Abstract
In sarcoma, the activity of NF-κB (nuclear factor κB) reduces the abundance of the microRNA (miRNA) miR-29. The tumor suppressor A20 [also known as TNFAIP3 (tumor necrosis factor–α–induced protein 3)] inhibits an upstream activator of NF-κB and is often mutated in lymphomas. In a panel of human sarcoma cell lines, we found that the activation of NF-κB was increased and, although the abundance of A20 protein and mRNA was decreased, the gene encoding A20 was rarely mutated. The 3′ untranslated region (UTR) of A20 mRNA has conserved binding sites for both of the miRNAs miR-29 and miR-125. Whereas the expression of miR-125 was increased in human sarcoma tissue, that of miR-29 was decreased in most samples. Overexpression of miR-125 decreased the abundance of A20 mRNA, whereas reconstituting miR-29 in sarcoma cell lines increased the abundance of A20 mRNA and protein. By interacting directly with the RNA binding protein HuR (human antigen R; also known as ELAVL1), miR-29 prevented HuR from binding to the A20 3′UTR and recruiting the RNA degradation complex RISC (RNA-induced silencing complex), suggesting that miR-29 can act as a decoy for HuR, thus protecting A20 transcripts. Decreased miR-29 and A20 abundance in sarcomas correlated with increased activity of NF-κB and decreased expression of genes associated with differentiation. Together, the findings reveal a unique role of miR-29 and suggest that its absence may contribute to sarcoma tumorigenesis.
Introduction
Classical proinflammatory signaling by the transcription factor NF-κB (nuclear factor κB) is inhibited by IκB (inhibitor of κB) family members, A20 [also known as TNFAIP3 (tumor necrosis factor–α–induced protein 3)], and the cylindromatosis protein encoded by CYLD1, all of which promote the retention of NF-κB in the cytoplasm (1). In response to TNFα, the kinase RIP1 (receptor-interacting serine/threonine-protein kinase 1) is activated through K63-linked ubiquitination, which activates the IκB kinase (IKK) complex that phosphorylates IκBα and IκBβ, causing their proteolysis and the subsequent nuclear translocation and activation of NF-κB (2). IκBα and A20 are immediate transcriptional targets of NF-κB that assist in the negative feedback control of the NF-κB signaling pathway. IκBα binds and promotes the nuclear export of NF-κB subunits (3). A20 removes K63-linked ubiquitin chains from RIP1 and catalyzes its K48-linked polyubiquitination through its deubiquitinase and E3 ubiquitin ligase activities, respectively, and thus promotes the proteasomal degradation of RIP1 (4). Failure to modulate this signaling circuitry leads to constitutive NF-κB activity, which is a common hallmark of numerous chronic diseases, including cancer (5). For example, deletion of A20 in mice abrogates homeostatic inhibition of NF-κB, resulting in systemic inflammation and severe cachexia (5), and mutations in A20 are associated with the constitutive activation of NF-κB in B cell lymphomas (6–8).
Previously, we showed that NF-κB is activated in rhabdomyosarcoma (RMS), a pediatric cancer with origins in skeletal muscle (9). RMS is representative of a subclass of soft tissue sarcomas that are rare but, when classified as a high-grade tumor, have a 40 to 60% incidence of metastasis and mortality (10). Increased NF-κB signaling in these tumors epigenetically suppresses the expression of the microRNA (miRNA) miR-29 through the Polycomb repressor complex PRC2, causing RMS cells to become less differentiated (9). Analogous to other tumor types (11), miR-29 may function as a tumor suppressor in RMS because exogenous addition of this miRNA reduces proliferation and enhances differentiation in RMS cells by targeting the myogenic inhibitor Yin Yang 1 (9).
MiRNAs suppress mRNA stability by directing the RNA-induced silencing complex (RISC) to the 3′ untranslated region (UTR) of its targeting mRNA (12). The stability of mRNAs can also be affected by RNA binding proteins (RBPs) that control posttranscriptional gene expression by binding AU-rich elements in the 3′UTR of target mRNAs (13, 14). HuR (human antigen R), which is ubiquitously expressed in mammalian cells, is an RBP that regulates the stability of transcripts encoded by genes involved in inflammation, cell growth, differentiation, and angiogenesis (15, 16). Under normal physiological conditions, HuR increases their half-life, whereas in tumors, HuR commonly switches functions to mediate their decay (17–19). Although not well studied, HuR is capable of mediating mRNA destabilization through the recruitment of the tumor suppressor miRNA let-7 (20), and HuR crosstalk with RISC and the Argonaute 2 (Ago2) subunit has also been described (21). Here, we identified a mechanism involving the interaction of miR-29 and HuR that stabilizes transcripts of A20 and inhibits NF-κB activity. We propose that the loss of this interaction contributes to sarcoma pathogenesis.
Results
Loss of A20 in sarcomas is associated with increased RIP1 abundance and NF-κB activation
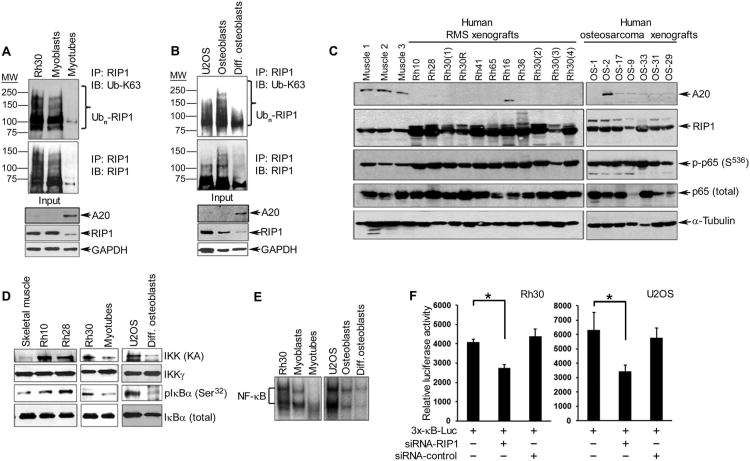
To gain insight into the mechanism underlying NF-κB activity in RMS, we examined the upstream mediators of this signaling pathway. In the human RMS cell line Rh30, the K63-linked polyubiquitination of RIP1 was greater compared with nontransformed immortalized murine C2C12 myoblasts and postmitotic differentiated myofibers (Fig. 1A). Similar results were observed with a second human RMS line, Rh28, as well as that of the murine alveolar RMS cell line 21459 (22) (fig. S1A). Additionally, the K63-linked ubiquitination of RIP1 was markedly increased in the osteosarcoma cell line U2OS and proliferating osteoblasts compared with differentiated bone cells (Fig. 1B), indicating that the observations were not unique to muscle-derived sarcomas. Because K63-linked ubiquitination of RIP1 is removed by A20 (23), we examined its correlation with the abundance of A20 in sarcoma cells compared with normal counterparts. Decreased abundance of A20 correlated with increased abundance and ubiquitination of RIP1 in RMS and U2OS cell lines (Fig. 1, A and B, and fig. S1A). This correlation was also observed in the majority of human RMS- or osteosarcoma-derived xenograft tumors derived from primary sarcomas (Fig. 1C). However, the abundance of RIP1 mRNA (fig. S1B) did not correlate with patterns of A20 abundance (Fig. 1A), which is consistent with previous findings that A20 mediates the proteolysis of RIP1, and suggests that an inverse relationship exists between RIP1 and A20 abundance in RMS and osteosarcoma. It is conceivable that this relationship is maintained in other sarcoma subtypes.
Fig. 1. RIP1 activates NF-κB in RMS and osteosarcoma in association with loss of A20.
(A) Immunoprecipitation (IP) for RIP1, followed by Western blotting (IB) for K63-linked ubiquitin (Ub-K63), RIP1, and A20 in lysates from human Rh30 cells and nontransformed mouse C2C12 muscle cells (undifferentiated myoblasts and differentiated myotubes). Blots were stripped and reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. (B) Same as in (A) except that reactions were performed with U2OS cell lines and human primary undifferentiated and differentiated osteoblasts. (C) Western blotting for A20, RIP1, phosphorylated p65 (Ser536), and total p65 (against α-tubulin as a loading control) in lysates from human RMS and osteosarcoma xenograft tumors compared with normal human skeletal muscle tissue. (D) Kinase assays followed by Western blotting to assess IKK activity in lysates from xenograft tumors or sarcoma cell lines, differentiated myotubes or osteoblasts, and murine skeletal muscle tissue used as control. (E) Electrophoretic mobility shift assay to measure the DNA binding activity of NF-κB in Rh30 and U2OS cell lines compared with their respective nontransformed control cell lines. (F) NF-κB reporter assays in Rh30 and U2OS sarcoma cell lines in the presence of either RIP1 small interfering RNA (siRNA) or a control siRNA. Data are means ± SD of relative luciferase activity, normalized to β-galactosidase to control for transfection efficiency, from a representative of three independent experiments; *P < 0.05, two-tailed Student′s t test. All blots (A to E) are representative of three independent experiments.
Because RIP1 is an upstream activator of IKK and NF-κB, we predicted that cells with increased RIP1 abundance would also exhibit greater NF-κB activity. In both RMS and osteosarcoma xenograft tumors, there were greater amounts of the transcriptionally active form of the p65 sub-unit of NF-κB (phosphorylated at Ser536) (24) in 17 of 18 tumors compared with normal human skeletal muscle tissue (Fig. 1C). Consistent with its role in activating NF-κB, the kinase activity of IKK (measured by kinase assays and phosphorylation of its substrate, IκBα) appeared to be increased in a subset of RMS tumors and Rh30 and U2OS cell lines compared with normal differentiated cells (Fig. 1D). In addition, the DNA binding activity of NF-κB was enhanced in both sarcoma cells and undifferentiated myoblasts and osteoblasts compared with that of normal or differentiated cell types (Fig. 1E). As expected, RIP1 knockdown in Rh30 and U2OS cells decreased the transcriptional activity of an NF-κB reporter (Fig. 1F), and its overexpression increased reporter activity (fig. S1C). However, the RIP1-induced increased in NF-κB reporter activity was partially inhibited by overexpression of A20 (fig. S1C). Together, the results suggested that increased NF-κB activity in RMS and osteosarcoma tumor cells involved increased K63-linked ubiquitination of RIP1, resulting from the loss of A20.
The abundance of A20 is regulated by opposing functions of miR-29 and miR-125
To determine how a reduction in A20 abundance is promoted in sarcomas, we analyzed the steady-state abundance of A20 mRNA. In human RMS and osteosarcoma cell lines, the abundance of both A20 mRNA and protein was low compared to their abundance in the control (fig. S2). To determine whether this regulation on A20 could be extended to additional sarcoma subtypes, we measured the abundance of A20 mRNA in a panel of 40 patient biopsies consisting of five histologically confirmed sarcoma subtypes: 4 pleomorphic RMS, 8 osteosarcoma, 12 myxoid liposarcoma, 8 well-differentiated liposarcoma, and 8 synovial sarcoma. We identified neoplastic (tumor) and morphologically “normal” nontumor areas by hematoxylin and eosin (H&E) staining and obtained samples for quantitative real-time polymerase chain reaction (qRT-PCR), thereby enabling each patient to be used as their own internal control (Fig. 2A). In 34 of 40 cases (88%), including all osteosarcoma and myxoid samples tested the abundance of A20 mRNA was reduced in the tumor tissue compared with that in the adjacent normal tissue (Fig. 2B), suggesting that the abundance of A20 mRNA and potentially the protein is reduced in multiple sarcoma subtypes.
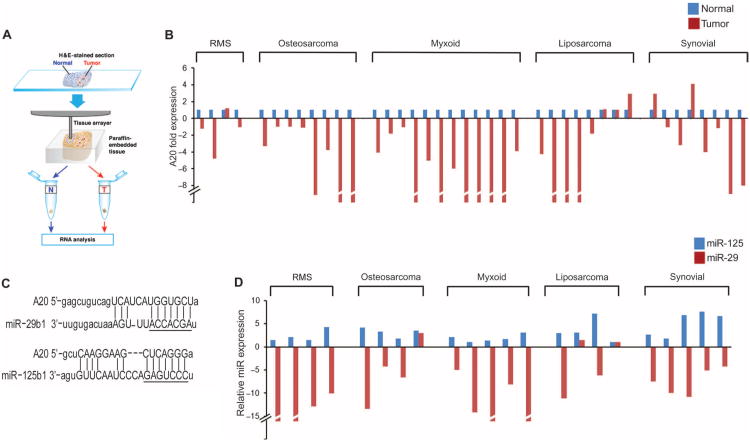
Fig. 2. Abundance of A20 and miR-29 is decreased in human sarcomas and is inversely correlated with that of miR-125.
(A) Schematic of the extraction of tumor and adjacent normal tissue from paraffin-embedded blocks. (B) qRT-PCR for A20 (normalized to GAPDH) in adjacent normal and tumor tissue sections of sarcoma biopsies. Data are representative of three technical repeats per sample; P = 6.055 × 10−5, two-tailed parametric t test. (C) TargetScan/MicroRNA prediction analysis for complementary sequences to the 3′UTR of A20. The seed regions of miR-29 and miR-125 sequences are underlined. (D) qRT-PCR for miR-29 and miR-125 in patient tumor material as in (B).
Reduced A20 mRNA can result from mutations in the A20 gene, including exons 2 and 5, such as those found in large B cell lymphomas (6—8); however, we did not find similar mutations in these exons or in the UTRs of the gene in six confirmed sarcoma cases compared with the A20 gene sequence in two commercially sourced normal human DNA samples (fig. S3). This implied that the reduction of A20 in sarcomas is not mediated by DNA mutations; thus, we examined whether the reduction in A20 mRNA was associated with promoter silencing. However, Rh30 and U2OS cells transfected with an A20 promoter luciferase reporter plasmid paradoxically had increased basal A20 promoter activity compared to that in nontransformed myoblasts (fig. S4A). Furthermore, A20 promoter activity was stimulated by TNFα (fig. S4B), which indirectly induces the expression of A20 by activating NF-κB (25). Collectively, these data indicated that the reduction in A20 mRNA abundance in sarcomas was not mediated at the transcriptional level.
To search for factors that may be involved in the posttranscriptional control of A20, we considered the role of miRNAs. TargetScan and miRanda algorithms, which search for RNA targets that contain sites complementary to the seed sequence of a particular miRNA, found sites in the 3′UTR of A20 that are highly conserved for the oncogenic miRNA miR-125b1 (26) as well as the tumor suppressor miRNA miR-29b1 (11) (Fig. 2C and fig. S5A). We assessed the abundance of miR-29b1 and miR-125b1 (herein called miR-29 and miR-125) in a similar panel of sarcoma cases that showed reduced abundance of A20 mRNA. Consistent with earlier observations in RMS (9), the amount of miR-29 was reduced in all four RMS tissue samples that were tested, in addition to other sarcoma subtypes (19 of 22 total cases; Fig. 2D). Also consistent with observations in other cancers (27, 28), the abundance of miR-125 was consistently increased in the sarcoma samples tested (22 of 22 cases; Fig. 2D), supporting the respective tumor-suppressing and tumor-promoting roles for these two miRNAs. Pearson association analysis of the abundance of miR-29 or miR-125 with A20 mRNA revealed a positive correlation between A20 and miR-29 and a negative correlation between A20 and miR-125, although the latter was not statistically significant (fig. S5, B and C).
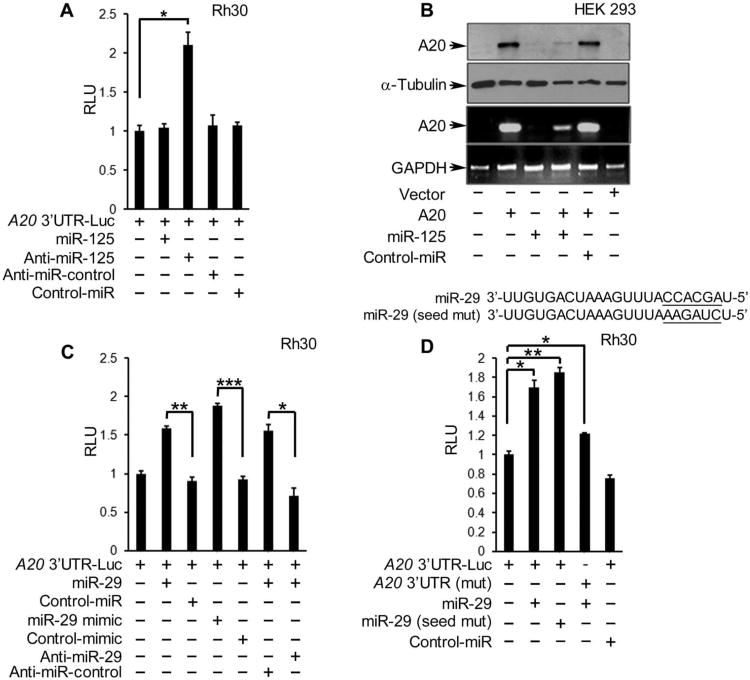
To assess the functional roles of these miRNAs in relation to the abundance of the A20 transcript, we cotransfected Rh30 and U2OS cells with an A20 3′UTR luciferase reporter and RNA oligonucleotides that either increased or silenced the expression of miR-29 and miR-125. Whereas overexpression of miR-125 had no affect on A20 reporter activity, an antagomir (anti-sense sequence) of miR-125 increased its activity (Fig. 3A). The lack of effect observed with overexpression of miR-125 may reflect saturating amounts of this miRNA in sarcoma cells. In human embryonic kidney (HEK) 293 cells, which had a basal abundance of miR-125 that is lower than that in Rh30 cells (fig. S5D), A20 3′UTR reporter activity was significantly decreased by addition of miR-125 (fig. S5E), and the abundance of both A20 protein and mRNA (exogenously expressed) was reduced compared with that in cells transfected with control miRNA (Fig. 3B). These findings are consistent with a report showing that the abundance of A20 is suppressed by miR-125 (29).
Fig. 3. A20 abundance and reporter activity is regulated by miR-29 and miR-125.
(A) A20 3′UTR luciferase reporter activity in Rh30 cells cotransfected with the indicated miRNAs, normalized to that in nontransfected cells and those transfected with the reporter alone. RLU, relative light units. (B) Western blotting and qRT-PCR in HEK 293 cells cotransfected with an A20 expression plasmid and miR-125 or a control (control-miR). Blots are representative of three experiments. (C and D)A20 3′UTR luciferase reporter assays in Rh30 cells cotransfected with (C) a wild-type (WT) UTR reporter construct and the indicated miRNAs or (D) a reporter containing WT or mutated miR-29 binding site [A20 3′UTR (mut)] and either WT or seed sequence-mutated miR-29. Graphs in (A), (C), and (D) are means ± SD from a representative of at least three independent experiments. *P < 0.05, **P < 0.01, and ***P< 0.005, two-tailed Student's t test.
However, when A20 3′UTR reporter assays were performed with exogenously expressed mature miR-29 or the precursor miR-29 mimic, we obtained a surprising result. In contrast to most miRNAs that function by suppressing the activity of the 3′UTR, the addition of mature or mimic forms of miR-29 significantly increased the 3′UTR activity of A20 in Rh30 and U2OS cells, and cotreatment with an antagomir to miR-29 inhibited this increase in the same cells (Fig. 3C and fig. S5F). In addition, mutating either the miR-29 binding sites in the A20 3′UTR or the seed sequence in miR-29 also increased the activity of the A20 3′UTR reporter in Rh30 (Fig. 3D) and U2OS cells (fig. S5G), suggesting that miR-29 was not binding directly to the A20 transcript to affect its stability. However, this regulation of A20 mRNA by miR-29 was specific to the 3′UTR of A20 because experiments in Rh30 cells with an equivalent reporter containing the A20 promoter, instead of the 3′UTR element, showed no change in activity when cotransfected with miR-29, miR-125, or a control miRNA sequence (fig. S5H). Together, these data indicated that miR-29 maintains the stability of A20 at the posttranscriptional level through a mechanism involving the A20 3′UTR, but that this mechanism is indirect and independent of the seed sequence of miR-29.
Expression of miR-29 increases A20 abundance and inhibits RIP1-mediated activation of NF-κB
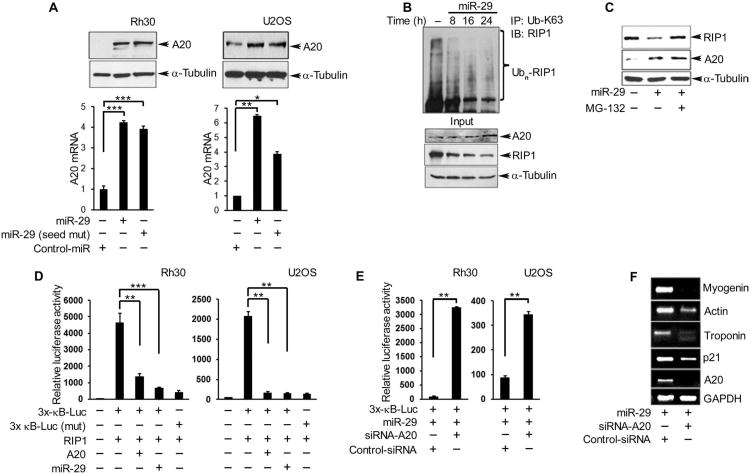
Because increased expression of a tumor suppressor gene by a tumor suppressor miRNA is an event not commonly described (30), we pursued the biological implication of this intriguing observation. Exogenous expression of miR-29 in sarcoma cells increased the abundance of both A20 mRNA and protein in Rh30 and U2OS cells, and the increases were independent of the miR-29 seed sequence (Fig. 4A). By 24 hours after transfection with miR-29 in both human Rh30 cells and mouse C2C12 myoblasts, RIP1 abundance and ubiquitination was decreased when A20 was abundant (Fig. 4B and fig. S6, A and B). The miR-29–induced decrease in RIP1 abundance in Rh30 cells was blocked by treatment of the cells with the proteasome inhibitor MG-132 (N-carbobenzyloxy-L-leucyl-L-leucyl-L-leucinal) (Fig. 4C). Neither a reduction in RIP1 ubiquitination and abundance nor an increase in A20 abundance was observed when cells were transfected with a control miRNA (fig. S6C). The inverse correlation between A20 and RIP1 abundance and ubiquitination may reflect A20-mediated RIP1 proteolysis, suggesting that miR-29 is capable of rescuing a form of A20 that is enzymatically active.
Fig. 4. MiR-29 rescues A20 abundance and promotes a differentiated phenotype in RMS tumor cells.
(A) Western blotting and qRT-PCR for A20 protein and mRNA, respectively, in Rh30 and U2OS cells 24 hours after transfection with miR-29, miR-29 (seed mut), or control-miR. (B) Immunoprecipitation and Western blotting for K36-linked ubiquitinated RIP1 in Rh30 cells performed at the indicated times after transfection with miR-29. (C) Western blotting for A20 and RIP1 in Rh30 cells transfected with miR-29 treated with or without MG-132 for 6 hours. (D) NF-κB promoter reporter luciferase assays in Rh30 and U2OS cells cotransfected with different combinations of RIP1, A20, and miR-29. (E) NF-κB reporter assays in Rh30 cells cotransfected with miR-29 and A20 or control siRNA. (F) RT-PCR for skeletal muscle differentiation markers in Rh30 cells cotransfected with miR-29 and A20 or control siRNA. Blots are representative of a minimum of two repeats, and data are means ± SEM from three independent experiments; *P < 0.05, **P < 0.01, and ***P < 0.005.
We examined whether the rescue of A20 stability by miR-29 inhibited the activation of NF-κB in sarcoma cells. Ectopic expression of RIP1 in Rh30 and U2OS cells significantly enhanced the activity of a wild-type, but not a mutated, NF-κB reporter (Fig. 4D). This activity was inhibited by overexpression of either miR-29 or A20 (Fig. 4D). However, the activity of the NF-κB reporter was not inhibited when miR-29 was overexpressed in the presence of an siRNA targeting A20 (Fig. 4E). These findings support the hypothesis that miR-29 promotes A20-mediated inhibition of RIP1 and NF-κB.
We assessed if the miR-29–mediated enhancement of A20 abundance affected the differentiation of sarcoma cells. Consistent with our previous findings (9), exogenous addition of miR-29 in Rh30 cells induced the expression of the gene that encodes the cyclin-dependent kinase inhibitor p21 and other genes that encode markers of myogenic terminal differentiation (fig. S6D). However, the miR-29–mediated induction of myogenic differentiation markers depended on A20 because exogenous expression of miR-29 in the presence of siRNA-mediated A20 knockdown was less effective at inducing myogenic genes in Rh30 cells (Fig. 4F).
NF-κB activity is reduced in differentiated skeletal muscle or myotubes (31), and this may be mediated by an increase in A20. Consistent with this possibility, when we transfected C2C12 myoblasts with an A20 3′UTR luciferase reporter in the absence or presence of an antagomir against miR-29 and allowed the cells to differentiate, the basal activity of the A20 reporter in the differentiated myotubes was significantly reduced when miR-29 was antagonized (fig. S6E). Thus, similar to RMS, the regulation of NF-κB in normal muscle differentiation may depend on the abundance of A20 under the control of miR-29.
MiR-29 stabilizes A20 by acting as an RNA decoy for HuR
Having identified a functional connection between A20 transcript stability and miR-29, we investigated the mechanism by which miR-29 increased A20 transcript and protein abundance in sarcoma cells. Because the miR-29 binding site in the 3′UTR of A20 was not strictly required to increase A20 reporter expression (Fig. 3D and fig. S5G), we postulated that miR-29 might protect A20 by binding a destabilizing factor (Fig. 5A). This model predicts that in the absence of miR-29, A20 mRNA is susceptible to decay, thereby leading to reduced A20 synthesis and the subsequent activation of RIP1 and NF-κB. However, in the presence of miR-29, sequences from this miRNA would mimic the 3′UTR of A20, thereby serving as an RNA decoy binding the destabilizing factor and protecting A20 mRNA from degradation.
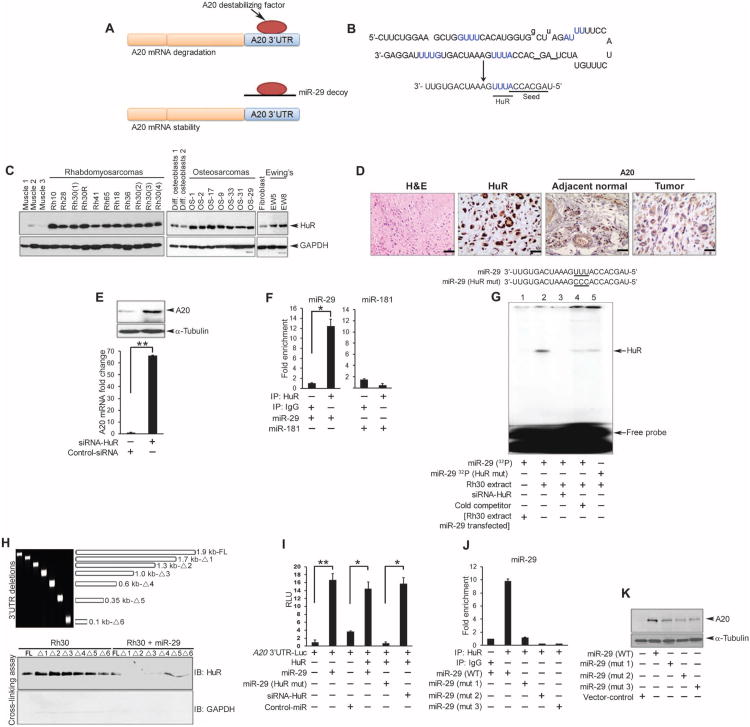
Fig. 5. MiR-29 functions as a decoy for HuR.
(A) Model of miR-29 as a decoy against an RNA-destabilizing factor that contributes to A20 mRNA turnover. (B) Sequences of precursor and mature miR-29. Predicted HuR binding sites are in blue. The seed sequence and HuR binding site are underlined in mature miR-29. (C) Western blot for HuR in human xenograft sarcoma tumors compared with control murine muscle tissue, human differentiated osteoblasts, or murine fibroblasts. (D) H&E staining and immunohistochemistry for A20 and HuR in formalin-fixed paraffin-embedded osteosarcoma. Images are representative of four samples, stained in duplicate; scale bars, 4 μm. (E) Western blotting and qRT-PCR for A20 in lysates from Rh30 cells transfected with control or HuR siRNA. (F) Immunoprecipitation for HuR or IgG followed by qRT-PCR for miR-29 or miR-181 in Rh30 cell lysates. (G) RMSA with a 32P-labeled miR-29 probe and extracts from Rh30 cells transfected with HuR (lanes 2 to 5), miR-29 (lane 1), HuR siRNA (lane 3), unlabeled miR-29 (lane 4, cold competitor), or miR-29 containing a mutated HuR binding site (lane 5, HuR mut). (H) Top: PCR fragments were amplified from the full-length (FL) A20 3′UTR or deletion constructs (Δ1 to Δ6) (table S1). Bottom: Western blotting for HuR in ultraviolet–cross-linked lysates from Rh30 cells transfected with or without miR-29 and the indicated fragment. Data are representative of two experiments. (I) A20 3′UTR luciferase reporter activity in Rh30 cells cotransfected as indicated. (J) Immunoprecipitation for HuR and qRT-PCR for miR-29 in lysates from U2OS cells transfected with plasmids expressing WT or mutated (mut 1 to mut 3, fig. S9A) precursor miR-29. (K) Western blotting for A20 in U2OS lysates transfected with WT or mutated m i R - 2 9. Blots are representative of at least two independent repeats, and data are means ± SEM from three independent experiments; *P < 0.05, **P < 0.01.
To test this model, we screened the RNA-Binding Protein Database (RBPDB) for RBPs that have consensus binding sites matching the precursor sequence in miR-29. This screen identified ELAVL1 [ELAV (embryonic lethal, abnormal vision, Drosophila)–like 1; also known as HuR] as the top candidate (fig. S7A). Sequence alignment showed that HuR recognized the consensus sites 5′-GUUU-3′ and 5′-AUUU-3′ within pre-miR-29, but only the 5′-AUUU-3′ site that is next to the seed sequence is present in mature miR-29 (Fig. 5B). Therefore, we investigated the involvement of HuR in sarcomas. The abundance of HuR was increased in subcutaneous xenograft sarcoma tumors (RMS, osteosarcoma, and Ewing's sarcoma) compared with normal control samples (Fig. 5C). Additionally immunohistochemical staining of primary human osteosarcoma tumors confirmed that HuR abundance was observed in neoplastic cells (Fig. 5D). In contrast, compared with the abundance of A20 in normal cells that were localized to the vasculature, A20 was undetectable in a section from the same tumor that was positive for HuR. To determine whether loss of A20 in tumor cells is mediated by HuR, we silenced HuR in Rh30 cells (fig. S7B). Compared with cells transfected with a nontargeted control siRNA, HuR knockdown increased the abundance of both A20 mRNA and protein (Fig. 5E). These findings suggested that HuR reduces the abundance of A20 in sarcomas.
To determine whether the A20 3′UTR was involved in HuR-mediated regulation of A20 abundance, we cotransfected Rh30 cells with HuR and an A20 3′UTR luciferase reporter plasmid and showed that A20 reporter activity was significantly decreased in cells overexpressing HuR (fig. S7C), supporting the model that HuR destabilizes A20 mRNA.
Our model (Fig. 5A) predicted that miR-29 protected A20 mRNA through a direct interaction between miR-29 and a destabilizing factor, which our findings suggested was HuR (fig. S7, A and C, and Fig. 5, B to E). We performed RNA immunoprecipitation of HuR [or immunoglobulin G (IgG) as a control] from lysates of sarcoma cells transfected with miR-29 or a control miRNA (miR-181, selected because it has no sequences that match HuR), followed by qRT-PCR. RNAs immunoprecipitated with HuR were significantly enriched for miR-29 but not miR-181 (Fig. 5F). To validate these findings, we performed RNA electrophoretic mobility shift assays (RMSAs) with a radiolabeled miR-29 probe and Rh30 cytoplasmic extracts. We identified that HuR bound to miR-29 (Fig. 5G, lane 2), and its binding was diminished either in extracts from cells in which endogenous HuR was knocked down (Fig. 5G, lane 3) or in the presence of nonradioactive miR-29 to compete with the radioactive miR-29, or with extracts containing transfected miR-29 (Fig. 5G, lanes 1 and 4). Likewise, a miR-29 oligomer that contained a mutated HuR binding site (5′-AUUU-3′ to 5′-ACCC-3′; Fig. 5G, lane 5) also showed a markedly reduced interaction with HuR. Together, the data indicated that miR-29 interacted directly with HuR.
To assess whether miR-29 can act as a decoy to prevent HuR from binding to A20 mRNA, we took two approaches. We performed cross-linking assays in cytoplasmic extracts of Rh30 cells transfected with or without miR-29 in the presence of in vitro–transcribed truncated fragments of the full-length A20 3′UTR (Fig. 5H), which is about 1.9 kb long and contains multiple putative HuR consensus binding sites (fig. S8A). Binding of endogenous HuR was detected with each of the A20 3′UTR fragments that contained at least one HuR binding site (full length and fragments Δ1 to Δ5). However, the amount of bound HuR appeared to be decreased when cross-linking reactions were repeated with the smallest 3′UTR fragment (Δ6), which lacked any HuR binding sites (Fig. 5H). Overexpression of miR-29 inhibited the binding of HuR to the full-length A20 3′UTR or many of the fragments (Fig. 5H). We also inferred the stability of the A20 3′UTR by measuring the effect of transfected HuR, miR-29, or both on the activity of the A20 3′UTR reporter in Rh30 cells. Consistent with our proposed model (Fig. 5A), overexpression of miR-29 blocked the decrease in A20 reporter activity that was induced by HuR (fig. S8B). The increased A20 reporter activity in response to miR-29 was not blocked by HuR (Fig. 5I), suggesting that there was sufficient overexpressed miR-29 to bind both endogenous and the exogenously expressed HuR. HuR knockdown in the absence of miR-29 overexpression also stimulated A20 reporter activity (Fig. 5I). However, neither overexpression of miR-29 that contained a mutation within the HuR binding site (Fig. 5I) nor overexpression of miR-181 (fig. S8B) increased A20 reporter activity. Together, these results support the model that miR-29, when overexpressed in sarcomas, acts as a decoy miRNA to protect A20 transcripts from destabilization mediated by HuR, and this function may contribute to its tumor suppressor role.
Because the precursor sequence of miR-29 contains additional putative HuR binding sites (Fig. 5B and fig. S7A), we investigated whether such sites contributed to the decoy activity of miR-29. We transfected sarcoma cells with plasmids containing wild-type pre-miR-29 or one of the three pre-miR-29 mutants in which HuR binding sites within the precursor sequence of miR-29 were mutated (fig. S9A) and performed RNA immunoprecipitation reactions for HuR, followed by qRT-PCR that amplified regions corresponding to either the bottom strand (that becomes the mature form) or the upper complementary strand of miR-29. Amplification of the bottom strand revealed that HuR interacted with the wild-type miR-29 precursor, but this interaction was strongly reduced when a mutation was introduced into the AUUU binding site (mut 2) (Fig. 5J). Unexpectedly, we observed a similar reduction in HuR binding when amplifying the upper strand of precursor miR-29 containing a mutation in the GUUU binding site (mut 1). Similar reduction in binding was observed with mutations in both strands (mut 3). These results suggested that the precursor form of miR-29 is capable of binding HuR in cells. To validate this notion, we assessed the abundance of A20 protein from the same extracts in which RNA immunoprecipitation reactions for HuR were performed. Compared to the vector control, expression of wild-type precursor miR-29 increased the abundance of A20 in sarcoma cells, but none of the miR-29 HuR binding site mutants increased A20 to the same extent (Fig. 5K). Thus, these data implied that the decoy activity of miR-29 may be mediated by both its mature and precursor sequences when overexpressed.
MiR-29 prevents HuR interaction with Argonaute
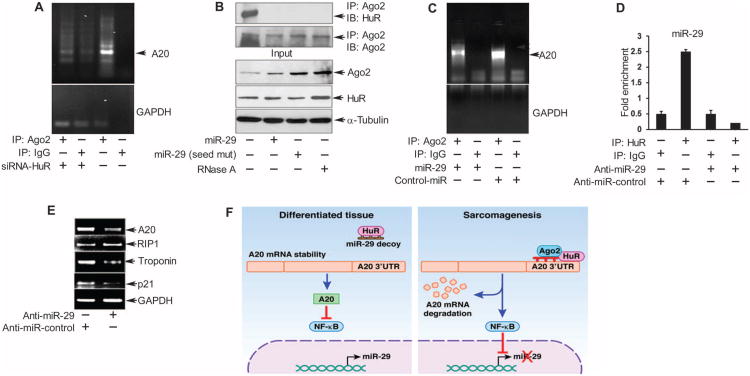
To promote RNA degradation, miRNAs and HuR interact with the RISC (20), which includes the protein Ago2 (12). Because both HuR (Fig. 5E) and miR-125 (Fig. 3B) reduced the abundance of A20 mRNA and protein, and silencing HuR (Fig. 4I) or antagonizing miR-125 (Fig. 3A) increased the activity of the A20 3′UTR reporter, suggesting that both of these may destabilize the A20 transcript, we performed RNA immunoprecipitation of Ago2 with Rh30 cells and found that miR-125 was enriched (fig. S9B). Knocking down HuR reduced the amount of miR-125 that immunoprecipitated with Ago2 (fig. S9B), suggesting that their association may be dependent on HuR. Thus, miR-125 may have a destabilizing effect on A20 transcripts, which would be consistent with this miRNA increasing in abundance in sarcoma (Fig. 2D); whether this effect requires HuR remains to be determined.
We also investigated whether RISC was part of the pathway through which miR-29 and HuR regulated the abundance of A20 transcripts. Results showed that in Rh30 cells, A20 mRNA immunoprecipitated with Ago2, but this interaction was decreased by silencing HuR (Fig. 6A). This suggested that the binding of HuR to the 3′UTR of A20 mRNA may be required to assemble Ago2-containing RISC onto A20 mRNA and mediate its degradation. Consistent with this notion and previous reports (21), endogenous HuR immunoprecipitated with Ago2, and this interaction was disrupted with ribonuclease A (RNase) (Fig. 6B), suggesting that an RNA product mediates or stabilizes their interaction. Because miR-29 functioned as a decoy for HuR, preventing its binding to A20 mRNA, we investigated whether miR-29 disrupted the interaction between HuR and Ago2-containing RISC and the loading of this complex onto A20 mRNA. Overexpression of mature miR-29 disrupted the interaction between HuR and Ago2 independent of the seed sequence in miR-29 (Fig. 6B), which was consistent with the ability of the miR-29 mutants to reduce A20 abundance (Fig. 5K). Furthermore, miR-29 also reduced the association of A20 mRNA with Ago2 (Fig. 6C), suggesting that miR-29 inhibited the interaction between HuR and Ago2 RISC that is necessary to degrade A20 mRNA.
Fig. 6. MiR-29 protects A20 by preventing a HuR-Ago2 complex.
(A to C) Immuno-precipitation for Ago2 in lysates from Rh30 cells transfected with (A) HuR siRNA, (B) WT or seed-mutant miR-29, or (C) miR-29 or control-miR, followed by RT-PCR for endogenous A20 mRNA (A and C) or Western blotting for HuR and Ago2. (D) Immunoprecipitation for HuR followed by qRT-PCR for miR-29 in lysates from C2C12 myoblasts transfected with anti–miR-29 or the indicated control and differentiated for 48 hours. (E) RT-PCR for A20, RIP1, p21, and troponin T2 fast against GAPDH in lysates of C2C12 myoblasts transfected as in (D) and 48 hours in differentiation medium. Data from gel and bar graphs in (A) to (E) are from a minimum of two and three independent experiments, respectively. *P < 0.05. (F) A model for how miR-29 protects A20 transcript stability and inhibits NF-κB activity in differentiated tissue compared with its absence in sarcoma tumors. Loss of A20 abundance perpetuates a feed-forward loop whereby increased NF-κB activity further silences miR-29, leading to a less differentiated state.
To determine whether the miR-29 decoy activity was important in a physiological context, we tested the effect of miR-29 inhibition on differentiation of C2C12 myoblasts because the abundance of miR-29 is increased during skeletal muscle differentiation in myogenesis (9). Inhibiting miR-29 with an antagomir in differentiated muscle cells reduced its interaction with HuR (Fig. 6D), as expected, and decreased the abundance of A20 mRNA and several differentiation markers (Fig. 6E). However, miR-29 depletion did not affect the abundance of RIP1 mRNA, which is consistent with A20 mediating a decrease in the abundance of RIP1 at the post-translational level (fig. S1). The results suggested that in skeletal muscle, miR-29 acted as a decoy for HuR, thereby increasing the abundance of A20 and promoting a differentiated phenotype.
Discussion
Studies investigating the regulation of A20 have mainly focused on the transcriptional activation of A20 by NF-κB in response to cytokine signaling (23, 32). Although mutations in A20 are present in large B cell lymphoma, we found no mutations in the A20 gene in sarcoma cells despite its reduced abundance. Our findings reveal, at least in sarcomas, a novel mechanism explaining A20 mRNA decay involving the ability of HuR to interact with an Ago2 complex in the absence of miR-29. As illustrated in Fig. 6F, our results support that in normally differentiated tissue, miR-29 is basally expressed as a result of A20-mediated inhibition of NF-κB. The expression of miR-29 acts as a decoy to inhibit HuR from binding to the 3′UTR of A20, thereby maintaining A20 mRNA stability and NF-κB repression, which in turn is needed to promote differentiation of skeletal muscle and potentially other mesenchymal cells. Evidence from our mutational analysis indicates that the decoy activity of miR-29 may include its precursor sequence, which contains several functional HuR binding sites. However, as we previously described (9), when NF-κB becomes activated during tumor initiation, miR-29 transcription is decreased through an epigenetic mechanism mediated by NF-κB. This allows HuR to bind A20 transcripts and, in association with Ago2 and perhaps miR-125 or others, degrade A20 mRNA. As a result, A20-mediated negative feedback on NF-κB is lost, enabling continued inhibition of miR-29 expression, thus perpetuating a regulatory circuit that may contribute to sarcomagenesis by maintaining tumor cells in a less differentiated state.
In addition, HuR is a protein often increased in human cancers (33), although the mechanism of this regulation has not been well described (16). Reports indicate that HuR can be directly regulated by NF-κB through functional binding sites within the HuR proximal promoter (34, 35). If this is the case, then constitutive NF-κB activation in sarcomas resulting from the loss of A20 is also likely to participate in the regulatory circuit described above, leading to increased expression of HuR.
Moreover, miR-29 is often repressed in human cancers and is thought to function as a tumor suppressor (11, 36). Until now, the tumor suppressor activity of miR-29, as well as that of most miRNAs described in the literature, has been ascribed to its seed sequence that directly binds to the 3′UTR sequence of tumor promoter mRNAs to limit their expression. In sarcomas, and possibly other malignancies not investigated here, we found that miR-29 binds to HuR in a seed sequence–independent manner, through a HuR binding motif present in mature miR-29. Although A20 mRNA contains miR-29 binding sites in its 3′UTR, A20 mRNA stability does not seem to be regulated directly by miR-29 because mutating these sites or mutating the seed sequence within mature miR-29 had no effect on the activity of an A20 reporter construct. Rather, we showed that by binding to HuR, miR-29 (at least by overexpression) prevents the interaction between HuR, Ago2, and A20 mRNA to shift the equilibrium away from A20 degradation toward its stabilization. As a result, activation of NF-κB is decreased through A20-mediated RIP1 proteolysis.
Although not entirely unique to miR-29, the ability of miRNAs to associate with proteins to mediate gene expression has only been rarely described (20, 37). In one example, miR-369 was found to direct the association of Ago2 and fragile × mental retardation–related protein 1 to the AU-rich elements of the TNFα mRNA to promote TNFα abundance (38). In a second example, Eiring and colleagues (39) described a similar decoy function of miR-328 against the heterogeneous nuclear ribonucleoprotein E2 to alleviate suppression of C/EBPα abundance in the blastic phase of chronic myeloid leukemia. However, even in this case, the decoy activity of miR-328 was selective to a 5′UTR C-rich intercistronic negative regulatory element within CEBPA mRNA, which is distinct from how we propose miR-29 protects A20 at its 3′UTR HuR binding elements. It is conceivable that the antagonizing activity of miR-29 against HuR may positively regulate the stability of AU-rich mRNAs, which encode other tumor suppressors that, similar to A20, are also less abundant in cancers. Many of the known AU-rich mRNAs encode proteins that are also associated with NF-κB regulation and have been shown to play an important role in different cancers and inflammation disorders (40, 41). Whether HuR and miR-29 engage in a similar interaction to regulate stability of these mRNAs in other cancer types or during inflammation as they appear to in sarcoma—and what role, if any, miR-125 or others might have in this mechanism—remains to be determined.
Materials and Methods
Tumor samples
Studies involving human samples were obtained after approval by the Institutional Review Board at Ohio State University (OSU), Department of Pathology (protocol number 2002H0089). All cases of the different sarcoma histologic subtypes used in this study were revalidated by an expert soft tissue and bone pathologist. Briefly, the neoplastic areas diagnostic of the sarcoma entity and the morphologically normal-appearing nontumoral areas were identified and marked on standard H&E-stained sections. These designated areas were subsequently matched to the tumor blocks, and with a tissue arrayer, 1.75-mm cores of neoplastic and normal-appearing areas were separately extracted from the paraffin blocks and put in corresponding separate sterile Eppendorf tubes as matched tumor and normal control specimens per case (as illustrated in Fig. 2A).
Reporter assays, site-directed mutagenesis, and DNA sequencing
A human A20 promoter (1.039 kb) was generated in the pLightSwitch_Prom reporter construct, whereas a human A20 3′UTR (2.164 kb) (NM_006290) was generated in a pLightSwitch_3UTR reporter plasmid, obtained from Switch Gear Genomics. A20 3′UTR reporter assays were performed in 24-well tissue culture plates. Cells were transfected with reporter constructs, siRNAs, or miRNAs with Lipofectamine 2000 reagent (Invitrogen). Luciferase assays were generally performed 24 hours after transfection, with the exception of C2C12 myoblasts, which were allowed to differentiate for 72 hours to myotubes before performing the assay. The amounts of transfected plasmids and miRNAs were kept proportionally equal by introducing a nonspecific miRNA or an empty vector. Luciferase assay reagents were obtained from Switch Gear Genomics. Plates were assayed for reporter activity with a Veritas Microplate Luminometer (Turner Biosystems).
Mutations in predicted miR-29 and miR-125 binding sites within the A20 3′UTR were produced with a second generation QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). Primer pairs used to generate these mutations were as follows (mutated nucleotides are indicated in uppercase letters): miR-125, forward: 5′-gcaagaagctcaaggaagAtTaAg-Taaaatggacgtattcag-3′, reverse: 5′-ctgaatacgtccattttAcTtAaTcttccttgag-cttcttgc-3′; and miR-29, forward: 5′-gctgtcagtcatcaGggCgAtatcctctgagc-3′, reverse: 5′-gctcagaggataTcGccGtgatgactgacagc-3′. Mutations were confirmed by DNA sequencing.
Somatic mutational analysis of exons 2 and 5, as well as the 5′UTR (accession no. BR756443) and 3′UTR (NM_006290) of the A20 gene were analyzed in sarcoma cases by direct Sanger sequencing. Normal human healthy genomic DNA (11691112001, Roche) was used as a control. Exons and UTRs were amplified with the REDTaq Genomic DNA Polymerase (Sigma-Aldrich) and the primers listed in table S2. After PCR amplification, samples were treated with ExoSAP-IT (Affymetrix). DNA sequencing was performed at the OSU Comprehensive Cancer Center Sequencing Core, and DNASTAR software was used for subsequent analysis. Deletion fragments of the 3′UTR were generated with the primers listed in table S1.
Bioinformatics and RNA reagents
MiRNA target prediction for A20 was performed with publicly available algorithms: TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/). MiRbase (http://www.mirbase.org/) was used for miR-29 sequences and annotation. Predictions for the RBPs were performed with the RBPDB database (http://rbpdb.ccbr.utoronto.ca/) (42). Synthesized miR-29 HuR binding site mutant (mutation sites are in uppercase): 5′-uag-caccaCCCgaaaucaguguu-3′ and miR-29 seed sequence mutant: 5′-uCUA-GAAauuugaaaucaguguu-3′ were acquired from Dharmacon Inc. MiR-29, miR-29 mimic, anti–miR-29, miR-125, anti–miR-125, miR-181, control-miR, and anti–miR-control were acquired from Life Technologies. miRIDIAN microRNA Mimic Control, which is based on cel-miR-67, was obtained from Thermo Scientific. GenScript's Custom Gene Service was used to determine functional HuR binding sites within precursor miR-29. Precursor miR-29 sequences were generated into a pRNAT-CMV3.1/hygro plasmid (table S3) and transfected into cells, which were subsequently lysed and subjected to RIP analysis with an antibody against HuR. Two custom-synthesized primer sets (Exiqon) were used to specifically amplify the region of interest from precursor miR-29 (upper strand and lower strand corresponding to the mature miR sequence). Amplification of miR-29 sequences was performed by qRT-PCR as recommended by the manufacturer. Knockdown of RIP1, A20, and HuR was performed with siRNA oligonucleotides obtained from Santa Cruz Biotechnology. Each siRNA consisted of a pool of three target-specific 19- to 25-nucleotide siRNAs designed for efficient knockdown of their respective targets.
qRT-PCR analysis
Total RNA was prepared from sarcoma cell lines, xenograft tumors, and normal tissues with standard TRIzol solution (Invitrogen). Complementary DNA was synthesized with Moloney murine leukemia virus reverse transcriptase (Invitrogen). PCR products were separated on an agarose gel and visualized by ethidium bromide staining by a gel documentation system (UltraLum Inc.).
Specifically for paraffin-embedded tissues, RNA was extracted with the Ambion RecoverAll Total Nucleic Acid Isolation Kit, as recommended by the manufacturer. The following primers were used for A20 qPCR: A20-1, forward: 5′-accccattgttctcggctat-3′, reverse: 5′-cggtctctgttaacaagtg-gaa-3′; A20-2, forward: 5′-tggcacaactcatctcatcaa-3′, reverse: 5′-gccatttcttg-tactcatgctg-3′. Human miR-125, miR-29, and RNU24 miRNA, used as an endogenous control, were obtained from Life Technologies. Reverse transcription reactions for miRNA expression were performed with a TaqMan McroRNA Reverse Transcription Kit, followed by qRT-PCR with a TaqMan Universal PCR Master Mix (Life Technologies). qPCR amplification reactions for A20 and RIP1 expression were performed with iTaq SYBR Green Supermix with ROX (Bio-Rad Laboratories). For both miRNA and gene amplifications, analysis was performed with the Applied Biosystems StepOnePlus Real-Time PCR system and preinstalled StepOne software. Unless mentioned, fold changes were calculated from the comparative ct (ΔΔct) method: ΔΔct = ΔctTargettumor − ΔctCalibratoradjacent normal. Semiquantitative PCR was performed with human A20 primers: forward, 5′-aactccaagccgggccctga-3′; reverse, 5′-aggcttggcacttcccggga-3′. Primers for human genes, p21, actin, myosin, troponin, and GAPDH, were previously described (9). Primers for mouse A20 and RIP1 are listed in table S4.
RIP and RMSAs
RNA immunoprecipitations were performed with a total of 1 mg of protein. All reagents were prepared in diethyl pyrocarbonate–treated water. After being washed with NT2 buffer, protein A–agarose was precoated with 15 μg of antibody (HuR, Ago2, or IgG) for 1 hour. Cells were lysed in NT2 buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40, RNase, proteinase, and phosphatase inhibitors. Beads and protein lysates were then incubated for 1 hour at 4°C. A thorough wash of the beads was performed in NT2 wash buffer [50 mM tris-HCl (pH 7.4), 50 mM NaCl, 1 mM MgCl2, 1.5 mM dithiothreitol, and 0.05% NP-40]. This was followed by deoxyribonuclease (DNase) treatment (TURBO DNase, Ambion) for 15 min at 30°C After DNase treatment, a quick wash was performed in NT2 wash buffer. Beads were then suspended in 100 μl of NT2 buffer containing 0.1% SDS and proteinase K (0.5 mg/ml) (15 min at 55°C). Phenol–chloroform–isoamyl alcohol mixture (100 μl) was added to the beads, and the suspension was mixed by vortexing and centrifuged for 1 min to separate phases. After the recovery of the upper phase, a mixture containing 10 μl of yeast transfer RNA (1 mg/ml) or glycogen (20 mg/ml; Roche), 12 μl of 3 M sodium acetate, and 250 μl of ethanol was added per 100 μl of collected aqueous phase, and the solution was mixed. This was followed by an ethanol precipitation step at −20°C overnight. After centrifugation, 5 to 10 ng of recovered RNA were used for each miRNA-specific reverse transcription reaction.
For RMSAs, 32P-labeled miR-29 wild-type (5′-uagcaccauuugaaauca-gug-3′) and mutant (5′-uagcaccaCCCgaaaucagug-3′) probes were generated (mutated sites are shown as capitalized letters). RNA oligonucleotides were acquired from Integrated DNA Technology, and labeling was performed with T4 polynucleotide kinase (Thermo Scientific). Radiolabeled probes, at 105 cpm, were incubated with 10 μg of cytoplasmic extract prepared from Rh30 cells either untransfected or transfected with miR-29, and incubations proceeded for 30 min at room temperature. For competition assays, 1000× molar excess of single-stranded oligonucleotides was added to the reaction. Samples were separated by 5% native polyacrylamide gel electrophoresis in 0.5% tris-glycine-EDTA buffer and subjected to autoradiography.
Immunohistochemistry and antibodies
For immunohistochemistry, representative sections of formalin-fixed tissues from human osteogenic sarcomas were cut at 4 μm and placed on positively charged slides. The slides were placed in a 60°C oven for 1 hour, cooled, deparaffinized, and rehydrated through xylene and graded ethanol solutions. Endogenous peroxidase was blocked by incubating the slides in 3% hydrogen peroxide aqueous solution. Endogenous biotin was also blocked with an avidin/biotin blocking kit (Dako). Antigen retrieval was performed by heat-induced epitope retrieval, in which the slides were placed in a 1× Target Retrieval Solution (Dako; pH 6) for 25 min at 96°C with a vegetable steamer (Black and Decker) and cooled for 15 min. Sections were then incubated with the primary antibodies for 60 min. For HuR, the Dako LSAB+ Detection System was used following the manufacturer's instructions. For A20, a biotinylated goat anti-mouse secondary antibody (Vector) was incubated for 30 min, followed by 30 min of incubation with Vectastain Elite (Vector). The chromogen used was 3,3′-diaminobenzidine (DAB+, Dako). A negative control consisted of omitting the primary antibody.
Ubiquitin Lys63 antibody was acquired from Millipore; RIP1 was from BD Biosciences; A20 was from Imgenex; and p65, phospho-p65 (Ser536), phospho-IκBα (Ser32), Ago2, HuR, GAPDH, A20, IKKγ, and IκBα were from Santa Cruz Biotechnology. Antibody against HuR (clone 4G8, for immunohistochemistry) was from Lifespan Biosciences, and antibodies against α-tubulin and IgG were from Sigma.
Statistical analysis
Data are presented as averages ± SEM. Student's t test was applied to perform pairwise comparisons between tumor and normal samples. Association between miRNA and A20 expression in tumors was analyzed by Pearson's correlation test. All the expression data were log-transformed before the analysis. Statistical analyses were performed with R software version 2.15.1 (http://www.r-project.org/). P values <0.05 were considered statistically significant.
Supplementary Material
www.sciencesignaling.org/cgi/content/full/6/286/ra63/DC1
Fig. S1. RIP1 activation and loss of A20 are associated with sarcoma cell lines.
Fig. S2. Abundance of A20 mRNA and protein is reduced in sarcomas.
Fig. S3. A20 is not frequently mutated in sarcomas.
Fig. S4. A20 regulation in sarcoma cells is not mediated at the transcriptional level.
Fig. S5. A20 is regulated by miR-29 and miR-125.
Fig. S6. Endogenous regulation of A20 and RIP1 by miR-29 promotes a differentiated phenotype in Rh30 cells.
Fig. S7. HuR inhibits the activity of the A20 3′UTR reporter.
Fig. S8. Putative HuR binding sites are present in the A20 3′UTR.
Fig. S9. Precursor miR-29 contains multiple HuR binding sites, and miR-125 association with Ago2 is dependent on HuR in sarcoma cells.
Table S1. Primers for the A20 3′UTR.
Table S2. Primers used to sequence A20 exons and UTRs.
Table S3. pRNAT-CMV3.1/hygro plasmids expressing wild-type or mutated miR-29 precursor sequences.
Table S4. Primers used for RT-PCR analysis.
Acknowledgments
We thank A. Ting (Mount Sinai Medical Center) for providing the RIP1 expression plasmid. We also acknowledge V. Shettigar, P. Gaspaprini, J. Shintaku, W. He, J. Peterson, J. Tomsic, J. M. Gu, and S. Liyanarachchi for their technical assistance and T. Vojt for his assistance with graphic illustrations.
Funding: This work was supported by OSU Comprehensive Cancer Center, “Up on the Roof” and “Pelotonia” fellowships awarded to M.Y.B., and NIH support R01CA163800 to D.P. and CA R01CA143082 to C.K. and D.C.G.
Footnotes
Author contributions: M.Y.B. performed the experiments; O.H.I., N.B., K.J.L., and D.C.G. contributed in the experimentation; M.Y.B., O.H.I., P.J.H., C.A.L., D.P., C.M.C., C.K., and D.C.G. contributed to experimental design and data interpretation; O.H.I., D.S.C., P.J.H., C.A.L., and W.K. provided resources and samples considered vital to the study; and M.Y.B. and D.C.G. wrote the manuscript.
Competing interests: C.K. received honoraria for presentations from Novartis, GlaxoSmithKline, and Takeda.
References
- 1.Didonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T. Catalysis by a multiprotein IκB kinase complex. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 3.Huang TT, Kudo N, Yoshida M, Miyamoto S. A nuclear export signal in the N-terminal regulatory domain of lκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc Natl Acad Sci U S A. 2000;97:1014–1019. doi: 10.1073/pnas.97.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S, Mori S, Ishikawa Y, Okamoto K, Tobinai K, Nakagama H, Nakahata T, Yoshino T, Kobayashi Y, Ogawa S. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, Stanelle J, Siebert R, Küppers R. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien D, Jacob AG, Qualman SJ, Chandler DS. Advances in pediatric rhabdomyosarcoma characterization and disease model development. Histol Histopathol. 2012;27:13–22. doi: 10.14670/hh-27.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 12.Bouasker S, Simard MJ. Structural biology: Tracing Argonaute binding. Nature. 2009;461:743–744. doi: 10.1038/461743a. [DOI] [PubMed] [Google Scholar]
- 13.Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 14.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 16.Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest. 2010;40:370–374. doi: 10.1111/j.1365-2362.2010.02279.x. [DOI] [PubMed] [Google Scholar]
- 17.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: Cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 24.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Jäättelä M, Mouritzen H, Elling F, Bastholm L. A20 zinc finger protein inhibits TNF and IL-1 signaling. J Immunol. 1996;156:1166–1173. [PubMed] [Google Scholar]
- 26.Klusmann JH, Li Z, Böhmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O'Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci U S A. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci U S A. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarma V, Lin Z, Clark L, Rust BM, Tewari M, Noelle RJ, Dixit VM. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. J Biol Chem. 1995;270:12343–12346. doi: 10.1074/jbc.270.21.12343. [DOI] [PubMed] [Google Scholar]
- 33.Shultz JC, Chalfant CE. The flip-flop HuR: Part of the problem or the solution in fighting cancer? J Clin Invest. 2011;122:16–19. doi: 10.1172/JCI61677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135:2030–2042.e3. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Rhee WJ, Ni CW, Zheng Z, Chang K, Jo H, Bao G. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2010;107:6858–6863. doi: 10.1073/pnas.1000444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 38.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 42.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: A database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencesignaling.org/cgi/content/full/6/286/ra63/DC1
Fig. S1. RIP1 activation and loss of A20 are associated with sarcoma cell lines.
Fig. S2. Abundance of A20 mRNA and protein is reduced in sarcomas.
Fig. S3. A20 is not frequently mutated in sarcomas.
Fig. S4. A20 regulation in sarcoma cells is not mediated at the transcriptional level.
Fig. S5. A20 is regulated by miR-29 and miR-125.
Fig. S6. Endogenous regulation of A20 and RIP1 by miR-29 promotes a differentiated phenotype in Rh30 cells.
Fig. S7. HuR inhibits the activity of the A20 3′UTR reporter.
Fig. S8. Putative HuR binding sites are present in the A20 3′UTR.
Fig. S9. Precursor miR-29 contains multiple HuR binding sites, and miR-125 association with Ago2 is dependent on HuR in sarcoma cells.
Table S1. Primers for the A20 3′UTR.
Table S2. Primers used to sequence A20 exons and UTRs.
Table S3. pRNAT-CMV3.1/hygro plasmids expressing wild-type or mutated miR-29 precursor sequences.
Table S4. Primers used for RT-PCR analysis.