tiangen). The GmRCAβ promoter was amplified from genomic DNA with the following primers: sense 5′-TGGCAGTAGCTGTTTCTAGTGATGG-3′ and anti-sense 5′-GCCTACTCGTTTTTACATCCCCTTA-3′. PCR was conducted in a 50-µl reaction volume using KOD polymerase (Toyobo, Japan) following the manufacturer’s recommendations, using a PTC-225 thermal cycler (MJ Research, Watertown, MA, USA). The cycling program consisted of one cycle at 94 °C for 2min, 35 cycles at 94 °C for 15 s and 68 °C for 3min, and one cycle at 4 °C for 10min. Amplification products were separated by electrophoresis on 1% agarose, and the band of expected size was excised and purified with a PCR gel purification kit AP-GX-4 (Axygen, China). Three sample PCR products were purified from soybean landraces and sequenced at Invitrogen (Shanghai, China). All sequences were checked manually and all singletons observed were verified by sequencing newly amplified fragments. Then the sequence was aligned using CLUSTALX version 1.83 (Thompson et al., 1997). A manual check was performed in every case to ensure sequencing and alignment quality. Polymorphism data was analysed using DnaSP version 4.10 (Rozas et al., 2003) to identify sequence variation. Identified SNPs with a minor allele frequency at least 10% were genotyped in the natural population used for the candidate-gene association analysis.
Bioinformatics analysis
Multiple sequence alignment was performed using CLUSTALX version 1.83. Phylogenetic trees were constructed using both neighbour-joining and Bayesian approaches. In the neighbour-joining method, the phylogenetic analysis was conducted using MEGA version 4.1(Kumar et al., 2008). The parameter setups were as follows: model = p-distance; bootstrap = 1000 replicates; and gap/missing data = pairwise deletion. In the Bayesian method, the analyses were conducted using MrBayes version 3.1 (Ronquist and Huelsenbeck, 2003) with the Jones, Taylor, Thornton substitution model (Jones et al., 1992), four chains, 1 million generations, and two runs. Trees were sampled every 100 generations, discarding a burn-in of 250 000 generations. The putative cis-acting regulatory elements were identified through search against both the PlantCARE and PLACE databases (Prestridge, 1991; Higo et al., 1999; Lescot et al., 2002).
Vector construction and Agrobacterium-mediated transformation
Two types of promoter sequences of GmRCAβ (type 2 and type 4) which belonged to promotor group A and group B, respectively (see Results) were cloned in soybean Suxie No. 1 and Kefeng No. 1, respectively. These 2300-bp fragments were fused with the β-glucuronidase (GUS) gene as a reporter in the binary vector pCAMBIA1381Z (Cambia, Australia) digested with corresponding restriction enzymes. The binary vector constructs, promoterless control (pCAMBIA1381Z), and CaMV 35S promoter control (pCAMBIA1301) were transformed into Agrobacterium tumefaciens EHA105 by the freeze–thaw method. The constructs were introduced into the soybean cotyledonary node via Agrobacterium-mediated transformation following Zhang et al. (1999).
Histochemical staining
GUS activity was determined in soybean cotyledonary nodes based on a previously described method (Jefferson et al., 1987) with some modifications. Plant materials were immersed in 50mM sodium phosphate buffer (pH 7.0) containing 0.1% Triton X-100, 10mM EDTA, 0.5mg/ml 5-bromo-4-chloro-3-indolyl glucuronide, 1mM K3[Fe(CN)6], and 1mM K4[Fe(CN)6] for 24h at 37 °C in darkness. The staining solution was then removed and the samples were dehydrated using an ethanol series of 70, 80, 90, and 100%, with exposure to each for a minimum of 60min. GUS staining was observed under an Olympus SZX12 stereomicroscope and photographed with a digital camera (CoolSNAP, RS photometrics).
Leaf protein extraction and Western blot analysis
For immunoblot analysis, the proteins were extracted using previously described procedures (Yin et al., 2010b). Protein extracts were separated by 12% SDS-PAGE and then transferred onto a nitrocellulose membrane (Bio-Rad) as described by Mitsuhashi and Feller (1992) using a Trans Blot system (Bio-Rad). Then, the nonspecific binding of antibodies was blocked with 5% nonfat dried milk in phosphate-buffered saline (pH 7.4) for 2h at room temperature. The membranes were then incubated overnight at 4 °C with polyclonal anti-cotton RCA antibodies (AS10700, Agrisera) diluted to 1:10 000 in phosphate-buffered saline plus 1% nonfat milk. The immune complexes were detected using goat anti-rabbit IgG-horseradish peroxidase (sc-2004; Santa Cruz Biotechnology) and developed with a solution containing 3,3′-diaminobenzidine tetrahydrochloride as the peroxidase substrate and then the membranes were scanned.
Trait measurement and phenotypic data collection
The data on seed weight and seed yield were collected at maturity. Seeds of six plants per genotype were hand harvested and dried to constant weight (Yin et al., 2010b).
Empirical chlorophyll transient parameters, such as Fv/Fm, ΦPSII, qP, and NPQ (see Table 2 for definitions), were measured using the upper-third leaf of the soybean plants at the R2 development stage. All measurements were performed at 25 °C using a PAM fluorometer (PAM2100, Heinz Walz, Effeltrich, Germany) using a previously described procedure (Yin et al., 2010a).
Table 2.
Correlation coefficients among GmRCAα and GmRCAβ expression and yield components in a natural soybean population
| Trait | Fv/Fm | ΦPSII | qP | NPQ | Seed weight | Seed yield |
|---|---|---|---|---|---|---|
| GmRCAβ expression | 0.107 | 0.219* | 0.319** | –0.042 | 0.180* | 0.303** |
| GmRCAα expression | 0.087 | 0.198* | 0.304** | –0.031 | 0.138 | 0.251** |
Fv/Fm, maximum quantum yield of PSII primary photochemistry in the dark-adapted state; ΦPSII, actual quantum yield in the light-adapted state; qP, photochemical quenching coefficient; NPQ, non-photochemical quenching parameter describing the regulated dissipation of excess energy. *, P<0.05; **, P<0.01.
Expression levels were determined by quantitative real-time PCR (qRT-PCR). Because of the high homology between GmRCAα and GmRCAβ (Yin et al., 2010b), specific primers and Taq Man-MGB probe (ABI, America) were applied to detect the specific expression levels. Expression of the soybean endogenous reference gene Tubulin (GenBank accession number AY907703.1) was used to normalize the transcript level in each sample. The primers and Taq Man-MGB probe used were as follows: Tub-F (5′ GGAGTTCACAGAGGCAGAG-3′) and Tub-R (5′-CACTTACGCATCACATAGCA-3′) for Tubulin; RCAa-F (5′-GATGGGCGTATGGAGAAGTTCT-3′), RCAα-R (5′-TGCGGAAAATTCCATTGCA-3′), and RCAα-MGB (5′-ACG ATCGTGTTGGCG-3′) for GmRCAα; RCAβ-F (5′-GGGACCAG CTTGAAGAA GGTTA-3′), RCAβ-R (5′-TGCTGGGTCTCTTCA ATCTCTTT-3′), and RCAβ-MGB (5′-CAGCAAGGTTTCCGG TG-3′) for GmRCAβ.
Statistical analysis and association mapping
Statistical analyses were performed using SAS system 9.0 for Windows and Microsoft Excel 2003. Analysis of variance was performed using SAS PROC GLM. The mean values of RCA expression for each landrace were calculated using SAS PROC MEANS. The regressive coefficients among the traits were calculated using SAS PROC REG. The Pearson phenotypic correlations among the traits were calculated using SAS PROC CORR. The significant differences in the gene expression levels of two RCA genes were determined using the two-tailed Student’s t-test of Microsoft Excel 2003, and differences at P ≤ 0.01 were considered significant. The association of each SNP in the promoter region with expression levels was examined by the candidate-gene association analysis method (Harjes et al., 2008; Rafalski, 2010). eQTLs for the expression of GmRCAβ were identified using genome-wide association analysis with the same association mapping panel and the same method used in a previous study (Hao et al., 2012).
Results
Expression and protein levels
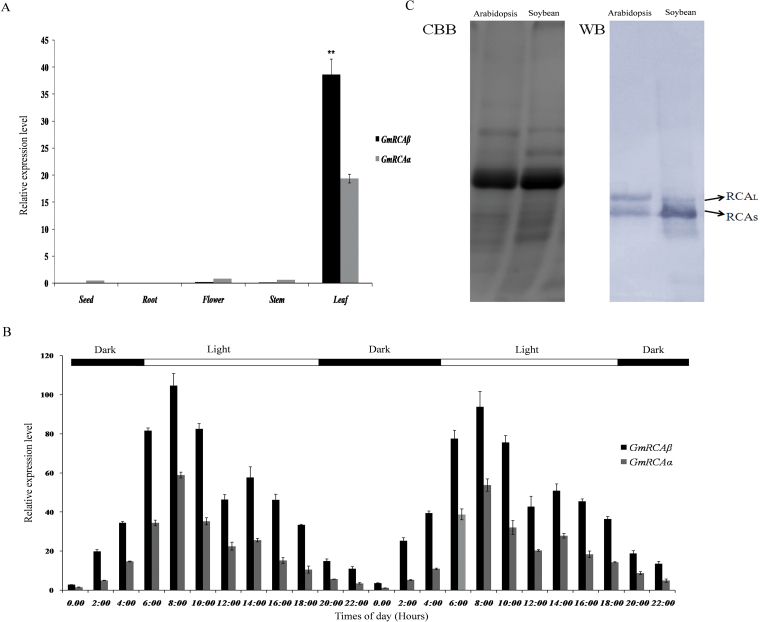
Quantitative real-time PCR analysis was conducted to investigate the expression of RCA genes in soybean. RCA expression was high in leaves where photosynthesis takes place, but low in flowers, stems, seeds, and roots (Fig. 1A). To examine whether RCA gene expression in soybean followed a diurnal pattern in addition to being affected in a tissue-specific manner, qRT-PCR analysis was performed with total RNA extracted from soybean leaves harvested at different time points during a 48-h cycle. The maximal level of RCA gene expression was detected at 08:00, 2h after the beginning of the light period. Following this peak in expression, the levels were significantly decreased at noon and then slightly increased in the early afternoon, but the overall decreasing tendency remained (Fig. 1B). The minimum observed levels were detected at midnight. These results suggested that the expression of RCA followed a diurnal pattern and was highest in the morning.
Fig. 1.
Expression pattern and Western blot analysis of RCA genes in soybean. (A) Quantitative real-time PCR analysis of organ-specific RCA expression in soybean; total RNA was isolated from flowers, roots, stems, and leaves harvested at R2 stage (flowering) and from seeds harvested at R6 stage (full seed); error bars represent standard errors of three independent repetitions. (B) Diurnal pattern of RCA mRNA accumulation in soybean leaves; total RNA was extracted at different time points during a 48-h cycle from the upper-third leaves of soybean at R2 stage; error bars represent standard errors of three independent repetitions. (C) Western blot analysis of the two RCA isoforms from Arabidopsis and soybean; extracts of total soluble protein from Arabidopsis and fully expanded young soybean leaves (4.2 µg) were separated by 12% SDS-PAGE; protein bands were detected by Coomassie blue staining (CBB) or Western blot (WB) probed with antibodies to RCA and visualized using alkaline phosphatase conjugated to a secondary antibody.
Because the two RCA isoforms in soybean are derived from separate genes, and are not produced by alternative splicing of a single RCA gene (Yin et al., 2010b), they might differ in expression and protein levels. Thus, the current work investigated the correlation between the two RCA isoforms at the mRNA and protein levels. As shown in Fig. 1A and B, GmRCAβ mRNA accumulated to a higher level than GmRCAα mRNA. Further analysis indicated that the expression levels of GmRCAα and GmRCAβ were significantly different (Fig. 1A). To test whether the relatively low expression of GmRCAα was in line with changes in protein levels, Western blot analysis was conducted. The results indicated that soybean plants contained two isoforms, with a higher concentration of the small isoform (Fig. 1C). In contrast, Arabidopsis contained two isoforms in almost equal amount, as reported by other study (Salvucci et al., 1987).
RCA promoter sequences
The promoter sequences of GmRCAα and GmRCAβ were analysed to determine whether the two different expression levels could arise from the regulatory elements and promoter sequence differences. The alignment of the 2300-bp promoter sequences of GmRCAα and GmRCAβ showed that two sequences matched poorly, with only 45.2% identity (Supplementary Fig. S2). Bioinformatics analysis showed that several important cis-acting elements, including the elements responsive to light, biotic stress and phytohormone and other basal element in the upstream regulatory region were similar for the two promoters, while the specific cis-elements and their copy numbers were different between GmRCAα and GmRCAβ promoter (Table 1). Moreover, heat stress-responsive elements were observed in the promoter region of GmRCAα but not in GmRCAβ, which was consistent with the previous study that the isoforms have physiological significance in thermal sensitivity under heat-stress conditions (Sanchez de Jimenez et al., 1995; Crafts-Brandner and Salvucci, 1997, 2004; Law and Crafts-Brandner, 2001; Portis, 2003). These results indicated that expression and regulation of GmRCAα and GmRCAβ was a complex process and that the difference in numbers of specific cis-elements and some common cis-elements might be the reason for the different expression of the two isoforms.
Table 1.
Comparison of cis-acting elements in the promoter regions of GmRCAα and GmRCAβ
| Element type | Name | Copy number in promoter | Motif sequence (5′–3′) | Function | |
|---|---|---|---|---|---|
| GmRCAα | GmRCAβ | ||||
| Light | GA motif | 1 | 0 | AAAGATGA | Part of a light-responsive element |
| I-box | 1 | 0 | CTCTTATGCT | Part of a light-responsive element | |
| 1 | 0 | TATTATCTAGA | |||
| ATCT motif | 1 | 0 | AATCTAATCT | Part of a conserved DNA module involved in light responsiveness | |
| Sp1 | 1 | 0 | CC(G/A)CCC | Light-responsive element | |
| TCT motif | 1 | 0 | TCTTAC | Part of a Light-responsive element | |
| chs-CMA1a | 1 | 0 | TTACTTAA | Part of a light-responsive element | |
| GTGGC motif | 1 | 0 | CATCGTGTGGC | Part of a light-responsive element | |
| AT1 motif | 0 | 1 | AATTATTTTTTATT | Part of a light-responsive module | |
| GAG motif | 0 | 1 | AGAGATG | Part of a light-responsive element | |
| GT1 motif | 0 | 1 | GGTTAA | Light-responsive element | |
| G-box | 0 | 3 | CACGTT | cis-Acting regulatory element involved in light responsiveness | |
| 1 | 0 | CACGTG | |||
| 1 | 0 | TACGTG | |||
| 1 | 2 | CACGTG | |||
| 0 | 1 | CACGTA | |||
| 0 | 1 | ACACGTGT | |||
| Box 4 | 3 | 3 | ATTAAT | Part of a conserved DNA module involved in light responsiveness | |
| Box II | 1 | 0 | CCACGTGGC | Part of a light-responsive element | |
| 0 | 1 | ACACGTAGA | |||
| Circadian | Circadian | 2 | 2 | CAAAGATATC | cis-Acting regulatory element involved in circadian control |
| Phytohormone | |||||
| AuxRR core | 0 | 1 | GGTCCAT | cis-Acting regulatory element involved in auxin responsiveness | |
| TGA element | 0 | 2 | AACGAC | Auxin-responsive element | |
| TCA element | 0 | 1 | GAGAAGAATA | cis-Acting element involved in salicylic acid responsiveness | |
| P-box | 0 | 1 | CCTTTTG | Gibberellin-responsive element | |
| GARE motif | 0 | 1 | TCTGTTG | Gibberellin-responsive element | |
| 1 | 0 | AAACAGA | |||
| ABRE | 1 | 0 | TACGTG | cis-Acting element involved in the abscisic acid responsiveness | |
| 1 | 2 | CACGTG | |||
| 1 | 0 | ACGTGGC | |||
| Biotic stress | HRE | 2 | 0 | AAAAAATTTC | cis-Acting element involved in heat stress responsiveness |
| Box-W1 | 1 | 0 | TTGACC | Fungal elicitor-responsive element | |
| MBS | 0 | 1 | CGGTCA | MYB binding site | |
| 0 | 2 | TAACTG | MYB binding site involved in drought inducibility | ||
| ARE | 0 | 1 | TGGTTT | cis-Acting regulatory element essential for anaerobic induction | |
| Basal element | CAAT box | 23 | 21 | CAATT/CAAT/CCAAT | Common cis-acting element in promoter and enhancer regions |
| TATA box | 42 | 45 | TATA/ATATAT/TTTTA | Core promoter element around –30bp of transcription start | |
| 5′-UTR pyrimidine-rich stretch | 1 | 2 | TTTCTTCTCT | cis-Acting element conferring high transcription levels | |
| Other | CAT box | 1 | 1 | GCCACT | cis-Acting regulatory element related to meristem expression |
| GCN4 motif | 0 | 4 | CAAGCCA | cis-Regulatory element involved in endosperm expression | |
| HD zip | 0 | 1 | CAAT(A/T)ATTG | Element involved in differentiation of the palisade mesophyll cells | |
| 0 | 1 | CAAT(G/C)ATTG | Element involved in the control of leaf morphology development | ||
| Skn-1 motif | 2 | 2 | GTCAT | cis-Acting regulatory element required for endosperm expression | |
| ATGCAAAT motif | 1 | 0 | ATACAAAT | cis-Acting regulatory element associated to the TGAGTCA motif | |
| O2 site | 1 | 0 | GATGACATGA | cis-Acting regulatory element involved in zein metabolism regulation | |
ABRE, abscisic acid-responsive element; ARE, anaerobic induction- responsive element; GARE, GA-responsive element; HD zip, homeodomain leucine zipper; HRE, heat stress-responsive element; MBS, MYB binding site; UTR, untranscribed region.
Correlation between gene expression and chlorophyll fluorescence and yield in a natural population
Empirical chlorophyll transient parameters, such as Fv/Fm, ΦPSII, qP, and NPQ, have been widely accepted as reflecting the structure and function of the photosynthetic apparatus (Papageorgiou, 2004). To further investigate the relationship between the effect of RCA gene expression on soybean photosynthesis and productivity, gene expression, chlorophyll fluorescence parameters, and yield components was measured in a natural population. As shown in Supplementary Table S1, the expression levels of the two RCA genes displayed high diversity in soybean germplasm, ranging from 0.47 to 66.41 for GmRCAα expression and from 2.38 to 196.49 for GmRCAβ expression. Variation analysis indicated that these wide ranges might mainly be caused by genotypic differences. Further analysis of gene expression in the natural population indicated that GmRCAβ accumulated more mRNA in all landraces than GmRCAα (Supplementary Fig. S3A), and the levels of GmRCAα and GmRCAβ expression were significantly different (Supplementary Fig. S3B). Correlation analysis suggested that the expression levels of both genes were positively correlated with ΦPSII, qP, and seed yield, but the correlations between GmRCAβ expression level and ΦPSII, qP, and seed yield were slightly higher than for GmRCAα. In addition, GmRCAβ expression level displayed a positive correlation with seed weight (Table 2). Overall, these results suggested that soybean landraces exhibited considerable natural variation in RCA gene expression level and that both isoforms could play an important role in modulating soybean photosynthetic capacity and seed yield.
eQTL analysis of GmRCAβ expression
Since RCA genes were positively correlated with chlorophyll fluorescence parameters and seed yield, changes in RCA expression at the transcript level may have a potential applicability in breeding for enhanced soybean productivity. The present study analysed the genetic regulation of GmRCAβ expression and evaluated the associations with 1142 SNPs by genome-wide association analysis. A total of 13 SNPs were identified as having significant marker-trait associations at P ≤ 0.01 (–logP ≥ 2.00) for the expression of the GmRCAβ (Table 3). Here, a cis-eQTL was defined as being located within 5Mb of the target gene; otherwise, it was termed as a trans-eQTL (Morley et al., 2004; Ghazalpour et al., 2006; Gatti et al., 2009, 2011; Swanson-Wagner et al., 2009; Grundberg et al., 2010). Among the 13 SNPs, BARC-040479-07752 was located in the same chromosome as GmRCAβ (chromosome 18) and was 1.62Mb away from GmRCAβ, suggesting that this SNP might be a cis-eQTL, while BARC-021337-04040 was also located in chromosome 18 and was 7.87Mb away from GmRCAβ, suggesting that this SNP might act in trans, but GmRCAβ was located between the two SNPs. The other 11 SNPs were located in other chromosomes, indicating that they may influence the expression levels of GmRCAβ via trans-regulation. These results indicated that GmRCAβ expression might be controlled by a combination of both cis-acting and trans-acting eQTL. In addition, the cis-acting eQTL that had the stronger association (–logP = 5.64) with expression may mainly control the expression of GmRCAβ.
Table 3.
Expression quantitative trait loci mapping of GmRCAβ in a natural population
| SNP | Chromosome | Position | −logP |
|---|---|---|---|
| BARC-032333-08950 | 14 | 5 286 848 | 2.00 |
| BARC-042189-08197 | 4 | 43 683 671 | 2.18 |
| BARC-028177-05786 | 6 | 13 550 805 | 2.64 |
| BARC-028177-05785 | 6 | 13 550 805 | 2.26 |
| BARC-021247-04012 | 6 | 47 038 336 | 2.02 |
| BARC-017541-03068 | 17 | 38 843 283 | 3.06 |
| BARC-025663-04986 | 15 | 9 360 685 | 2.10 |
| BARC-040479-07752 | 18 | 1 228 510 | 5.64 |
| BARC-021337-04040 | 18 | 10 714 500 | 2.32 |
| BARC-019775-04370 | 12 | 7 494 368 | 2.03 |
| BARC-039961-07622 | 12 | 15 574 190 | 2.33 |
| BARC-018911-03272 | 10 | 3 519 980 | 2.74 |
| BARC-031677-07213 | 10 | 43 841 452 | 2.36 |
Significant association: P ≤ 0.01, –logP ≥ 2.00. SNP, single-nucleotide polymorphism.
Allelic variation in the GmRCAβ promoter
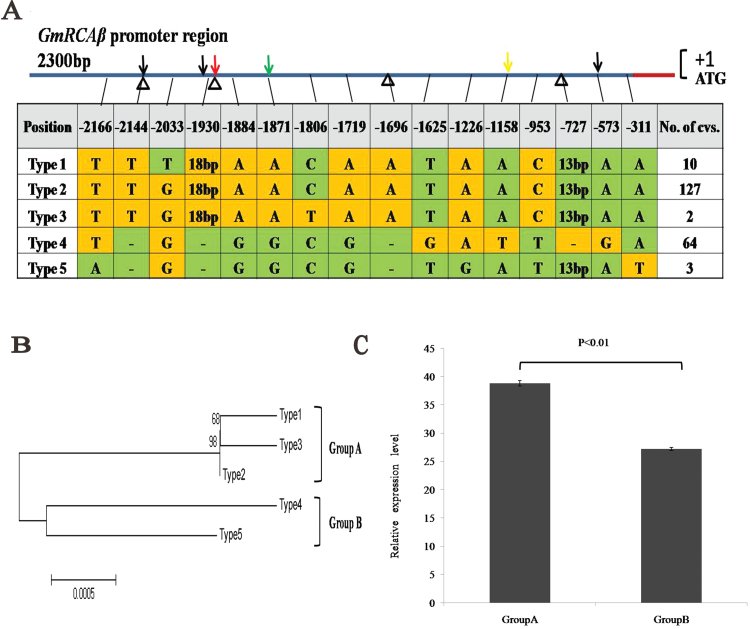
As the expression level of GmRCAβ is the result of interactions of multiple cis-and trans-acting genetic factors, and because cis-acting eQTLs may mainly control GmRCAβ expression, the GmRCAβ promoter sequence, which may affect both the cis- and trans-acting eQTLs (Vuylsteke and van Eeuwijk, 2008), might be a potential factor in expression level diversity. To identify whether sequence polymorphisms in the promoter region of GmRCAβ were related to the observed expression level, a 2300-bp region of the promoter, which was shown to be sufficient for the proper regulation of expression in plant (Orozco and Ogren, 1993; Liu et al., 1996; Qu et al., 2011; Yang et al., 2012), was cloned and sequenced, including the promoter and the 5′-UTR region, in a natural population including 219 landraces of different geographic origins (Supplementary Figs S4 and S5). Multiple sequence alignment revealed 16 polymorphic sites and five types of sequences (Fig. 2A). Phylogenetic trees were constructed using neighbour-joining and Bayesian approaches (Fig. 2B and Supplementary Fig. S6), which indicated that these sequence types were classified into two groups: types 1, 2, and 3 in group A and types 4 and 5 in group B. Genes with group A promoters were expressed at significantly higher levels than those with group B promoters (Fig. 2C), suggesting an association between promoter sequence type and expression level. Of these polymorphisms, five polymorphic sites, including SNPs and indels, were located in core regulatory elements such as the GAG motif, the AT-1 motif, and the TATA box (Table 4).
Fig. 2.
Functional and polymorphic analysis of the GmRCAβ promoter region in a natural population of soybean. (A) Nucleotide polymorphisms in the GmRCAβ promoter region; polymorphic nucleotides are indicated by different colours, red arrow indicates the AT1 motif, black arrows indicate the TATA boxes, green arrow indicates the ARE, and the yellow arrow indicates the GAG motif; arrowheads indicate deletion and insertion sites. (B) Neighbour-joining phylogenetic analysis of GmRCAβ promoter sequences. (C) GmRCAβ mRNA levels in landraces with group A and B promoters; leaves were harvested at R6 stage (full seed) and analysed by quantitative real-time PCR; error bars represent standard errors of three independent repetitions (this figure is available in colour at JXB online).
Table 4.
Polymorphic sites in the promoter region of GmRCAβ
| Position | Polymorphic type | Regulatory element change | Function of cis-elements | Frequency (%) |
|---|---|---|---|---|
| –2166 | T/A | – | – | 1.46 |
| –2144 | 1-base indel | TATA box | Core promoter element around –30 of transcription start | 67.48 |
| –2033 | T/G | – | – | 4.85 |
| –1930 | 18-base indel | AT1 motif | Part of a light-responsive module | 67.48 |
| TATA box | Core promoter element around –30 of transcription start | |||
| –1884 | A/G | – | – | 67.48 |
| –1871 | A/G | ARE | cis-Acting regulatory element essential for anaerobic induction | 67.48 |
| –1806 | C/T | – | – | 0.97 |
| –1719 | A/G | – | – | 67.48 |
| –1696 | 1-base indel | – | – | 67.48 |
| –1625 | T/G | – | – | 68.93 |
| –1226 | A/G | – | – | 1.46 |
| –1158 | A/T | GAG motif | Part of a light responsive element | 68.93 |
| –953 | C/T | – | – | 67.48 |
| –727 | 13-base indel | – | – | 68.93 |
| –573 | A/G | TATA box | Core promoter element around –30 of transcription start | 68.93 |
| –311 | A/T | – | – | 1.46 |
–, not located in regulatory elements.
Candidate-gene association analysis of the SNPs in the promoter region of GmRCAβ
To validate whether the sequence polymorphisms in the promoter region were the causal sites for expression level variation of GmRCAβ, candidate-gene association analysis was performed. All SNPs with a minor allele frequency above 10% were considered. Significantly associated SNPs (including indels) were identified by EM analysis (expectation-maximization, an algorithm for solving mixed models in association analysis; Yu et al., 2005) using TASSEL software. For GmRCAβ, there were seven SNPs (including an 18-bp indel) significantly associated with the expression of GmRCAβ (Table 5). Interestingly, three SNPs were located in the regulatory elements respectively, such as the AT-1 motif, the TATA box, and the anaerobic induction-responsive element. In addition, all associated SNPs existed in a linked manner (in the same linkage disequilibrium block) with the same P-value.
Table 5.
Single-nucleotide polymorphisms in the GmRCAβ promotor region significantly associated with gene expression
| Position | Polymorphic type | Regulatory element change |
|---|---|---|
| –2144 | 1-base indel | TATA box |
| –1930 | 18-base indel | AT1 motif, TATA box |
| –1884 | A/G | – |
| –1871 | A/G | ARE |
| –1719 | A/G | – |
| –1696 | 1-base indel | – |
| –953 | C/T | – |
All significant associations: P = 1.38. –, not located in regulatory elements.
GUS expression driven by different promoter sequence types
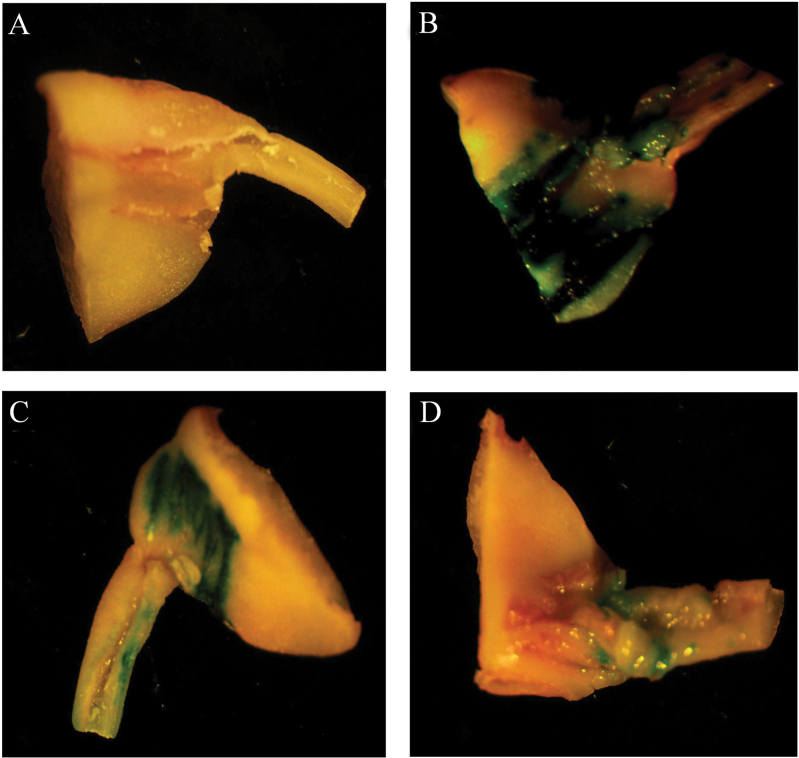
Although several SNPs in the promoter region of GmRCAβ were identified and associated with the expression of GmRCAβ, these SNPs were mainly classified into two groups. Therefore, GUS expression behind group A and group B promoters was compared using a transient assay to determine whether these different promoter types could result in differences in expression level. The type 2 promoter sequence of group A and type 4 promoter sequence of group B were cloned (Supplementary Fig. S7A) and fused with the GUS gene as a reporter (Supplementary Fig. S7B). Histochemical staining indicated that both group A and group B promoters were able to drive the GUS expression, but GUS expression driven by the group A promoter was stronger than that by the group B promoter (Fig. 3). These results indicated that the promoter sequence type of GmRCAβ could influence gene expression level.
Fig. 3.
Histochemical staining of soybean cotyledonary nodes driven by the different promoter sequence types of GmRCAβ: (A) promoterless control; (B) CaMV 35S promoter; (C) group A type promoter; (D) group B type promoter (this figure is available in colour at JXB online).
Discussion
Different RCA isoform expression patterns in soybean
In most plants, the expression of the RCA gene is tissue specific and light inducible and exhibits diurnal and circadian oscillations in addition to being affected by development and phytohormones (Zielinski et al., 1989; Pilgrim and McClung, 1993; Zhang and Komatsu, 2000; Sharma and Komatsu, 2002). In this study, expression pattern analysis of RCA indicated that GmRCAα and GmRCAβ in soybean were organ specific and followed a diurnal pattern (Fig. 1A, B) but that their expression levels were significantly different (Fig. 1A and Supplementary Fig. S3A). Consistent with these results, soybean leaves contained unequal amounts of the two isoforms (Fig. 1C), with higher levels of the small isoform (Salvucci et al., 1987). It is not clear why the different patterns of RCA isoform expression in soybean and other species such as rice and wheat result in the accumulation of a greater amount of β-RCA than α-RCA, whereas certain plant species such as Arabidopsis, camelinam and spinach express equal amounts of α- and β-RCA (Salvucci et al., 1987; Wang et al., 2010; Fukayama et al., 2012). Recent studies on species differences in the regulatory properties of RCA have indicated that the redox regulation of α-RCA, which provides fine control over the sensitivity of the holoenzyme to inhibition by ADP, does not seem to be essential in all plant species. Similarly, ADP inhibition of β-RCA varies among plant species and even occurs in species that also express a α-RCA (Carmo-Silva and Salvucci, 2013). Hence, the different patterns of RCA isoform expression observed in the current study may be necessary for control over enzyme activity and increasing overall soybean photosynthesis and productivity.
What causes the significantly different expression levels of the two RCA genes in soybean? This study compared the sequences of the two promoters, which play a key role in gene expression and regulation. Some cis-elements were specifically present in their promoter region, and the copy numbers of each cis-element were different between two RCA genes (Table 1). It has been reported that promoter inducibility and strength varies depending on cis-element copy numbers and, more specifically, spacing of motifs relative to the TATA box (Rushton et al., 2002; Gurr and Rushton, 2005). The differences in copy numbers of some important cis-elements, such as G-box and pyrimidine-rich 5′-UTR, might cause different expression of the two RCA genes. In addition, I-box elements, which have been shown to be involved in light-regulated and/or circadian clock-regulated expression of photosynthetic genes (Borello et al., 1993; Terzaghi and Cashmore, 1995; Agius et al., 2005), were only present in GmRCAα. However, it has been demonstrated that I-box elements downregulate gene expression in strawberry and melon fruit, suggesting a new function of the I-box elements as a negative regulator (Yamagata et al., 2002; Agius et al., 2005).
Expression levels of GmRCAα and GmRCAβ may modulate seed yield
Although different patterns of RCA gene expression were observed in soybean, the effect of this on yield components remains unknown. A direct link between growth and gene expression has been confirmed in rice, in which different levels of expression that are positively correlated with these traits of this agronomically important grain are dependent on sequence variation in the promoter of the GS5 gene (Li et al., 2011). Studies have shown that endogenous levels of RCA expression are of importance to plant photosynthesis under both optimal (Martínez-Barajas et al., 1997) and supraoptimal temperatures (Ristic et al., 2009). Thus, the present study investigated the effect of expression on soybean yield by evaluating the expression levels of RCA in a natural population. The expression levels of both genes were positively correlated with chlorophyll fluorescence parameters and seed yield, suggesting that changes in RCA expression has a potential applicability in breeding for enhanced soybean productivity. Moreover, the expression levels displayed high diversity in the germplasm, and the phenotypic values in natural population ranged from 0.47 to 66.41 for GmRCAα expression and from 2.38 to 196.49 for GmRCAβ expression. However, the regression analysis indicated that a given change in GmRCAα was associated with a greater change in photosynthesis or yield (Supplementary Table S2). Therefore, further studies need to compare GmRCAα and GmRCAβ in transgenic soybean.
New insights into the genetic regulation of GmRCAβ expression
Gene expression is believed to be a composite reflection of multiple genetic cis- or trans-acting factors, according to eQTL analysis (Delker and Quint, 2011). Moreover, target gene expression can be controlled by a combination of both cis- and trans-acting elements (Druka et al., 2010). A previous study using a recombinant inbred line of soybean detected only two trans-acting eQTLs for GmRCAβ (Yin et al., 2010b), suggesting that expression of this gene might be a consequence of trans-regulation. However, it must be emphasized that all QTL studies rely on natural genetic variation in the population under study (Druka et al., 2010). Only those eQTLs detected in different materials or under multiple environments are valuable for breeding (Yin et al., 2010b). In the present study, further eQTL mapping was conducted in a collection of 219 soybean landraces that represented the genetic diversity of landraces from different geographic origins in China (Supplementary Fig. S1). The eQTL mapping indicated that the expression of GmRCAβ could be controlled by a combination of both cis-acting and trans-acting eQTLs. In agreement with this finding, the complex expression pattern of RCA reflects the interactions of multiple trans-acting protein factors with multiple cognate cis-acting DNA elements (Liu et al., 1996). However, no coincident eQTLs were observed between the current and the previous eQTL study (Yin et al., 2010b). A possible reason for this might be the differences in the mapping populations, since the natural population has higher diversity and has undergone natural selection in the process of evolution, which may be more favourable for QTL mapping of the minor or larger effects. Other possible reasons might be the low marker density, phenotyping error, or statistical defects associated with genome-wide association analysis (Zhao et al., 2011). In future studies for further understanding the genetic mechanisms of the expression levels of GmRCAβ in soybean, more diverse germplasms with lower linkage disequilibrium and more candidate gene-based markers will be chosen to detect the possible causal genes.
Although this study identified multiple eQTLs that were responsible for expression variation, the mechanism of how gene expression is regulated by eQTLs remains unknown. The promoter of GmRCAβ that affects both cis-acting and trans-acting eQTLs (Vuylsteke and van Eeuwijk, 2008) might be a potential factor generating diversity at the expression level. Moreover, it is well known that promoter elements play a key role in gene regulation and any changes in their sequences will dramatically influence gene expression resulting in variable trait expression. In the current study, the SNPs in the GmRCAβ promoter were identified and shown to correlate with expression level. Further transient expression showed that GUS expression driven by the group A promoter was stronger than that by the group B promoter, suggesting that promoter sequence types could influence the expression level of GmRCAβ. A similar result has been observed in rice, in which the type of Hd3a promoter contributed to diversity in flowering time and Hd3a expression level (Takahashi et al., 2009).
However, such diversity of expression level may not be solely determined by promoter sequence, because introns and 3′-UTRs also have significant effect on transcript synthesis and accumulation, which in turn alter the trait expression (Kumar et al., 2010). DeRidder et al. (2012) recently showed that AtRCAβ transcripts lacking their native 3′-UTR were significantly more stable than their full-length counterparts, indicating post-transcriptional regulation might participate in controlling of RCA expression in Arabidopsis. In addition, acclimation of RCA expression in Arabidopsis appeared to be due to changes in mRNA stability, not to increased rates of transcription. Hence, changes in mRNA levels may be the result either from transcriptional or post-transcriptional regulation, or both.
SNPs associated with GmRCAβ expression and their applications for crop improvement
The identification of alleles that affect key phenotypes is of utmost importance for the utilization of genetic resources in crop improvement (Holloway and Li, 2010). In human, more than 100 cis-regulatory polymorphisms were identified and shown to associate with phenotypic variation (Wray, 2007). In the present study, seven SNPs were identified as significantly associated with the expression of GmRCAβ (Table 5). Interestingly, three SNPs were located in regulatory elements such as the AT1 motif and the TATA box, which are important for the light regulation of gene expression, and data gathered in several species indicate that the expression of RCA is highly regulated by light (Qu et al., 2011). These SNPs in regulatory regions may impact the phenotype by altering the activity of the promoter and changing the expression levels of the RCA genes (Kumar et al., 2010). In addition, all associated SNPs existed in a linked manner (in the same linkage disequilibrium block) with the same P-value, indicating that any of the SNPs can be used as an allele-specific molecular marker (Kumar et al., 2010) to optimize gene expression. The identification of these allele-specific molecular markers would improve the understanding of how variation within regulatory factors affects transcript accumulation, enabling the prediction of the mechanisms by which genetic variation within these genetic elements shapes phenotypic diversity in natural populations, which would, in turn, facilitate the introgression of novel alleles through marker-assisted breeding.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. The geographic distributions of landraces used in this study
Supplementary Fig. S2. Multiple sequence alignment of promoter sequences of GmRCAα and GmRCAβ.
Supplementary Fig. S3. Expression levels of GmRCAα and GmRCAβ in the natural population.
Supplementary Fig. S4. Sequence analysis of the GmRCAβ promoter in soybean.
Supplementary Fig. S5. Sequence analysis of the GmRCAα promoter in soybean.
Supplementary Fig. S6. Phylogenetic analysis of GmRCAβ promoter sequence.
Supplementary Fig. S7. PCR products and restriction analysis of 1381Z group A and group B promoters.
Supplementary Table S1. Descriptive statistics and variance analysis of GmRCAα and GmRCAβ expression in natural population.
Supplementary Table S2. Regression coefficients among GmRCAα and GmRCAβ expression and yield components in a natural soybean population.
Supplementary Table S3. Putative cis-acting elements and functions for GmRCAβ.
Supplementary Table S4. Putative cis-acting elements and functions for GmRCAα.
Acknowledgements
The authors thank Mr Deyuan Ma from the Jiangpu Experimental Station of Nanjing Agricultural University for technical assistance in experiments. They also thank Dr Xiaohong He for advice with the statistical analysis. This work was supported in part by the National Basic Research Program of China (973 Program; 2010CB125906, 2009CB118400), the National Natural Science Foundation of China (31000718, 31171573, 31201230), and the Jiangsu Provincial Support Program (BE2012328, BK2012768).
References
- Agius F, Amaya I, Botella MA, Valpuesta V. 2005. Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. Journal of Experimental Botany 56, 37–46 [DOI] [PubMed] [Google Scholar]
- Ainsworth E, Yendrek C, Skoneczka J, Long S. 2012. Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell and Environment 35, 38–52 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. 2009. What has natural variation taught us about plant development, physiology, and adaptation? The Plant Cell 21, 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Ceccarelli E, Giuliano G. 1993. Constitutive, light-responsive and circadian clock-responsive factors compete for the different I box elements in plant light-regulated promoters. The Plant Journal 4, 611–619 [DOI] [PubMed] [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB. 2010. Polygenic and directional regulatory evolution across pathways in Saccharomyces . Proceedings of the National Academy of Sciences, USA 107, 5058–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva AE, Salvucci ME. 2013. The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiology 161, 1645–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Salvucci ME. 2004. Analyzing the impact of high temperature and CO2 on net photosynthesis: biochemical mechanisms, models and genomics. Field Crops Research 90, 75–85 [Google Scholar]
- Crafts-Brandner SJ, van de Loo FJ, Salvucci ME. 1997. The two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase differ in sensitivity to elevated temperature. Plant Physiology 114, 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Quint M. 2011. Expression level polymorphisms: heritable traits shaping natural variation. Trends in Plant Science 16, 481–488 [DOI] [PubMed] [Google Scholar]
- DeRidder BP, Shybut ME, Dyle MC, Kremling KA, Shapiro MB. 2012. Changes at the 3′-untranslated region stabilize Rubisco activase transcript levels during heat stress in Arabidopsis . Planta 236, 463–476 [DOI] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Jiang N, Chen X, Kearsey M, Waugh R. 2010. Expression quantitative trait loci analysis in plants. Plant Biotechnology Journal 8, 10–27 [DOI] [PubMed] [Google Scholar]
- Fukayama H, Ueguchi C, Nishikawa K, Katoh N, Ishikawa C, Masumoto C, Hatanaka T, Misoo S. 2012. Overexpression of Rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing Rubisco content in rice leaves. Plant and Cell Physiology 53, 976–986 [DOI] [PubMed] [Google Scholar]
- Gatti DM, Harrill AH, Wright FA, Threadgill DW, Rusyn I. 2009. Replication and narrowing of gene expression quantitative trait loci using inbred mice. Mammalian Genome 20, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti DM, Lu L, Williams RW, Sun W, Wright FA, Threadgill DW, Rusyn I. 2011. MicroRNA expression in the livers of inbred mice. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 714, 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ. 2006. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genetics 2, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Kwan T, Pastinen TM. 2010. Analysis of the impact of genetic variation on human gene expression. Methods in Molecular Biology 628, 321–339 [DOI] [PubMed] [Google Scholar]
- Gurr SJ, Rushton PJ. 2005. Engineering plants with increased disease resistance: what are we going to express? Trends in Biotechnology 23, 275–282 [DOI] [PubMed] [Google Scholar]
- Hao D, Cheng H, Yin Z, Cui S, Zhang D, Wang H, Yu D. 2012. Identification of single nucleotide polymorphisms and haplotypes associated with yield and yield components in soybean (Glycine max) landraces across multiple environments. Theoretical and Applied Genetics 124, 447–458 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET. 2008. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B, Li B. 2010. Expression QTLs: applications for crop improvement. Molecular Breeding 26, 381–391 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences: CABIOS 8, 275–282 [DOI] [PubMed] [Google Scholar]
- King M-C, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 [DOI] [PubMed] [Google Scholar]
- Kumar GR, Sakthivel K, Sundaram R, Neeraja C, Balachandran S, Rani NS, Viraktamath B, Madhav M. 2010. Allele mining in crops: prospects and potentials. Biotechnology Advances 28, 451–461 [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RD, Crafts-Brandner SJ. 2001. High temperature stress increases the expression of wheat leaf ribulose-1,5-bisphosphate carboxylase/oxygenase activase protein. Archives of Biochemistry and Biophysics 386, 261–267 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J. 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics 43, 1266–1269 [DOI] [PubMed] [Google Scholar]
- Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L, Lin F, Luan W, Chen P, Yan Z. 2008. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theoretical and Applied Genetics 117, 857–871 [DOI] [PubMed] [Google Scholar]
- Liu Z, Taub CC, McClung CR. 1996. Identification of an Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiology 112, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Barajas E, Molina-Galán J, Sánchez de Jiménez E. 1997. Regulation of Rubisco activity during grain-fill in maize: possible role of Rubisco activase. Journal of Agricultural Science 128, 155–161 [Google Scholar]
- Mitsuhashi W, Feller U. 1992. Effects of light and external solutes on the catabolism of nuclear-encoded stromal proteins in intact chloroplasts isolated from pea leaves. Plant Physiology 100, 2100–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430, 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Ogren WL. 1993. Localization of light-inducible and tissue-specific regions of the spinach ribulose bisphosphate carboxylase/oxygenase (rubisco) activase promoter in transgenic tobacco plants. Plant Molecular Biology 23, 1129–1138 [DOI] [PubMed] [Google Scholar]
- Papageorgiou GC. 2004. Chlorophyll a fluorescence: a signature of photosynthesis. Dordrecht, The Netherlands: Kluwer Academic Publishers [Google Scholar]
- Pilgrim ML, McClung CR. 1993. Differential involvement of the circadian clock in the expression of genes required for ribulose-1,5-bisphosphate carboxylase/oxygenase synthesis, assembly, and activation in Arabidopsis thaliana . Plant Physiology 103, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis AR. 2003. Rubisco activase—Rubisco’s catalytic chaperone. Photosynthesis Research 75, 11–27 [DOI] [PubMed] [Google Scholar]
- Prestridge DS. 1991. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Computer Applications in the Biosciences: CABIOS 7, 203–206 [DOI] [PubMed] [Google Scholar]
- Qu D, Song Y, Li W, Pei X, Wang Z, Jia S, Zhang Y. 2011. Isolation and characterization of the organ-specific and light-inducible promoter of the gene encoding rubisco activase in potato (Solanum tuberosum). Genetics and Molecular Research 10, 621–631 [DOI] [PubMed] [Google Scholar]
- Rafalski JA. 2010. Association genetics in crop improvement. Current Opinion in Plant Biology 13, 174–180 [DOI] [PubMed] [Google Scholar]
- Raines CA. 2011. Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiology 155, 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z, Momčilović I, Bukovnik U, Prasad PV, Fu J, DeRidder BP, Elthon TE, Mladenov N. 2009. Rubisco activase and wheat productivity under heat-stress conditions. Journal of Experimental Botany 60, 4003–4014 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE. 2002. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. The Plant Cell 14, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, van de Loo FJ, Stecher D. 2003. Two isoforms of Rubisco activase in cotton, the products of separate genes not alternative splicing. Planta 216, 736–744 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Werneke JM, Ogren WL, Portis AR. 1987. Purification and species distribution of Rubisco activase. Plant Physiology 84, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez de Jimenez E, Medrano L, Martinez-Barajas E. 1995. Rubisco activase, a possible new member of the molecular chaperone family. Biochemistry 34, 2826–2831 [DOI] [PubMed] [Google Scholar]
- Sharma A, Komatsu S. 2002. Involvement of a Ca2+-dependent protein kinase component downstream to the gibberellin-binding phosphoprotein, RuBisCO activase, in rice. Biochemical and Biophysical Research Communications 290, 690–695 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner RA, DeCook R, Jia Y, Bancroft T, Ji T, Zhao X, Nettleton D, Schnable PS. 2009. Paternal dominance of trans-eQTL influences gene expression patterns in maize hybrids. Science 326, 1118–1120 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. 2009. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences, USA 106, 4555–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. 1995. Light-regulated transcription. Annual Review of Plant Biology 46, 445–474 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuylsteke M, van Eeuwijk F. 2008. The use of general and specific combining abilities in a context of gene expression relevant to plant breeding. Euphytica 161, 115–122 [Google Scholar]
- Wang D, Li XF, Zhou ZJ, Feng XP, Yang WJ, Jiang DA. 2010. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiologia Plantarum 139, 55–67 [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics 8, 206–216 [DOI] [PubMed] [Google Scholar]
- Yamagata H, Yonesu K, Hirata A, Aizono Y. 2002. TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of thecucumisin gene. Journal of Biological Chemistry 277, 11582–11590 [DOI] [PubMed] [Google Scholar]
- Yang Z, Lu Q, Wen X, Chen F, Lu C. 2012. Functional analysis of the rice rubisco activase promoter in transgenic Arabidopsis . Biochemical and Biophysical Research Communications 418, 565–570 [DOI] [PubMed] [Google Scholar]
- Yin Z, Meng F, Song H, He X, Xu X, Yu D. 2010a. Mapping quantitative trait loci associated with chlorophyll a fluorescence parameters in soybean (Glycine max (L.) Merr.). Planta 231, 875–885 [DOI] [PubMed] [Google Scholar]
- Yin Z, Meng F, Song H, Wang X, Xu X, Yu D. 2010b. Expression quantitative trait loci analysis of two genes encoding rubisco activase in soybean. Plant Physiology 152, 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB. 2005. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Komatsu S. 2000. Molecular cloning and characterization of cDNAs encoding two isoforms of ribulose-1,5-biosphosphate carboxylase/oxygenase activase in rice (Oryza sativa L.). Journal of Biochemistry 128, 383–389 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xing A, Staswick P, Clemente TE. 1999. The use of glufosinate as a selective agent in Agrobacterium-mediated transformation of soybean. Plant Cell, Tissue and Organ Culture 56, 37–46 [Google Scholar]
- Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J. 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa . Nature Communications 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE, Werneke JM, Jenkins ME. 1989. Coordinate expression of Rubisco activase and Rubisco during barley leaf cell development. Plant Physiology 90, 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.