Abstract
Staphylococcus aureus is a prominent cause of human infections globally. The high prevalence of infections is compounded by antibiotic resistance—a significant problem for treatment. Methicillin-resistant S. aureus (MRSA) is endemic in hospitals and healthcare facilities worldwide, and is an increasingly common cause of community-associated bacterial infections in industrialized countries. Although much focus is placed on the role of S. aureus as a human pathogen, it is in fact a human commensal organism that has had a relatively long coexistence with the human host. Many S. aureus infections can be explained by host susceptibility or other predisposing risk factors. On the other hand, the emergence/re-emergence of successful S. aureus clones (referred to as epidemic waves) suggests a rapid bacterial adaption and evolution, which includes the emergence of antibiotic resistance and increased virulence and/or transmissibility. It is within this context that we review our understanding of selected S. aureus epidemic waves, and highlight the use of genome sequencing as a means to better understand the evolution of each lineage.
Keywords: Staphylococcus aureus, MRSA, Epidemic, Genome sequencing, Antimicrobial resistance
1. Introduction
Staphylococcus aureus is a remarkably successful pathogen. Despite the availability of effective antimicrobial agents to treat S. aureus infections, it continues to be a major cause of morbidity and mortality worldwide (Lowy, 1998). Epidemics due to the spread of successful clones continue to be reported from virtually every geographic region (Chambers and DeLeo, 2009). As a result, S. aureus has proven to be among the most persistent of pathogens in the healthcare and community setting. In both venues the emergence of antimicrobial resistant strains, especially those that are resistant to methicillin, has been a constant feature. Many factors appear to contribute to the success of S. aureus as a pathogen, however its capacity to persist as a commensal, its frequent resistance to multiple antimicrobial agents and its armamentarium of virulence determinants, often with redundant functions, are among the most important (Fluit et al., 2001; Foster, 2005; Otto, 2010).
Methicillin-resistant S. aureus (MRSA) are well established in both the healthcare setting and in the community. They are among the most common causes of such nosocomial infections as intravenous catheter associated infections, ventilator associated pneumonias and surgical wound infections in some industrialized countries such as the United States (Klevens et al., 2006; Rosenthal et al., 2012). More recently nosocomial infective endocarditis due to MRSA has become a major concern (Benito et al., 2009). In the community, MRSA are the most common cause of skin and soft tissue infections in the United States (US). Five to ten percent of community-based infections are invasive and potentially life threatening (Fridkin et al., 2005; Klevens et al., 2007; Moran et al., 2006). The emergence of community-associated methicillin-resistant S. aureus (CA-MRSA) as an important pathogen has occurred over the past 15–20 years (Chambers, 2001; David and Daum, 2010; DeLeo et al., 2010). In the United States, a single epidemic clone—USA300, emerged as the predominant isolate accounting for the vast majority of cutaneous infections. These community-based strains appear to have enhanced virulence as well as an enhanced capacity to colonize multiple body sites and to survive on environmental surfaces (DeLeo et al., 2010; Otto, 2010; Uhlemann et al., 2011). As a result they are also more easily spread from person to person.
CA-MRSA infections were initially defined by a set of criteria established by the CDC (Buck et al., 2005). These definitions excluded infections arising from traditional healthcare-associated risks such as residence in a long-term care facility, recent hospitalization or the need for hemodialysis. The infection was further defined as starting in the community and, for hospitalized patients, isolation of the clinical isolate within 48 h of admission. Alternatively investigators identified the strains associated with community-based outbreaks and infections based on the molecular characteristics of the isolates. These included the pulsed field gel electrophoresis (PFGE) profile, multilocus sequence type (MLST or ST) or staphylococcal protein A (spa) type as well as the staphylococcal chromosomal cassette mec (SCCmec) type (see below for more details). The SCCmec type defines the mobile element that carries the gene responsible for methicillin resistance. A final distinction between the community and healthcare-associated (HA) staphylococci was their antibiotic susceptibility with the healthcare-associated isolates having a broad range of resistance to different antibiotic families while the CA-MRSA isolates, while resistant to methicillin, were susceptible to many other antibiotics (David and Daum, 2010; Miller et al., 2007).
Many of the discriminating features that distinguished the community from the HA strains are no longer valid (David et al., 2008; Miller et al., 2007). Increasingly, the highly successful community-based clones have invaded the healthcare setting and are now successful nosocomial pathogens. In many medical centers they have become a common cause of healthcare-associated bacteremias (Jenkins et al., 2009; Seybold et al., 2006). Along the way they have also become increasingly antibiotic resistant. As a result, PFGE types such as USA300 are now commonly found in both settings.
A unique feature of the repeated worldwide MRSA epidemics has been the discrete number of staphylococcal clones associated with these events (Chambers and DeLeo, 2009). Rather than a diversity of strains causing disease as is seen with methicillin-susceptible S. aureus, the MRSA outbreaks have been limited to a relatively small number of staphylococcal lineages. This is true for both the strains classically associated with healthcare, as well as community-based infections. These different staphylococcal lineages have evolved over time, accumulating mutations that alter gene expression and function, and perhaps most importantly, acquiring new genetic elements via horizontal gene exchange (Malachowa and DeLeo, 2010). These new elements have been critical in altering animal species specificity, the nature of invasive infections, and the type antimicrobial resistance associated with the strains. The capacity to acquire novel elements is essential to the success of these clones.
The recent emergence of whole genome sequencing as a technique that can be efficiently and inexpensively performed on a large scale has provided a powerful new tool to explore the evolution of these highly successful clones. Insights have included a better understanding of the contribution of selected genes to virulence, the epidemiology of outbreaks and strain transmission and an understanding of the molecular changes that facilitate the spread of these strains from one animal species to another (DeLeo et al., 2011; Harris et al., 2010; Uhlemann et al., 2012b). In this paper, we review the evolution of three highly successful community and healthcare-associated lineages, clonal complex 30 (CC30), ST239, and ST8 (USA300) (Table 1). These unique clones demonstrate remarkable plasticity in their ability to adapt to environmental changes. The contribution of the new sequencing methods to our understanding of the evolution of these clones is also discussed.
Table 1.
Characteristics of selected epidemic S. aureus clones
| Strain (clonal complex) | SCCmec type | Time period | Geographic locations | Characteristics |
|---|---|---|---|---|
| Phage-type 80/81 (CC30) | NA | 1950s–1960s | Australia, New Zealand, North America (United States, Canada), Europe (United Kingdom, Denmark) | HA and CA infections. Notorious for causing fatal infections in neonates. |
| EMRSA-16 (CC30/ST36) | II | 1992–present | Europe (especially the United Kingdom), North America (United States), South America (Brazil) | HA-MRSA. Leading cause of MRSA infections in the United Kingdom in the 1990s and early 2000s. This clone contains inactivating mutations in genes encoding alpha-hemolysin and accessory gene regulator subunit C. |
| Southwest Pacific Clone (CC30/USA) | IV | Australia, New Zealand, Samoa, Europe (primarily the United Kingdom), Asia, North America, South America | CA-MRSA. | |
| ST239, Brazilian or Hungarian clone, EMRSA-1 (CC8) | III | 1980s–present | Asia (includes Saudi Arabia, Russia, China, Korea), Australia, Europe (includes Portugal, Spain, United Kingdom, Germany), North America (United States), South America (Brazil), Southeast Asia (e.g., Malaysia, Thailand, Singapore) | Pandemic clone. A recombinant strain comprised of genomic DNA from CC8 and CC30 lineages. |
| USA300 (CC8/ST8) | IV | Early 2000s–present | Primarily North America & South America; but also present in Europe | Epidemic CA-MRSA strain. Most prominent cause of community bacterial infections in the United States. Now also a cause of HA-MRSA infection. |
NA, not applicable; HA, hospital-associated; CA, community-associated; SSTI, skin and soft tissue infections.
2. Evolution of healthcare-associated MRSA: the CC30 lineage
As a first step toward understanding the evolution of healthcare-associated S. aureus, it is important to provide a cursory description of methods used to categorize or type S. aureus. Phage-typing was among the first successful methods used to understand S. aureus epidemiology and outbreaks (Williams et al., 1953). The method utilizes the differential susceptibility of S. aureus clones and isolates to cytolysis by different bacteriophages. Although phage typing was an important advance for early epidemiological studies of S. aureus, it has since given way to more precise molecular typing methods. MLST (or ST) is a widely used typing method that indexes variation accumulating slowly over time, and can thus be used to measure evolution over an extended period (Enright et al., 2000; Enright and Spratt, 1999). By comparison, PFGE indexes variation that accumulates rapidly and is highly discriminatory; typing by this method is appropriate for studies of outbreaks or short-term epidemiological studies (Enright and Spratt, 1999). The US Centers for Disease Control and Prevention (CDC) established a national database of PFGE types to monitor the prevalence of MRSA (McDougal et al., 2003). A third widely used typing method, known as spa typing, is suitable for analysis of local or widespread S. aureus outbreaks (Shopsin et al., 1999). Based on MLST, PFGE, and spa typing, there are multiple healthcare-associated MRSA (HA-MRSA) lineages and clones worldwide (Chambers and DeLeo, 2009; Grundmann et al., 2010).
Early studies of MRSA evolution by Enright et al. used MLST to assign the majority of MRSA isolates, including all major MRSA clones, to five clonal complexes (CCs) (Enright et al., 2002). Some MRSA lineages are dispersed globally (for example, STs belonging to CC30), whereas others appear in specific geographic locations (Chambers and DeLeo, 2009; Enright et al., 2002). It should also be noted that molecular data on MRSA are still incomplete or lacking for many regions of the world. Although current typing methods are highly useful for tracking S. aureus outbreaks, and they provide information important for our understanding of pathogen evolution, they index variation at a relatively small number of nucleotides. By comparison, whole-genome sequencing approaches have the ability to provide the full extent of genetic diversity among S. aureus isolates, and this method has become increasingly employed to better understand the evolution of MRSA.
There is simply no space here to provide a comprehensive review of the evolution of all HA-MRSA lineages. Therefore, we focus our discussion on selected HA-MRSA lineages for which recent genome-scale analyses have revealed new insights into the emergence and success of each.
2.1. EMRSA-16/ST36 and evolution of the phage-type 80/81 lineage
2.1.1. Phage-type 80/81 S. aureus
Strains that belong to CC30 are among the most prominent causes of S. aureus human infections. A penicillin-resistant S. aureus clone known as phage-type 80/81 S. aureus, which based on modern typing methods is a CC30 clone, was pandemic in the 1950s and 1960s (Blair and Carr, 1958; Bynoe et al., 1956; Gillespie and Alder, 1957; Rountree and Freeman, 1955). The pandemic caused by phage-type 80/81 S. aureus was one of the major bacterial pandemics of the 20th century and caused infections—many of them fatal—in both hospital and community settings (Hassall and Rountree, 1959). Infections manifested initially as pustules, boils, abscess, or carbuncles, which then frequently led to invasive syndromes such as cellulitis, sepsis, and pneumonia (Blair and Carr, 1958; Hassall and Rountree, 1959). Moreover, the strain was penicillin-resistant, a major problem for treatment. Rountree and Freeman first reported outbreaks in newborns at Royal Prince Alfred Hospital in Sydney, Australia (Rountree and Freeman, 1955). In 1954, 43 members of the Royal Prince Alfred Hospital staff acquired infections (primarily boils) caused by this strain, and a number of long-time hospitalized patients suddenly developed skin lesions from which the same strain was isolated (Rountree and Freeman, 1955). Rountree and Freeman reported that infections with phage-type 80/81 S. aureus need not be accompanied by nasal carriage, albeit carriage in healthcare staff and in newborns was noted frequently (Light et al., 1975; Rountree and Beard, 1958; Rountree and Freeman, 1955).
Phage-type 80/81 S. aureus was known for its exceptional virulence and high infectivity and transmissibility (Blair and Carr, 1958; Gillespie and Alder, 1957; Hassall and Rountree, 1959). Indeed, Gillespie and Alder reported in 1958 that this clone was the predominant S. aureus phage-type responsible for hospital-acquired infections in the United States (Gillespie and Alder, 1957), findings in accordance with data from Canada (Bynoe et al., 1956). However, by the mid-to-late 1960s, nasal carriage and the number of infections caused by phage-type 80/81 S. aureus had declined dramatically (Jessen et al., 1969; Light et al., 1975). Jessen et al. (1969) suggested that the decline of this epidemic clone might be due to its susceptibility to newly introduced antibiotics such as methicillin, although there is no conclusive evidence for this hypothesis. Thus, an explanation for the emergence, spread, and ultimate decline of the phage-type 80/81 S. aureus “epidemic wave” remained largely unknown.
2.1.2. Epidemic MRSA-16 (EMRSA-16)
Approximately 25 years after the decline of phage-type 80/81 S. aureus (∼1991), another CC30 strain—this time an MRSA strain—emerged as a significant cause of infections in hospitals in Northhamptonshire, England (Cox et al., 1995). This newly emerged strain, known as EMRSA-16 (later identified as an ST36 strain), spread rapidly among hospitals in England and ultimately became one of the most successful HA-MRSA strains in United Kingdom (Murchan et al., 2004). Notably, EMRSA-16 and EMRSA-15 (ST22, not discussed here) isolates accounted for 95% of MRSA bacteremias in the United Kingdom during the period of 1998–2000 (Johnson et al., 2001). The first reported infections were in elderly patients with risk factors for infection (e.g., post-surgery) or those who are seriously ill (Cox et al., 1995). These patients were typically colonized with EMRSA-16 in the nose, throat, and areas of broken skin, and the organism caused a range of infections, including septicemia, which was often fatal. In addition, most isolates were resistant to multiple antibiotics, including penicillin, methicillin, erythromycin, and ciprofloxacin (Cox et al., 1995; Holden et al., 2004). Murchan et al. (2004) reported that virtually all EMRSA-16 isolates were positive for genes encoding staphylococcal enterotoxins A, (sea), G (seg), and I (sei), and toxic shock syndrome toxin-1 (tst).
Phage-typing and PFGE were used routinely to track hospital outbreaks of EMRSA-16 (Murchan et al., 2004). This strain ultimately became the second most abundant cause of HA-MRSA infections in the United States, and was named USA200 according to the PFGE nomenclature developed by the CDC (McDougal et al., 2003). Consistent with findings in the United Kingdom, USA200/EMRSA-16 was a leading cause of persistent bacteremia in the healthcare setting in the United States (Xiong et al., 2009), and an abundant colonizer and cause of infection in many other countries (Montesinos et al., 2006). Thus, EMRSA-16, much like phagetype 80/81 S. aureus (except for its virtual absence as a cause of community-associated infections), became a major pandemic MRSA strain. Although EMRSA-16 remains a cause of healthcare-associated infections, there has been a clear decline in its abundance—especially in the United Kingdom—since 2003 (Ellington et al., 2010b). The similarity in the decline of EMRSA-16 and phage-type 80/81 S. aureus is striking and a number of questions needed to be addressed. For example, was EMRSA-16 simply a re-emerged MRSA version of phage-type 80/81 S. aureus? How closely related were these lineages and why was EMRSA-16 largely restricted to the healthcare setting, whereas phage-type 80/81 S. aureus caused infections in both the community and healthcare settings? Phage-type of EMRSA-16 was distinct from that of phage-type 80/81 (Cox et al., 1995), suggesting that these strains were not clones.
2.1.3. Concurrent emergence of the Southwest Pacific clone
There was an additional twist to the evolution of the CC30 lineage. During the period in which EMRSA-16 was abundant, there emerged a community-associated MRSA (CA-MRSA) strain known as the Southwest Pacific (SWP) clone (Adhikari et al., 2002; Vandenesch et al., 2003). This strain was later typed as ST30 and was reported initially in Australia, New Zealand, and Western Samoa (Vandenesch et al., 2003). Notably, the SWP clone contained genes encoding Panton–Valentine leukocidin (PVL), a leukocyte toxin also present in phage-type 80/81 S. aureus (but not EMRSA-16) (Vandenesch et al., 2003). Robinson and colleagues subsequently reported that the SWP clone and phage-type 80/81 S. aureus were ST30 and closely related (Robinson et al., 2005). Moreover, this group proposed that the SWP clone was a direct descendant of phage-type 80/81 S. aureus (Robinson et al., 2005). Although these findings had limited bearing on the evolution of HA-MRSA, the proposed model for the evolution of the CC30 was intriguing. However, new approaches were needed to better understand the full extent of similarity or diversity between these strains, and track their evolution.
2.1.4. A genome-sequence based approach to understanding the evolution of the CC30 lineage
Holden et al. (2004) reported the complete genome sequence for an EMRSA-16 clinical isolate named MRSA252. These data were an important step toward a comprehensive understanding of the evolution of the CC30 lineage, and provided a framework for future studies that utilize comparative whole-genome sequencing. One of the noteworthy observations emerging from the EMRSA-16 genome sequence was that 6% of the genome was novel compared to the other genomes published at the time (Holden et al., 2004). This novel genetic material included two genomic islands, indicating that mobile DNA is exchanged readily by S. aureus, thereby providing a means for rapid evolution (Holden et al., 2004). Indeed, Holden et al. noted that all but one of the antibiotic resistance determinants of EMRSA-16 reside on mobile genetic elements (MGEs).
To gain an enhanced understanding of the evolution of the CC30 lineage, DeLeo et al. (2011) used comparative whole genome sequencing to determine the phylogenetic relationship of phage-type 80/81 isolates, the SWP clone, and selected contemporary CC30 hospital isolates with EMRSA-16. Analysis of the core genomes of these isolates segregated the representative isolates into three sublineages within CC30—phage-type 80/81 S. aureus, SWP clone, and contemporary healthcare-associated CC30 isolates, including EMRSA-16 (Fig. 1). More notably, the analyses indicate that neither EMRSA-16 nor SWP clone are direct descendants of phage-type 80/81 S. aureus (DeLeo et al., 2011). Rather, the data indicated there was coincident divergence from a recent common ancestor. Using a larger set of CC30 isolates for genome sequencing, McAdam et al. verified the existence of the three independent lines of descent for these sublineages (McAdam et al., 2012). These researchers also identified a fourth (minor) CC30 sublineage related to EMRSA-16—isolates reported as being recovered from community-associated infections—and these are either MSSA isolates or contain SCCmec IV (McAdam et al., 2012). Collectively, the findings by DeLeo et al. and McAdam et al. resolve the long-standing controversy about the evolution of the CC30 lineage. That is, neither the SWP clone nor EMRSA-16 and related strains are simply re-emerged, methicillin-resistant versions of the phage-type 80/81 clone (Fig. 1).
Fig. 1.
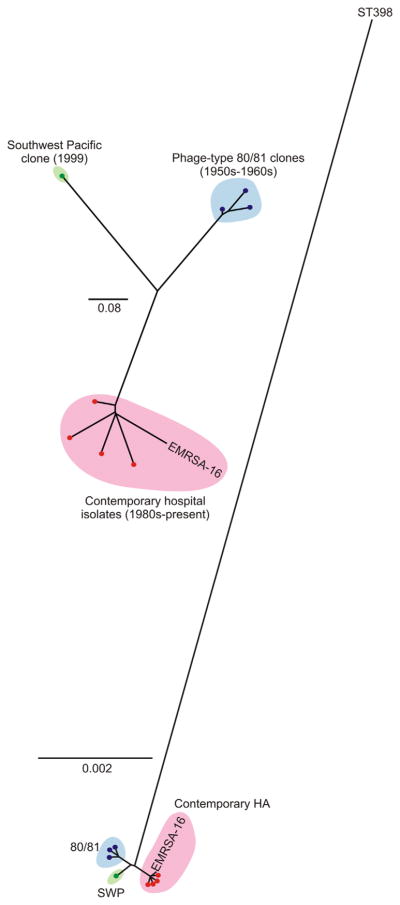
Phylogenetic analyses of selected CC30 strains/isolates. Left panel is a phylogenetic analysis of CC30 strains (indicated by dots or labeled as EMRSA-16) based upon analysis of a contiguous 1.4 Mb region of the genome. Right panel is a phylogenetic analysis of CC30 strains based upon concatenated SNPs in the core genome. SWP, Southwest Pacific clone; 80/81, phage type 80/81 S. aureus; Contemporary HA, contemporary hospital CC30 isolates. Adapted from DeLeo et al. (2011).
In addition to performing phylogenetic analyses, DeLeo et al. identified SNPs or other mutations in EMRSA-16 and related contemporary CC30 isolates relative to phage-type 80/81 S. aureus that likely impact interaction with the human host. For example, the vast majority of healthcare-associated CC30 isolates contained identical SNPs in the genes encoding alpha-hemolysin (hla) and accessory gene regulator C (agrC) (DeLeo et al., 2011). These mutations, which result in a premature stop codon in Hla and a non-conservative amino acid substitution in AgrC (Gly → Arg), were absent from phage-type 80/81 S. aureus and the SWP clone. Re-creating these mutations in a phage-type 80/81 isolate eliminated production of RNAIII (agrCG55R mutant) and Hla (both mutants) in vitro, and more importantly, the isogenic mutant strains had reduced capacity to cause lethal bacteremia or pneumonia in mouse infection models (DeLeo et al., 2011). In addition, McAdam et al. reported a nonsense mutation in the gene encoding squalene desaturase (crtM) in healthcare-associated CC30 isolates relative to pandemic phage-type 80/81 S. aureus (McAdam et al., 2012). The mutation in crtM results in the formation of a pseudogene and likely disrupts production of staphyloxanthin carotenoid—a pigment that confers resistance to neutrophil oxidants (Liu et al., 2005). Taken together, these findings provide strong support to the idea that nucleotide changes in hla, agrC, and crtM in EMRSA-16 and related strains reduces virulence capacity, and thereby largely restricts this sublineage to healthcare settings in which individuals are sick or have underlying risk factors for infection. This hypothesis is compatible with the known clinical epidemiology of EMRSA-16 and related clones (see above).
The finding that contemporary healthcare-associated CC30 strains such as EMRSA-16 have reduced virulence capacity in animal models seems at variance with the known high abundance of bloodstream infections caused by these strains (Cox et al., 1995; Fowler et al., 2007; Johnson et al., 2001). This might in part be a reflection of suboptimal infection control measures, which thereby lead to an increased reservoir of MRSA carriers and subsequent infections. However, it is possible that decreased virulence capacity of this sublineage compared with community-associated clones makes them better suited for long-term colonization and persistence in humans, which in turn, increases the chance of bacteremia in the context of specific predisposing conditions. It is also possible (or perhaps likely) that the ability of S. aureus to attenuate virulence is key to its long-term success as a human commensal organism. Such a phenomenon might have contributed to the decline of epidemic S. aureus clones such as phage-type 80/81 S. aureus. Although progress has been made, more information is needed to understand the basis for the emergence, success and ultimate decline of EMRSA-16 and related strains.
2.1.5. Regulatory mechanisms balancing methicillin resistance versus virulence
CC30 strains with comparatively low virulence such as EMRSA-16 commonly exhibit high minimal inhibitory concentrations (MICs) toward methicillin, i.e. high levels of methicillin resistance. In contrast, the recently emerged high-virulence community-associated MRSA strains such as USA300 have significantly lower MICs. These findings suggest that a simultaneous strong production of determinants of virulence and methicillin resistance comes with a considerable metabolic burden for the bacteria. In support of this hypothesis, recent reports indicate that there are regulatory mechanisms by which the expression of virulence determinants limits that of mecA, the penicillin binding protein (PBP) 2a responsible for methicillin resistance, and vice versa. For example, Collins et al. (2010) reported that CC30 strains harboring SCCmec type II have reduced virulence capacity (as measured by their ability to lyse T cells) and lower relative fitness than their methicillin-sensitive relatives. Rudkin et al. (2012) subsequently showed that the mecA gene is responsible for this regulatory phenomenon. Expression of PBP2a leads to cell wall changes that inhibit induction of the global virulence regulator Agr by the Agr-encoded extracellular autoinducing peptide, which is a crucial part of the Agr quorumsensing circuit controlling virulence factor expression (Ji et al., 1997). Furthermore, Cheung et al. showed that the Agr system, which is strongly expressed in CA-MRSA strains and may in part be responsible for high CA-MRSA virulence (Li et al., 2009), inhibits expression of mecA in the CA-MRSA strain USA300 (Memmi et al., 2012). These findings provide an explanation on the molecular level for the observation that pronounced virulence and high resistance levels to methicillin appear to be mutually exclusive in MRSA strains.
2.2. St239
2.2.1. Epidemiology of ST239
Research on MRSA outbreaks and epidemic waves has for the longest time focused on Europe, North America, and Japan. However, MRSA infections have become a significant problem in countries such as China, India, and parts of Southeast Asia, where the fast economic development has been accompanied by better healthcare and increasing hospitalization rates. China, for example, reports that within the last 10 years, S. aureus infections in hospitals were caused by MRSA strains in >60% of cases (Xiao et al., 2011). In Asia, HA-MRSA clones are for the most part quite different from those found in the Western industrialized countries. ST239 is (or has been) the most abundant ST recovered from hospital infections in Asia (except Japan) and eastern Australia (Aires de Sousa et al., 2003; Kim et al., 2011; Ko et al., 2005; Liu et al., 2009; Nimmo et al., 2008; Soo Ko et al., 2005; Xu et al., 2009). In addition, many MRSA isolates recovered from hospital patients in South America, Eastern Europe, and the Middle East belong to ST239 (Brazilian, Portuguese, Hungarian, and Viennese clones) (Alp et al., 2009; Amorim et al., 2002; Bartels et al., 2008; Grundmann et al., 2010; Havaei et al., 2011; Mayor et al., 2007; Melter et al., 2003; Vivoni et al., 2006; Wisplinghoff et al., 2005). Given the high population in the countries where it predominates, ST239 may represent the most successful MRSA lineage worldwide.
Recently, ST239 moved into the focus of molecular MRSA research, owing to an outbreak in London that could be attributed to that ST (Edgeworth et al., 2007). The London outbreak strain (an ST239 strain named TW20) was primarily associated with vascular access device-related bacteremia, suggesting that TW20 has increased ability to cause bacteremia. Whole-genome sequencing of TW20 revealed the presence of several mobile genetic elements, comprising antibiotic resistance among many other genes with unknown function (Holden et al., 2010). Notably, comparison of a series of global ST239 isolates by whole-genome sequencing indicated that TW20 originated from Asia, as it was more similar to ST239 isolates from Thailand than those from South America or Europe (Harris et al., 2010).
2.2.2. The surface protein SasX
The association of TW20 with bacteremia and the fact that the “Asian clade” of ST239 is a primary cause of hospital-associated MRSA infections in Asia prompted the investigation of molecular factors potentially underlying the epidemiological success of Asian ST239 strains. Holden et al. described the presence of a ϕSPβ-like prophage in TW20 that was not found in other S. aureus strains and only known in a highly similar form from S. epidermidis RP62A (Holden et al., 2010). Concluding from their genome comparisons, Harris et al. reported that the phage is associated with the Asian clade of ST239 (Holden et al., 2010). The ϕSPβ-like prophage contains genes potentially conferring aminoglycoside resistance, but only one gene that raised researchers' interest as potentially responsible for the increased virulence of the Asian ST239 strains, namely an LPXTG motif-harboring predicted surface protein encoded close to the 3′ end of the phage.
Li et al. (2012) investigated the recent epidemiology of MRSA infections in eastern China with a focus on the TW20 ϕSPβ-like prophage-encoded surface protein, which they called SasX. They found that the frequency of sasX-positive clones among invasive MRSA strains had increased significantly within the last ∼10 years. Furthermore, the sasX gene, which is apparently always linked to the ϕSPβ-like prophage, was found in increasing numbers among other STs, indicating horizontal gene transfer. Potentially explaining the epidemiological success of sasX-containing strains, Li et al. (2012) described that SasX increases nasal colonization by enhanced adherence to epithelial cells. Moreover, SasX caused bacterial aggregation, which in turn likely caused the observed sasX-dependent resistance to neutrophil phagocytosis. Finally, presence of sasX significantly increased pathogen survival in skin and lung infection models.
The mobile genetic element-encoded SasX protein represents the first molecular determinant linked to pathogenic success during an MRSA outbreak. It explains at least in part the success of ST239 in Asia, because the study suggested that many ST239 strains in Asia appear to harbor the sasX gene. However, it should be noted that the features of the TW20 outbreak in London, namely the association with bacteremia and the observed decreased rather than increased colonization capacities compared to other MRSA strains (Holden et al., 2010), are difficult to interpret based on sasX. It is likely that the epidemiological success of the Asian clade of ST239 is caused by a variety of factors, some of which are still unknown. These factors may also vary in importance in a specific geographical situation, where different subsets of competing MRSA clones are present.
2.2.3. Evolutionary History of ST239
Robinson and Enright first reported the remarkable evolutionary history of ST239 (Robinson and Enright, 2004). Their study revealed that ST239 is a hybrid of ST8 and ST30, founded by a single large chromosomal replacement (Robinson and Enright, 2004). Whole genome sequencing of TW20 further characterized the likely mechanism of DNA transfer from CC30 into a CC8 strain. Although TW20 belongs to CC8 based on MLST, and ∼82% of the protein coding sequences matched those in CC8 strains, ∼86% of the coding sequences matched those in MRSA252, the sequenced EMRSA-16 (CC30) isolate described above (Holden et al., 2004, 2010). Interestingly, the additional CC30-like genetic material is present on MGEs, suggesting that a major recombination event lead to the formation of a chimeric strain containing genetic material from CC8 (∼80%) and CC30 (∼20%) strains (Holden et al., 2010). Virtually all ST239 isolates harbor an SCCmec type III element, which is present in the DNA transferred from CC30, although curiously this SCCmec type has not been reported in CC30 isolates (Holden et al., 2010). In an effort to better understand the population structure of the ST239 lineage, Smyth et al. (2010) used 111 isolates recovered worldwide to identify three major clades and several minor sublineages or haplotypes of ST239 that are differentiated by point mutations. These authors estimate that the origin of the most recent common ancestor for these global isolates was between 1945 and 1961, a time period encompassing the introduction of penicillin and methicillin. Importantly, the original SCCmec type III element has apparently been maintained by ST239 and not reacquired multiple times by MSSA progenitors (Smyth et al., 2010). These observations suggest that a single clone of ST239 disseminated globally and underwent diversification, a finding at variance with the explanation for the worldwide abundance of ST5 MRSA, which arose in specific geographic locations by the repeated acquisition of SCCmec by ST5 MSSA strains (Nubel et al., 2008).
3. Community-associated MRSA
In the early 1990s, an increasing number of MRSA infections acquired from the community emerged in patients who did not have the traditional healthcare risk factors. These infections were first recognized in limited outbreaks from different parts of the world such as in the indigenous communities of Western Australia and in several regions of the United States (Udo et al., 1993). The death of four children from rural Minnesota and North Dakota without nosocomial exposures marked the formal recognition of this new entity of S. aureus infections (CDC, 1999). Phenotypic and molecular characterization of CA-MRSA isolates further demonstrated that they differed from the major circulating MRSA clones, varied between different countries and were only resistant to beta-lactam antibiotics.
CA-MRSA infections most frequently affect the skin and soft tissues, but they have also been linked to severe invasive disease, such as necrotizing pneumonia and sepsis, often in young and otherwise healthy individuals. Outbreaks of CA-MRSA have been associated with several common features including close contact, poor hygienic conditions, shared equipment/supplies, high risk of superficial skin abrasions, and lack of access to medical care to treat infections.
While the early stages of the CA-MRSA era were characterized by a diversity of clones, since then only a small number of predominant clones have become established, such as ST93 in Australia, ST80 in Europe and ST8 in the United States Here, we will focus our discussion on ST8-USA300, currently the most widespread CA-MRSA strain and compare its epidemiology and cardinal genotypic characteristics with other prominent CA-MRSA clones.
3.1. CC8
3.1.1. Epidemiology of USA300
By the early 2000s, a single clone named USA300 based on a newly established PFGE nomenclature (McDougal et al., 2003), was established as the most predominant clone in the United States (Tenover and Goering, 2009). Molecular typing further classified this strain as ST8, most commonly spa type t008 and harboring SCCmec type IV, genes encoding Panton–Valentine leukocidin (PVL), and the msr(A) erythromycin resistance genes.
The CDC first formally recognized USA300 in an outbreak of CA-MRSA infections among football players in Pennsylvania in 2000 (CDC, 2003a). Subsequently, this clone was also identified in an outbreak investigation of SSTIs in correctional facilities in Mississippi, Georgia, California, and Texas (CDC, 2001, 2003b), as well as among athletes in Colorado, Indiana and Los Angeles County (CDC, 2003a). No clear link between any of these outbreaks could be established. Subsequently, a single PFGE-type, USA300-0114, was recognized as the predominant strain in the United States and led to an astronomical increase in MRSA infections. For example, by 2005 it was estimated that ∼90% of all MRSA infections in San Francisco were community-associated (Liu et al., 2008).
The dominance of USA300 as the primary CA-MRSA cause of SSTIs was demonstrated in a nationwide study from urban emergency departments in the United States, as USA300 was isolated from ∼98% of SSTIs in some parts of the country (Moran et al., 2006; Talan et al., 2011). USA300 has now become firmly established in Canada. The international spread of USA300 to every continent except Antarctica is well documented, although it has not become as dominant in these countries yet (David and Daum, 2010).
Early after its first recognition it was already noted that USA300 rapidly contributed to the burden of MRSA infections acquired in the hospital setting (Liu et al., 2008). In 2004, this clone accounted for 28% of healthcare-associated bloodstream infections and 20% of nosocomial infections (Seybold et al., 2006). In parallel to its emergence in the healthcare setting, USA300 has increasingly accumulated resistance to a variety of antibiotics (Coombs et al., 2012; Talan et al., 2011), which may in part account for its increasing resilience in the hospital setting. Resistance has mainly been acquired through plasmids conferring resistance to clindamycin (ermA and ermC), tetracycline (tetK and tetM) (Tenover et al., 2006) and mupirocin (mupA) (Diep et al., 2008a). In addition, resistance to fluoroquinolones, conferred by chromosomal mutations, continues to be on the rise (Moran et al., 2006; Talan et al., 2011). Several USA300 isolates have also developed reduced susceptibility to vancomycin, and in some cases, to daptomycin (Graber et al., 2007; Hafer et al., 2012; Hageman et al., 2008), in addition to occasional resistance to gentamicin and trimethoprim–sulfamethoxazole. The prospect of increasing multi-drug resistance in USA300 strains with high virulence potential is a source of great concern.
Virulence alone is unlikely to account for the extraordinary dominance of USA300 infections. Although increased nasal colonization with USA300 was notably absent from the initial outbreak investigations of already antibiotic-treated patients, the first evidence for an increase in USA300 colonization on a population level was documented by the National Health and Nutrition Examination Survey (NHANES) (Gorwitz et al., 2008; Graham et al., 2006). This survey of nasal swabs collected from ∼5000 non-institutionalized people over 4-year period reported a statistically significant increase in USA300 during the 2001–2004 study period, and specifically in USA300-0114—the epidemic clone (Tenover et al., 2006). Although the percent of USA300 nasal colonization in non-institutionalized individuals increased during this time period, the overall level was relatively low (prevalence of MRSA nasal colonization was 1.5%, of which only 19.7% were USA300 or USA400) (Gorwitz et al., 2008), and perhaps could not account for the high overall burden of USA300 in the community. Direct skin-to-skin contact with infected or colonized individuals (in the absence of nasal colonization), or with contaminated objects, was suspected as the route of transmission (Kazakova et al., 2005). Indeed, it is now known that some MRSA and USA300 in particular appear to have an enhanced propensity to colonize extra nasal body sites, such as inguinal and perirectal skin regions (Miko et al., 2012; Peters et al., 2010; Senn et al., 2012). This enhanced ability to colonize additional body sites is an intriguing vehicle for the widespread transmission of USA300 in communities. Moreover, USA300 is also more frequently recovered from environmental surfaces in community households, which has been directly linked to transmission between household members (Knox et al., 2012; Uhlemann et al., 2011). This enhanced prevalence on fomites might reflect the increased burden of USA300 colonization of multiple body sites in individuals or rather an increased capacity of USA300 to survive on inert surfaces.
3.1.2. USA300 genomics and evolution
The complete genome sequence of a USA300 isolate, FPR3757, was reported by Diep et al. (2006); a second genome was published shortly thereafter by Highlander et al. (2007). By comparing the USA300 genome to ten other S. aureus genomes, a substantial number of novel genes were identified. These genes mainly cluster within five regions of the genome, including a novel pathogenicity island (SaPI5) encoding two enterotoxins Seq and Sek (Diep et al., 2006). Most remarkably, the sequence revealed the horizontal acquisition of a novel MGE, the arginine catabolic mobile genetic element (ACME), containing gene clusters for the arginine deaminase pathway and an oligopeptide permease system. This element was absent from other S. aureus strains but present at high prevalence in the skin commensal S. epidermidis (Diep et al., 2006).
A number of studies have since addressed the genomic evolution of USA300 (Kennedy et al., 2008; Tewhey et al., 2012; Uhlemann et al., 2012a). Kennedy et al. first compared the genomes of ten geographically diverse USA300 isolates and noted that all but two isolates differed by only a discrete number of SNPs, suggesting a recent divergence from a common ancestor (Kennedy et al., 2008). Importantly, the findings provide strong support to the idea that there has been clonal emergence of USA300 and subsequent spread throughout the United States, rather evolutionary convergence and the independent emergence of similar organisms. It is also noteworthy that two of these clonal isolates exhibited significantly decreased mortality in a mouse sepsis model, supporting the idea that minimal changes in the core genome can significantly impact S. aureus virulence.
To gain understanding of the adaptation of USA300 in communities, Uhlemann et al. (2012a) sequenced eight longitudinally sampled USA300 isolates collected over 15 months from three community households. The greatest genetic heterogeneity occurred between isolates from different households and within one heavily colonized household, although phylogenetic analysis suggested that all but one isolate likely arose from a recent common ancestor. Sequencing of persisting USA300 isolates identified a small number novel non-synonymous SNPs, deletions, and small genome rearrangements, preferentially in genes involved in the major functional categories aspects of adhesion, cell wall biosynthesis, virulence, and carbohydrate metabolism. The observed small genome rearrangements may provide an intriguing mechanism for rapid genome diversification and host niche adaptation.
A remarkable genetic similarity of USA300 isolates was also noted by Tewhey et al. (2012), who compared 18 USA300 isolates from SSTIs and 18 from severe infections collected over nearly 5 years in a defined geographic region (Tewhey et al., 2012). Non-synonymous mutations were more frequently accumulated in virulence genes of SSTI isolates, findings that may explain in part the ability of strains to cause mild versus severe disease.
Although progress has been made, many open questions remain regarding the evolution of USA300 as it has adapted to both community and healthcare settings.
3.1.3. Putative virulence factors of USA300
While many of the initial studies tried to link the enhanced virulence phenotype of USA300 to MGEs, in particular to prophage-encoded PVL, several lines of research in experimental animal models suggest that there is little or no contribution of MGEs to USA300 virulence, with the exception of necrotizing pneumonia and perhaps during the early stages of skin infections (DeLeo et al., 2010; Diep et al., 2008b, 2010; Kobayashi et al., 2011; Li et al., 2010; Lipinska et al., 2011; Montgomery et al., 2009; Voyich et al., 2006). Studies of human infections largely support these observations, except that PVL does not contribute to a worse outcome for patients with complicated SSTIs (Bae et al., 2009). In contrast, modulated expression of core-genome encoded α-toxin (Hla), phenolsoluble modulin-alpha peptides (PSMα) and accessory gene regulator (Agr) have a dramatic influence on virulence in several animal models of infection (Bubeck Wardenburg et al., 2007; Kobayashi et al., 2011; Li et al., 2010; Wang et al., 2007). PSMs are small cytolytic peptides that serve many different functions in S. aureus pathogenesis. They contribute to the structure as well as the dissemination of biofilms (Periasamy et al., 2012) and propagate inflammation (Li et al., 2010; Wang et al., 2007). A variant, PSM-mec, is encoded on select SCCmec elements and when present in high amounts relative to other PSMs, contributes significantly to S. aureus virulence (Queck et al., 2008). Furthermore, a previously unrecognized core-genome encoded toxin, staphylococcal enterotoxin-like toxin X (SEIX), contributes significantly to lethality in a rabbit necrotizing pneumonia model (Wilson et al., 2011).
To further elucidate the origin of USA300, Li et al. (2009) compared several major MRSA isolates in a combination of virulence and genotyping assays. These studies indicated that USA300 and USA500 have a high capacity for virulence and an enhanced ability to evade the host immune response. The noted enhanced virulence capacity was due to an increased expression of core genome-encoded virulence determinants, such as PSMs and α-toxin (Li et al., 2009). Based on sequence comparisons of seven housekeeping genes and seven surface protein encoding genes, this study also established USA500 as the progenitor of USA300, which ultimately acquired ACME, a SaPI that encodes enterotoxin K (sek) and enterotoxin Q (seq), and the prophage encoding lukSF-PV.
The fact that the virulence phenotype of USA300 was already established in its USA500 progenitor also argues that the acquisition of MGEs has played a limited role in the evolution of USA300 virulence, and suggests that these elements have a different role in S. aureus biology. In light of the incomparably higher spread of USA300 compared to USA500, it has been speculated that these MGEs are more important for transmission of USA300 rather than contributing to virulence.
3.1.4. ACME—a putative fitness factor of USA300
ACME has been the premier candidate as a MGE fitness factor, based on its putative involvement in pH homeostasis. However, until recently its exact contribution to the fitness of USA300, especially on skin, has remained unknown. ACME encompasses a 30.9 kb genomic region, encoding 33 genes, and is thought to have been acquired from coagulase negative staphylococci (Diep et al., 2006). ACME was first implicated as a virulence factor in a rabbit bacteremia model (Diep et al., 2008b), although a subsequent study found no evidence that ACME directly contributes to the virulence of USA300 using a rat model of necrotizing pneumonia and a mouse model of skin infection (Montgomery et al., 2009). These findings appear at variance, but differences in the animal species (rabbit versus rat and mouse) or infection models (bacteremia versus pneumonia or skin infection) used could account for the reported differences in the contribution of ACME to virulence.
Linking in the observation that S. aureus is uniquely hypersensitive to polyamines, which are encountered in high concentrations during infection and in inflamed skin, Joshi and colleagues demonstrated that USA300 exhibited complete resistance to polyamines (Joshi et al., 2011). This polyamine-resistant phenotype depended on the presence of a spermine/spermidine acetyl-transferase (speG) encoded within the ACME island. The abrogation of the host polyamine enabled a ΔspeG mutant to persist in infected wounds. The polyamine resistance of USA300 was also shown to provide a major fitness advantage during SSTIs but not in sepsis (Thurlow et al., 2013). However, speG needs to act in conjunction with other gene clusters within ACME. It was shown that the constitutive ACME-encoded arginine-deiminase system (Arc) converts arginine to ornithine and ammonia, which enables survival in an acidic environment comparable to the human skin. In parallel, this ACME-Arc system drives the excessive production of host polyamines, compounds uniquely toxic to S. aureus, necessitating its protection by speG (Thurlow et al., 2013).
To date, ACME has only infrequently been encountered in other S. aureus lineages and has not been ascribed to any other successful CA-MRSA clone. While initially all USA300 isolates were believed to carry ACME, more recent studies indicate that between 9% and 15% of USA300 isolates do not harbor this element (Montgomery et al., 2008; Uhlemann et al., 2011). It is currently unclear if these ACME-negative isolates represent a distinct sublineage or if their potential increase indicates that this element is not as stably maintained in USA300.
3.1.5. Comparison of USA300 with other major CA-MRSA clones
Despite the number of available genome sequences of CA-MRSA strains, no clear consensus exists regarding key features that enabled their collective emergence and success. A number of studies have compared virulence of select CA-MRSA (and HA-MRSA) strains in animal models of infection (e.g., Li et al., 2009, 2010; Voyich et al., 2005). Li et al. (2010) found that the most virulent strains in a rabbit abscess model consisted of USA300, USA500 and ST80, whereas USA400, an early dominant CA-MRSA in the United States, caused smaller abscesses by comparison. A study by Chua et al. (2011) reported that ST93 was the most virulent strain among those compared (including USA300, USA400, and ST239) in wax moth larvae and mouse skin infection models. Therefore, the virulence observed in animal models cannot fully explain the prominent distribution or success of certain CA-MRSA lineages. As another example, the highly virulent ST80 strain is one of the most frequent CA-MRSA strains in Europe and is occasionally encountered in Australia and North Africa, but it has caused very limited disease compared to USA300 or ST93 in their respective geographic regions. Like many CA-MRSA strains, ST80 usually harbors PVL and carries a SCCmec-type IV cassette. Recently, Stegger et al. (2012) reported the genome of a representative ST80 isolate from Denmark. This isolate contained a novel SCCmec-type IV cassette with an integrated partial plasmid, as well as three integrated pro-phages, encoding PVL and immunomodulatory genes, sak and scn, but no unique identifying features were reported.
The ST93 Queensland MRSA clone has emerged over the past decade in Australia, resulting in an increase in CA-MRSA infections (from 6.6% to 11.5% nationwide). In 2010, this strain constituted 41% of all CA-MRSA infections in Australia, 28% of all MRSA infections, and 4.9% of all S. aureus community-onset infections (SAP10, 2010). Infections with ST93-MRSA also predominantly manifest as SSTIs, but an enhanced clinical virulence as evidenced by reports of severe invasive infection—such as necrotizing pneumonia, deep-seated abscess, osteomyelitis, septic arthritis and septicemia—has been suggested (Coombs et al., 2009; Risson et al., 2007). There are isolated reports of ST93 infections in New Zealand and the UK, but many of these cases could be epidemiologically linked to Australia (Ellington et al., 2010a).
By MLST analysis ST93 represents a singleton and is distinct from other S. aureus clones, but also harbors SCCmec IV [2B] and PVL. Based on its genome sequence, ST93, like USA300, contains α-hemolysin, PVL and PSM-α (Chua et al., 2010) gain, no overt novel virulence determinant has been identified in ST93 that could easily explain its high virulence capacity, suggesting the contribution of altered gene expression or subtle genetic alterations in the core genome. Furthermore, it is tempting to speculate that, while ST93 is highly virulent, the lack of an obvious fitness factor, such as the ACME-arcA-speG system in USA300, might account for its relatively limited geographic distribution.
Lastly, the complete genome sequence of an ST59 CA-MRSA isolate, a lineage predominant in Taiwan, was reported recently (Huang et al., 2012a, 2012b). Since the original identification of ST59, these isolates have frequently been associated with multi-drug resistance. Compared to the USA300 strain FPR3757 and the USA4000 strain MW2, most of the open reading frames in the ST59 strain M013 were highly conserved (Huang et al. 2012a). However, genomic islands SaPI5 and νSa3 as well as prophages ϕSa3usa and ϕSa3 are absent in M013 (Huang et al., 2012a, 2012b). In addition, the majority of the ORFs in the pathogenicity-related genomic island νSaβ are deleted in the corresponding region of the M013 genome. ST59 also harbors PVL, but the genes encoding the toxin appear to have distinct sequence composition. A comparative genome sequence analysis of multi-drug resistant ST59 isolates identified truncated hsdM and hsdS genes on νSaβ, and these genes encode part of the restriction-modification system components (Huang et al., 2012a, 2012b). This defect may have contributed to the acquisition of IS1216V and MMESPM1, which are enterococcal DNA elements that confer multi-drug resistance (Hung et al. 2012b).
Taken together, despite a series of genome-scale and genome sequence analyses of diverse CA-MRSA strains, no single unique genetic locus has been identified that accounts for the success of these strains. While PVL is disproportionally present in many CA-MRSA lineages, its absence does not appear to diminish the virulence capacity of S. aureus. Perhaps it is not as much the acquisition of novel virulence factors, but rather a paucity of inactivating mutations in genes such as hla, agrC or crtM (as discussed above for the CC30 lineage) that enables CA-MRSA strains to maintain a high virulence phenotype. This model infers that as successful S. aureus clones are increasingly exposed to triggers such as the anti-biotic rich nosocomial environment, they will eventually accumulate inactivating mutations that reduce their capacity to cause severe infections, and this phenomenon thereby promotes survival of S. aureus as a commensal organism. Alternatively, an enhanced ability of successful CA-MRSA strains to up-regulate virulence factors or evade the innate immune response could contribute to their unique success as pathogens.
4. Concluding remarks
S. aureus is perhaps most widely known as a human pathogen and for its ability to acquire resistance to antibiotics. However, it is equally a commensal organism that asymptomatically colonizes humans and other mammals. It could be argued that the existence as a commensal microbe is better suited for long-term fitness and survival (as opposed to life as a pathogen). From that viewpoint, the emergence or re-emergence S. aureus/MRSA strains that cause widespread and sometimes fatal infections is somewhat puzzling. One explanation is that host susceptibility, as with sick or immune comprised individuals, plays a major role in the emergence of new strains. That is, strains that normally colonize humans asymptomatically have the ability to cause opportunistic infections in a more susceptible (e.g., sick) host. The ongoing emergence of antibiotic resistant strains is readily explained by selective pressure in the healthcare setting, and thus the combination of susceptible hosts and antibiotic use likely favor emergence of new MRSA strains. On the other hand, there is no clear explanation for the emergence and success of specific MRSA clones such as EMRSA-16. For example, why do such strains predominate in specific geographic locations and then often decline rapidly to make way for another unrelated clone? One possibility is that this phenomenon involves a polygenetic trait and/or the agr locus. Shopsin et al. (Shopsin et al., 2010) have shown that inactivating mutations in agr provide a short-term adaptive survival mechanism in the infected host, but they are deleterious for the long-term transmission of strains. These observations appear compatible with the findings of DeLeo et al. (2011) and McAdam et al. (2012) as discussed above. In addition, Holden et al. (2013) and Knight et al. (2012) suggested that the recent replacement of EMRSA-16 with EMRSA-15 as the dominant HA-MRSA strain in the United Kingdom involved accumulation of mutations associated with resistance to fluoroquinolones, and increased fitness as assessed by growth rate in vitro. The basis for the increased fitness of EMRSA-15 (CC22) is unknown, but Knight et al. propose that the smaller SCCmec type IV element of EMRSA-15 has a reduced fitness cost compared with the larger SCCmec type II element present in EMRSA-16 strains.
An explanation for the emergence of CA-MRSA also remains a mystery. The overall burden of antibiotics and widespread use of antibiotics—in and out of healthcare settings—could have contributed, but there is no conclusive evidence to support this notion or explain why CA-MRSA strains have been largely restricted to a select few lineages. It may be that these strains have arisen by rare stochastic events, or alternatively, they may be more susceptible to integration of specific genetic elements, such as SCCmec type IV or ACME (for USA300). In any case, more work is needed to understand the emergence and success of CA-MRSA. These questions are now being addressed using genome-scale approaches, which include expression microarrays and genome sequencing. Such data are an important step toward a comprehensive understanding of S. aureus epidemic waves and the evolution of MRSA in hospitals and the community.
Acknowledgments
This work was supported by the US National Institutes of Health (K08 AI090013 to ACU, and R01 AI077690 and R01 AI077690 to FDL), a Paul A. Marks Scholarship (to ACU), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, US National Institutes of Health (MO and FRD).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Adhikari RP, Cook GM, Lamont I, Lang S, Heffernan H, Smith JM. Phenotypic and molecular characterization of community occurring, Western Samoan phage pattern methicillin-resistant Staphylococcus aureus. The Journal of Antimicrobial Chemotherapy. 2002;50:825–831. doi: 10.1093/jac/dkf242. [DOI] [PubMed] [Google Scholar]
- Aires de Sousa M, Crisostomo MI, Sanches IS, Wu JS, Fuzhong J, Tomasz A, de Lencastre H. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. Journal of Clinical Microbiology. 2003;41:159–163. doi: 10.1128/JCM.41.1.159-163.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alp E, Klaassen CH, Doganay M, Altoparlak U, Aydin K, Engin A, Kuzucu C, Ozakin C, Ozinel MA, Turhan O, Voss A. MRSA genotypes in Turkey: persistence over 10 years of a single clone of ST239. The Journal of Infection. 2009;58:433–438. doi: 10.1016/j.jinf.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Amorim ML, Aires de Sousa M, Sanches IS, Sa-Leao R, Cabeda JM, Amorim JM, de Lencastre H. Clonal and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) from a Portuguese hospital over time. Microbial Drug Resistance. 2002;8:301–309. doi: 10.1089/10766290260469561. [DOI] [PubMed] [Google Scholar]
- Bae IG, Tonthat GT, Stryjewski ME, Rude TH, Reilly LF, Barriere SL, Genter FC, Corey GR, Fowler VG., Jr Presence of genes encoding the Panton-Valentine leukocidin exotoxin is not the primary determinant of outcome in patients with complicated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus: results of a multinational trial. Journal of Clinical Microbiology. 2009;47:3952–3957. doi: 10.1128/JCM.01643-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels MD, Nanuashvili A, Boye K, Rohde SM, Jashiashvili N, Faria NA, Kereselidze M, Kharebava S, Westh H. Methicillin-resistant Staphylococcus aureus in hospitals in Tbilisi, the Republic of Georgia, are variants of the Brazilian clone. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology. 2008;27:757–760. doi: 10.1007/s10096-008-0500-z. [DOI] [PubMed] [Google Scholar]
- Benito N, Miro JM, de Lazzari E, Cabell CH, del Rio A, Altclas J, Commerford P, Delahaye F, Dragulescu S, Giamarellou H, Habib G, Kamarulzaman A, Kumar AS, Nacinovich FM, Suter F, Tribouilloy C, Venugopal K, Moreno A, Fowler VG, Jr Investigators, I.-P. Health care-associated native valve endocarditis: importance of non-nosocomial acquisition. Annals of Internal Medicine. 2009;150:586–594. doi: 10.7326/0003-4819-150-9-200905050-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Carr M. Staphylococci in hospital-acquired infections; types encountered in the United States. Journal of the American Medical Association. 1958;166:1192–1196. doi: 10.1001/jama.1958.62990100020011d. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton–Valentine leukocidin in Staphylococcus aureus pneumonia. Nature Medicine. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- Buck JM, Como-Sabetti K, Harriman KH, Danila RN, Boxrud DJ, Glennen A, Lynfield R. Community-associated methicillin-resistant Staphylococcus aureus, Minnesota, 2000–2003. Emerging Infectious Diseases. 2005;11:1532–1538. doi: 10.3201/eid1110.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bynoe ET, Elder RH, Comtois RD. Phage-typing and antibiotic-resistance of staphylococci isolated in a general hospital. Canadian Journal of Microbiology. 1956;2:346–358. doi: 10.1139/m56-041. [DOI] [PubMed] [Google Scholar]
- CDC. From the Centers for Disease Control and Prevention Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus – Minnesota and North Dakota, 1997–1999. JAMA: The Journal of the American Medical Association. 1999;282:1123–1125. [PubMed] [Google Scholar]
- CDC. Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison – Mississippi, 2000. MMWR. Morbidity and mortality weekly report. 2001;50:919–922. [PubMed] [Google Scholar]
- CDC. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants – Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. MMWR. Morbidity and mortality weekly report. 2003a;52:793–795. [PubMed] [Google Scholar]
- CDC. Methicillin-resistant Staphylococcus aureus infections in correctional facilities – Georgia, California, and Texas, 2001–2003. MMWR. Morbidity and mortality weekly report. 2003b;52:992–996. [PubMed] [Google Scholar]
- Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature reviews. Microbiology. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Seemann T, Harrison PF, Davies JK, Coutts SJ, Chen H, Haring V, Moore R, Howden BP, Stinear TP. Complete genome sequence of Staphylococcus aureus strain JKD6159, a unique Australian clone of ST93-IV community methicillin-resistant Staphylococcus aureus. Journal of Bacteriology. 2010;192:5556–5557. doi: 10.1128/JB.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KY, Seemann T, Harrison PF, Monagle S, Korman TM, Johnson PD, Coombs GW, Howden BO, Davies JK, Howden BP, Stinear TP. The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct. PLoS One. 2011;6:e25887. doi: 10.1371/journal.pone.0025887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J, Rudkin J, Recker M, Pozzi C, O'Gara JP, Massey RC. Offsetting virulence and antibiotic resistance costs by MRSA. The ISME Journal. 2010;4:577–584. doi: 10.1038/ismej.2009.151. [DOI] [PubMed] [Google Scholar]
- Coombs GW, Goering RV, Chua KY, Monecke S, Howden BP, Stinear TP, Ehricht R, O'Brien FG, Christiansen KJ. The molecular epidemiology of the highly virulent ST93 Australian community Staphylococcus aureus strain. PLoS One. 2012;7:e43037. doi: 10.1371/journal.pone.0043037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs GW, Nimmo GR, Pearson JC, Christiansen KJ, Bell JM, Collignon PJ, McLaws ML Australian Group for Antimicrobial, R. Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients, 2006. Communicable Diseases Intelligence. 2009;33:10–20. doi: 10.33321/cdi.2009.33.2. [DOI] [PubMed] [Google Scholar]
- Cox RA, Conquest C, Mallaghan C, Marples RR. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16) The Journal of Hospital Infection. 1995;29:87–106. doi: 10.1016/0195-6701(95)90191-4. [DOI] [PubMed] [Google Scholar]
- David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clinical Microbiology Reviews. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David MZ, Glikman D, Crawford SE, Peng J, King KJ, Hostetler MA, Boyle-Vavra S, Daum RS. What is community-associated methicillin-resistant Staphylococcus aureus? The Journal of Infectious Diseases. 2008;197:1235–1243. doi: 10.1086/533502. [DOI] [PubMed] [Google Scholar]
- DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, Mathema B, Braughton KR, Whitney AR, Villaruz AE, Martens CA, Porcella SF, McGavin MJ, Otto M, Musser JM, Kreiswirth BN. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18091–18096. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Chambers HF, Graber CJ, Szumowski JD, Miller LG, Han LL, Chen JH, Lin F, Lin J, Phan TH, Carleton HA, McDougal LK, Tenover FC, Cohen DE, Mayer KH, Sensabaugh GF, Perdreau-Remington F. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Annals of Internal Medicine. 2008a;148:249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- Diep BA, Chan L, Tattevin P, Kajikawa O, Martin TR, Basuino L, Mai TT, Marbach H, Braughton KR, Whitney AR, Gardner DJ, Fan X, Tseng CW, Liu GY, Badiou C, Etienne J, Lina G, Matthay MA, DeLeo FR, Chambers HF. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton–Valentine leukocidin-induced lung inflammation and injury. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5587–5592. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. The Journal of Infectious Diseases. 2008b;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- Edgeworth JD, Yadegarfar G, Pathak S, Batra R, Cockfield JD, Wyncoll D, Beale R, Lindsay JA. An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2007;44:493–501. doi: 10.1086/511034. [DOI] [PubMed] [Google Scholar]
- Ellington MJ, Ganner M, Warner M, Boakes E, Cookson BD, Hill RL, Kearns AM. First international spread and dissemination of the virulent Queensland community-associated methicillin-resistant Staphylococcus aureus strain. Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010a;16:1009–1012. doi: 10.1111/j.1469-0691.2009.02994.x. [DOI] [PubMed] [Google Scholar]
- Ellington MJ, Hope R, Livermore DM, Kearns AM, Henderson K, Cookson BD, Pearson A, Johnson AP. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. The Journal of Antimicrobial Chemotherapy. 2010b;65:446–448. doi: 10.1093/jac/dkp448. [DOI] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. Multilocus sequence typing. Trends in Microbiology. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- Fluit AC, Wielders CL, Verhoef J, Schmitz FJ. Epidemiology and susceptibility of 3051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. Journal of Clinical Microbiology. 2001;39:3727–3732. doi: 10.1128/JCM.39.10.3727-3732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nature reviews. Microbiology. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Fowler VG, Jr, Nelson CL, McIntyre LM, Kreiswirth BN, Monk A, Archer GL, Federspiel J, Naidich S, Remortel B, Rude T, Brown P, Reller LB, Corey GR, Gill SR. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. The Journal of Infectious Diseases. 2007;196:738–747. doi: 10.1086/520088. [DOI] [PubMed] [Google Scholar]
- Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, Jernigan JA, Harriman K, Harrison LH, Lynfield R, Farley MM Active Bacterial Core Surveillance Program of the Emerging Infections Program, N. Methicillin-resistant Staphylococcus aureus disease in three communities. The New England Journal of Medicine. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- Gillespie WA, Alder VG. Control of an outbreak of staphylococcal infection in a hospital. Lancet. 1957;272:632–634. doi: 10.1016/s0140-6736(57)91091-7. [DOI] [PubMed] [Google Scholar]
- Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. The Journal of Infectious Diseases. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- Graber CJ, Wong MK, Carleton HA, Perdreau-Remington F, Haller BL, Chambers HF. Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerging Infectious Diseases. 2007;13:491–493. doi: 10.3201/eid1303.060960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, 3rd, Lin SX, Larson EL. A U.S. population-based survey of Staphylococcus aureus colonization. Annals of Internal Medicine. 2006;144:318–325. doi: 10.7326/0003-4819-144-5-200603070-00006. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW European Staphylococcal Reference Laboratory Working, G. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Medicine. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2012;56:5845–5851. doi: 10.1128/AAC.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman JC, Patel J, Franklin P, Miscavish K, McDougal L, Lonsway D, Khan FN. Occurrence of a USA300 vancomycin-intermediate Staphylococcus aureus. Diagnostic Microbiology and Infectious Disease. 2008;62:440–442. doi: 10.1016/j.diagmicrobio.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassall JE, Rountree PM. Staphylococcal septicaemia. Lancet. 1959;1:213–217. doi: 10.1016/s0140-6736(59)90047-9. [DOI] [PubMed] [Google Scholar]
- Havaei SA, Vidovic S, Tahmineh N, Mohammad K, Mohsen K, Starnino S, Dillon JA. Epidemic methicillin-susceptible Staphylococcus aureus lineages are the main cause of infections at an Iranian university hospital. Journal of Clinical Microbiology. 2011;49:3990–3993. doi: 10.1128/JCM.05445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiology. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlickova H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramirez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nubel U. A genomic portrait of the emergence, evolution and global spread of a methicillin resistant Staphylococcus aureus pandemic. Genome Research. 2013 doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Batra R, Parkhill J, Bentley SD, Edgeworth JD. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW) Journal of Bacteriology. 2010;192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TW, Chen FJ, Miu WC, Liao TL, Lin AC, Huang IW, Wu KM, Tsai SF, Chen YT, Lauderdale TL. Complete genome sequence of Staphylococcus aureus M013, a pvl-positive, ST59-SCCmec type V strain isolated in Taiwan. Journal of Bacteriology. 2012a;194:1256–1257. doi: 10.1128/JB.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung WC, Takano T, Higuchi W, Iwao Y, Khokhlova O, Teng LJ, Yamamoto T. Comparative genomics of community-acquired ST59 methicillin-resistant Staphylococcus aureus in Taiwan: novel mobile resistance structures with IS1216V. PLoS One. 2012b;7:e46987. doi: 10.1371/journal.pone.0046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TC, McCollister BD, Sharma R, McFann KK, Madinger NE, Barron M, Bessesen M, Price CS, Burman WJ. Epidemiology of healthcare-associated bloodstream infection caused by USA300 strains of methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Infection Control and Hospital Epidemiology: The Official Journal of the Society of Hospital Epidemiologists of America. 2009;30:233–241. doi: 10.1086/595963. [DOI] [PubMed] [Google Scholar]
- Jessen O, Rosendal K, Bulow P, Faber V, Eriksen KR. Changing staphylococci and staphylococcal infections. A ten-year study of bacteria and cases of bacteremia. The New England Journal of Medicine. 1969;281:627–635. doi: 10.1056/NEJM196909182811201. [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, Livermore DM, Cookson BD U.E. Participants. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) The Journal of Antimicrobial Chemotherapy. 2001;48:143–144. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Molecular Microbiology. 2011;82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, Boo T, McAllister S, Anderson J, Jensen B, Dodson D, Lonsway D, McDougal, Arduino M, Fraser VJ, Killgore G, Tenover FC, Cody S, Jernigan DB. A clone of methicillin-resistant Staphylococcus aureus among professional football players. The New England Journal of Medicine. 2005;352:468–475. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ES, Lee HJ, Chung GT, Lee YS, Shin DH, Jung SI, Song KH, Park WB, Kim NJ, Park KU, Kim EC, Oh MD, Kim HB. Molecular characterization of methicillin-resistant Staphylococcus aureus isolates in Korea. Journal of Clinical Microbiology. 2011;49:1979–1982. doi: 10.1128/JCM.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R National Nosocomial Infections Surveillance System. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992–2003. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2006;42:389–391. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK Active Bacterial Core surveillance, M.I. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA: The Journal of the American Medical Association. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Knight GM, Budd EL, Whitney L, Thornley A, Al-Ghusein H, Planche T, Lindsay JA. Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. The Journal of Antimicrobial Chemotherapy. 2012;67:2514–2522. doi: 10.1093/jac/dks245. [DOI] [PubMed] [Google Scholar]
- Knox J, Uhlemann AC, Miller M, Hafer C, Vasquez G, Vavagiakis P, Shi Q, Lowy FD. Environmental contamination as a risk factor for intra-household Staphylococcus aureus transmission. PLoS One. 2012;7:e49900. doi: 10.1371/journal.pone.0049900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KS, Lee JY, Suh JY, Oh WS, Peck KR, Lee NY, Song JH. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. Journal of Clinical Microbiology. 2005;43:421–426. doi: 10.1128/JCM.43.1.421-426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. The Journal of Infectious Diseases. 2011;204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. The Journal of Infectious Diseases. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, Otto M. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nature Medicine. 2012;18:816–819. doi: 10.1038/nm.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light IJ, Atherton HD, Sutherland JM. Decreased colonization of newborn infants with Staphylococcus aureus 80/81: Cincinnati General Hospital, 1960–1972. The Journal of Infectious Diseases. 1975;131:281–285. doi: 10.1093/infdis/131.3.281. [DOI] [PubMed] [Google Scholar]
- Lipinska U, Hermans K, Meulemans L, Dumitrescu O, Badiou C, Duchateau L, Haesebrouck F, Etienne J, Lina G. Panton–Valentine leukocidin does play a role in the early stage of Staphylococcus aureus skin infections: a rabbit model. PLoS One. 2011;6:e22864. doi: 10.1371/journal.pone.0022864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Graber CJ, Karr M, Diep BA, Basuino L, Schwartz BS, Enright MC, O'Hanlon SJ, Thomas JC, Perdreau-Remington F, Gordon S, Gunthorpe H, Jacobs R, Jensen P, Leoung G, Rumack JS, Chambers HF. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2008;46:1637–1646. doi: 10.1086/587893. [DOI] [PubMed] [Google Scholar]
- Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. The Journal of Experimental Medicine. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang H, Du N, Shen E, Chen H, Niu J, Ye H, Chen M. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrobial Agents and Chemotherapy. 2009;53:512–518. doi: 10.1128/AAC.00804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. The New England Journal of Medicine. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cellular and Molecular Life Sciences: CMLS. 2010;67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor L, Ortellado J, Menacho C, Lird G, Courtier C, Gardon C, Meugnier H, Bes M, Vandenesch F, Etienne J. Molecular characterization of methicillin-resistant Staphylococcus aureus isolates collected in Asuncion, Paraguay. Journal of Clinical Microbiology. 2007;45:2298–2300. doi: 10.1128/JCM.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam PR, Templeton KE, Edwards GF, Holden MT, Feil EJ, Aanensen DM, Bargawi HJ, Spratt BG, Bentley SD, Parkhill J, Enright MC, Holmes A, Girvan EK, Godfrey PA, Feldgarden M, Kearns AM, Rambaut A, Robinson DA, Fitzgerald JR. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9107–9112. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. Journal of Clinical Microbiology. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melter O, Aires de Sousa M, Urbaskova P, Jakubu V, Zemlickova H, de Lencastre H. Update on the major clonal types of methicillin-resistant Staphylococcus aureus in the Czech Republic. Journal of Clinical Microbiology. 2003;41:4998–5005. doi: 10.1128/JCM.41.11.4998-5005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmi G, Nair DR, Cheung A. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. Journal of Bacteriology. 2012;194:759–767. doi: 10.1128/JB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko BA, Uhlemann AC, Gelman A, Lee CJ, Hafer CA, Sullivan SB, Shi Q, Miller M, Zenilman J, Lowy FD. High prevalence of colonization with Staphylococcus aureus clone USA300 at multiple body sites among sexually transmitted disease clinic patients: an unrecognized reservoir. Microbes and Infection/Institut Pasteur. 2012;14:1040–1043. doi: 10.1016/j.micinf.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LG, Perdreau-Remington F, Bayer AS, Diep B, Tan N, Bharadwa K, Tsui J, Perlroth J, Shay A, Tagudar G, Ibebuogu U, Spellberg B. Clinical and epidemiologic characteristics cannot distinguish community-associated methicillin-resistant Staphylococcus aureus infection from methicillin-susceptible S. aureus infection: a prospective investigation. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2007;44:471–482. doi: 10.1086/511033. [DOI] [PubMed] [Google Scholar]
- Montesinos I, Delgado T, Riverol D, Salido E, Miguel MA, Jimenez A, Sierra A. Changes in the epidemiology of meticillin-resistant Staphylococcus aureus associated with the emergence of EMRSA-16 at a university hospital. The Journal of Hospital Infection. 2006;64:257–263. doi: 10.1016/j.jhin.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. The Journal of Infectious Diseases. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Daum RS. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infection and Immunity. 2009;77:2650–2656. doi: 10.1128/IAI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA E.M.I.N.S. Group. Methicillin-resistant S. aureus infections among patients in the emergency department. The New England Journal of Medicine. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- Murchan S, Aucken HM, O'Neill GL, Ganner M, Cookson BD. Emergence, spread, and characterization of phage variants of epidemic methicillin-resistant Staphylococcus aureus 16 in England and Wales. Journal of Clinical Microbiology. 2004;42:5154–5160. doi: 10.1128/JCM.42.11.5154-5160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo GR, Fong J, Paterson DL, McLaws ML. Changing epidemiology of meticillin-resistant S. aureus in Queensland, Australia, 2000–2006: use of passive surveillance of susceptibility phenotypes. The Journal of Hospital Infection. 2008;70:305–313. doi: 10.1016/j.jhin.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Nubel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annual Review of Microbiology. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Brooks JT, Limbago B, Lowery HK, McAllister SK, Mindley R, Fosheim G, Gorwitz RJ, Guest JL, Hageman J, Fridge J, Rimland D. Methicillin-resistant Staphylococcus aureus colonization in HIV-infected outpatients is common and detection is enhanced by groin culture. Epidemiology and Infection. 2010:1–11. doi: 10.1017/S0950268810002013. [DOI] [PubMed] [Google Scholar]
- Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Molecular Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risson DC, O'Connor ED, Guard RW, Schooneveldt JM, Nimmo GR. A fatal case of necrotising pneumonia due to community-associated methicillin-resistant Staphylococcus aureus. The Medical Journal of Australia. 2007;186:479–480. doi: 10.5694/j.1326-5377.2007.tb01002.x. [DOI] [PubMed] [Google Scholar]
- Robinson DA, Enright MC. Evolution of Staphylococcus aureus by large chromosomal replacements. Journal of Bacteriology. 2004;186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DA, Kearns AM, Holmes A, Morrison D, Grundmann H, Edwards G, O'Brien FG, Tenover FC, McDougal LK, Monk AB, Enright MC. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–1258. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- Rosenthal VD, Bijie H, Maki DG, Mehta Y, Apisarnthanarak A, Medeiros EA, Leblebicioglu H, Fisher D, Alvarez-Moreno C, Khader IA, Del Rocio Gonzalez Martinez M, Cuellar LE, Navoa-Ng JA, Abouqal R, Guanche Garcell H, Mitrev Z, Pirez Garcia MC, Hamdi A, Duenas L, Cancel E, Gurskis V, Rasslan O, Ahmed A, Kanj SS, Ugalde OC, Mapp T, Raka L, Yuet Meng C, Thu le TA, Ghazal S, Gikas A, Narvaez LP, Mejia N, Hadjieva N, Gamar Elanbya, Guzman Siritt ME, Jayatilleke K INICC Members. International Nosocomial Infection Control Consortium (INICC) Report, data summary of 36 countries, for 2004–2009. American Journal of Infection Control. 2009;40:396–407. doi: 10.1016/j.ajic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Rountree PM, Beard MA. Further observations on infection with phage type 80 staphylococci in Australia. Medical Journal of Australia. 1958;2:789–795. [PubMed] [Google Scholar]
- Rountree PM, Freeman BM. Infections caused by a particular phage type of Staphylococcus aureus. Medical Journal of Australia. 1955;42:157–161. [PubMed] [Google Scholar]
- Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O'Gara JP, Massey RC. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. The Journal of Infectious Diseases. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAP10. 2010 < http://www.agargroup.org/files/SAP2010MRSA.pdf>.
- Senn L, Basset P, Nahimana I, Zanetti G, Blanc DS. Which anatomical sites should be sampled for screening of methicillin-resistant Staphylococcus aureus carriage by culture or by rapid PCR test? Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18:E31–33. doi: 10.1111/j.1469-0691.2011.03724.x. [DOI] [PubMed] [Google Scholar]
- Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. The Journal of Infectious Diseases. 2010;202:1593–1599. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. Journal of Clinical Microbiology. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song JH, de Lencastre H, Robinson DA. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One. 2010;5:e8582. doi: 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo Ko K, Kim YS, Song JH, Yeom JS, Lee H, Jung SI, Jeong DR, Kim SW, Chang HH, Ki HK, Moon C, Oh WS, Peck KR, Lee NY. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrobial Agents and Chemotherapy. 2005;49:3583–3585. doi: 10.1128/AAC.49.8.3583-3585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegger M, Price LB, Larsen AR, Gillece JD, Waters AE, Skov R, Andersen PS. Genome sequence of Staphylococcus aureus strain 11819–97, an ST80-IV European community-acquired methicillin-resistant isolate. Journal of Bacteriology. 2012;194:1625–1626. doi: 10.1128/JB.06653-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ E.M.I.N.S. Group. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2011;53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. The Journal of Antimicrobial Chemotherapy. 2009;64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. Journal of Clinical Microbiology. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Cannavino CR, Leake JA, Bansal V, Topol EJ, Torkamani A, Bradley JS, Schork NJ. Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics. 2012;13:508. doi: 10.1186/1471-2164-13-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host & Microbe. 2013;13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. The Journal of Hospital Infection. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- Uhlemann AC, Kennedy AD, Martens C, Porcella SF, Deleo FR, Lowy FD. Toward an understanding of the evolution of Staphylococcus aureus strain USA300 during colonization in community households. Genome Biology and Evolution. 2012a;4:1275–1285. doi: 10.1093/gbe/evs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, Knox J, Miller M, Hafer C, Vasquez G, Ryan M, Vavagiakis P, Shi Q, Lowy FD. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One. 2011;6:e22407. doi: 10.1371/journal.pone.0022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio. 2012b;3 doi: 10.1128/mBio.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton–Valentine leukocidin genes: worldwide emergence. Emerging Infectious Diseases. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivoni AM, Diep BA, de Gouveia Magalhaes AC, Santos KR, Riley LW, Sensabaugh GF, Moreira BM. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. Journal of Clinical Microbiology. 2006;44:1686–1691. doi: 10.1128/JCM.44.5.1686-1691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. Journal of Immunology. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. Is Panton–Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? The Journal of Infectious Diseases. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nature Medicine. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Williams RE, Rippon JE, Dowsett LM. Bacteriophage typing of strains of Staphylococcus aureus from various sources. Lancet. 1953;1:510–514. doi: 10.1016/s0140-6736(53)92357-5. [DOI] [PubMed] [Google Scholar]
- Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, Morrison WI, Fitzgerald JR. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathogens. 2011;7:e1002271. doi: 10.1371/journal.ppat.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H, Ewertz B, Wisplinghoff S, Stefanik D, Plum G, Perdreau-Remington F, Seifert H. Molecular evolution of methicillin-resistant Staphylococcus aureus in the metropolitan area of Cologne, Germany, from 1984 to 1998. Journal of Clinical Microbiology. 2005;43:5445–5451. doi: 10.1128/JCM.43.11.5445-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YH, Giske CG, Wei ZQ, Shen P, Heddini A, Li LJ. Epidemiology and characteristics of antimicrobial resistance in China. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2011;14:236–250. doi: 10.1016/j.drup.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Xiong YQ, Fowler VG, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. The Journal of Infectious Diseases. 2009;199:201–208. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BL, Zhang G, Ye HF, Feil EJ, Chen GR, Zhou XM, Zhan XM, Chen SM, Pan WB. Predominance of the Hungarian clone (ST 239-III) among hospital-acquired meticillin-resistant Staphylococcus aureus isolates recovered throughout mainland China. The Journal of Hospital Infection. 2009;71:245–255. doi: 10.1016/j.jhin.2008.10.029. [DOI] [PubMed] [Google Scholar]


