Abstract
Despite the frequent detection of circulating tumor antigen–specific T cells, either spontaneously or following active immunization or adoptive transfer, immune-mediated cancer regression occurs only in the minority of patients. One theoretical rate-limiting step is whether effector T cells successfully migrate into metastatic tumor sites. Affymetrix gene expression profiling done on a series of metastatic melanoma biopsies revealed a major segregation of samples based on the presence or absence of T-cell-associated transcripts. The presence of lymphocytes correlated with the expression of defined chemokine genes. A subset of six chemokines (CCL2, CCL3, CCL4, CCL5, CXCL9, and CXCL10) was confirmed by protein array and/or quantitative reverse transcription-PCR to be preferentially expressed in tumors that contained T cells. Corresponding chemokine receptors were found to be up-regulated on human CD8+ effector T cells, and transwell migration assays confirmed the ability of each of these chemokines to promote migration of CD8+ effector cells in vitro. Screening by chemokine protein array identified a subset of melanoma cell lines that produced a similar broad array of chemokines. These melanoma cells more effectively recruited human CD8+ effector T cells when implanted as xenografts in nonobese diabetic/severe combined immunodeficient mice in vivo. Chemokine blockade with specific antibodies inhibited migration of CD8+ T cells. Our results suggest that lack of critical chemokines in a subset of melanoma metastases may limit the migration of activated T cells, which in turn could limit the effectiveness of antitumor immunity.
Introduction
Despite the expression of multiple defined tumor antigens by melanoma tumor cells, and the characterization of CD8+ T cells that can recognize those antigens and exert cytolytic activity, spontaneous immune-mediated elimination of melanoma remains uncommon. The identification of defined epitopes derived from shared melanoma antigens presented by dominant HLA alleles has enabled the development of antigen-specific vaccination or adoptive T-cell therapy as strategies to increase the frequency of tumor-reactive effector T cells in an attempt to promote immune-mediated rejection (1, 2). However, these interventions have met with limited success, despite a detectable increase in tumor antigen–specific CD8+ T cells in the blood in many studies (3–6). Although there are outstanding issues about the qualitative aspects of those induced T cells to be considered, these observations have together suggested that resistance mechanisms downstream from induction and expansion of tumor antigen–specific T cells may be dominant in many individual patients.
Numerous putative mechanisms by which tumors can evade immune attack despite the presence of antigen-specific CD8+ effector T cells have been proposed and supported by preclinical models (7). One hypothetical barrier is inadequate recruitment of activated T cells into metastatic tumor sites. Homing of effector T cells to inflamed tissues is thought to depend on adhesion molecules such as LFA-1 and VLA-4 (8, 9) and also on the activity of specific chemokines (10). A recent report has suggested that primary melanoma lesions that contain a T-cell infiltrate express higher levels of the chemokines CXCL9/Mig and CXCL10/IP10 (11). However, the chemokines expressed by metastatic melanoma tumors that might support recruitment of CD8+ effector cells is not known. Tumor antigen–specific T cells would not be expected to control tumor growth if they fail to enter the tumor microenvironment.
To begin to gain insight into the nature of the melanoma tumor microenvironment in individual patients, Affymetrix gene expression profiling was done on a series of melanoma metastases sampled by core biopsy or surgical excision. We found that groups of metastases were segregated largely based on immunologically relevant genes, and in particular, that a subset of tumors expressed both T-cell – specific transcripts and a broad array of chemokines. Confirmatory and functional assays together suggest that an optimal chemokine profile in the melanoma tumor microenvironment may be critical for improved recruitment of CD8+ effector T cells into metastatic tumor sites, and imply that one level of tumor escape from immune destruction may be the lack of expression of such chemokines.
Materials and Methods
Cell lines
The melanoma cell lines SKMel23, SKMel28, 537, 624, and 888 were maintained in RPMI containing 10% FCS. Primary melanocyte cell lines were purchased from Cascade Biologics.
Biopsy samples
Tumor samples were obtained by core biopsy or excisional biopsy or obtained from material resected from patients as part of routine clinical management. Tumor was grossly separated from nontumor tissue and a portion was quickly frozen in liquid nitrogen for preparation of RNA and for long-term storage. Samples were obtained from patients who signed written informed consent for tissue procurement clinical trials at the University of Chicago or the University of Virginia.
RNA extraction and quality control
Total RNA was extracted from tumor samples (80 mg each) using the GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich Corp.). If contaminating melanin pigment was seen, then an additional purification by cesium chloride centrifugation was done. RNA quality was evaluated by agarose gel electrophoresis and ethidium bromide staining. Quantitation of the RNA samples was determined by absorbance at 260 nm, and the purity and concentration were confirmed using a Gene Spec III (Miraibio).
Gene array analysis
Quality-controlled RNA samples were prepared as targets according to the Affymetrix GeneChip Expression Analysis Manual (Affymetrix). Briefly, 10 µg of total RNA were used to synthesize double-stranded cDNA using the Superscript Choice System (Invitrogen Corp.). First-strand cDNA synthesis was primed with a T7-(dT24) oligonucleotide. From the phase-log gel-purified cDNA, biotin-labeled antisense cRNA was synthesized using BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics). After precipitation with 4 mol/L LiCl, 20 µg of cRNA were fragmented in fragmentation buffer [40 mmol/L Tris-acetate (pH 8.1), 100 mmol/L KOAc, 30 mmol/L MgOAc] for 35 min at 94°C. Fragmented cRNA (12 µg) was hybridized to arrays (Affymetrix U133A) for 16 h at 45°C and 60 rpm in an Affymetrix Hybridization Oven 640. Arrays were washed and stained with streptavidin-phycoerythrin (PE) in an Affymetrix Fluidics Station 450 using the Affymetrix GeneChip protocol and scanned using the Affymetrix GeneChip Scanner 3000. The acquisition and initial quantification of array images were done using the GCOS (Affymetrix). Subsequent data analysis involved the normalization of array values followed by nonsupervised hierarchical clustering using dChip software.
Immunohistochemistry
Representative melanoma cases were immunostained with antibodies for CD8 (mouse, clone C8/144B, Neomarkers), CD20 (mouse, clone L26, DAKO), and CD68 (mouse, clone KP1, DAKO). Slides were deparaffinized in xylene and hydrated with alcohol before being placed in 0.3% H2O2/methanol blocking solution to quench endogenous peroxidase activity followed by subsequent antigen unmasking in EDTA buffer. Incubation with the primary antibodies was done for 1 h at room temperature. After TBS washing, the slides were incubated for 30 min at room temperature with goat anti-mouse IgG conjugated to a horseradish peroxidase–labeled polymer (Envision+ System, DAKO) or to an alkaline phosphatase–labeled polymer (MACH3TM, Biocare Medical). Reactions were developed with 3,3′-diaminobenzidine chromogen or Vulcan Red, respectively, and counterstained with hematoxylin. Appropriate negative controls for the immunostaining were prepared by omitting the primary antibody step and substituting it with nonimmune serum of the appropriate species. Images were photographed at ×100 magnification using an AxioCam (Zeiss) digital camera. Staining was scored semiquantitatively as “present (2+),” “rare (1+),” or “absent (0).”
Chemokine protein arrays
Tumor fragments were stored in liquid nitrogen until use. Samples were weighed, and 100 mg were homogenized and sonicated in lysis buffer containing protease inhibitors. Cleared tumor lysates after centrifugation were incubated with membranes coated with 38 antihuman chemokine antibodies according to the manufacturer’s protocol (RayBiotech, Inc.). The membranes were then incubated with biotinylated detection antihuman chemokine antibodies and a streptavidin-peroxidase conjugate sequentially. After the final wash, the membranes were developed using the enhanced chemiluminescence method (RayBiotech). Chemokine proteins were quantitated using UnScanit software, and the expression level of each chemokine was determined as the percentage of the positive control.
Real-time reverse transcription-PCR
Expression of some genes was confirmed using real-time reverse transcription-PCR (RT-PCR). Total RNA was DNase I treated, and cDNA was synthesized from 1 µg total RNA using MLV Reverse Transcriptase (Invitrogen). As a control, a mock batch of cDNA was made without adding reverse transcriptase. The reactions were run on an ABI PRISM 7700 Sequence Detection System machine and analyzed using Sequence Detector vl.7a software (Applied Biosystems). Results were only included in the analysis if the reaction with mock cDNA yielded a background signal. Prevalidated primers and probes specific for CXCL9 (assay ID Hs00171065_m1), CXCL10 (assay ID Hs00171042_m1), and CCL3 (assay ID Hs00234142_m1) were purchased from Applied Biosystems. Other custom primer/probe sets used are listed in Supplementary Methods.
Analysis of chemokine receptor expression
To evaluate the expression of chemokine receptors on CD8+ T-cell subsets, normal donor peripheral blood mononuclear cells (PBMC) were stained with anti-CCR7 PE-Cy7, anti-CD45RA PE-Cy5, anti-CD8 FITC, or anti-CCR1 APC, anti-CCR2 APC, anti-CCR5 APC, anti-CXCR3 APC, anti-CXCR4 APC, or an IgG APC isotype-matched control (BD Pharmingen). Cells were washed and fixed with 3% paraformaldehyde. Data were acquired on a multicolor FACS LSRII, and analysis was done using FlowJo software (Becton Dickinson). Gating was done on CCR7+CD45RA+ cells for the naive phenotype, and CCR7−CD45RA+ cells for the effector phenotype.
Measurement of migration to chemokines in vitro
CD8+ lymphocytes were selected from normal donor PBMC by positive selection using magnetic beads (Miltenyi Biotec). The selected cells were cultured with anti-CD3/anti-CD28 monoclonal antibody (mAb)-coated beads for 7 d to generate CD8+ effector cells. These cells were loaded into the top chamber of transwell inserts (5.0 µm pore size, Costar). In the bottom well, RPMI medium containing different levels of different chemokines, or culture supernatant from melanoma cell lines, was added. All recombinant chemokines including CCL2 (MCP1), CCL3 (MIP1-α), CCL4 (MIP1-β), CCL5 (RANTES), CXCL8 [interleukin (IL)-8], CXCL9 (MIG), and CXCL10 (IP-10) were purchased from R&D Systems. Plates were incubated at 37°C for 2 h; the contents of the lower chamber were collected; and the percentage of CD8+ cells present in the bottom chamber was determined by flow cytometry.
Analysis of T-cell recruitment to tumor xenografts in vivo
Human melanoma cell lines (3 × 106 cells) were implanted s.c. in the flank of 6-wk-old nonobese diabetic/severe combined immunodeficient (NOD/scid) mice (six mice per group). Tumors were allowed to grow until 0.5 to 1 cm in diameter (~ 4 wk) before adoptive transfer of T cells. CD8+ effector T cells were then labeled with CFSE (10 µmol/L for 7 min) and injected i.v. via the tail vein. Mice were sacrificed 24 to 48 h later; tissues were harvested; and analysis of single-cell suspensions was done by flow cytometry using a FACS LSRII. Gating was done by forward scatter and side scatter, and the percentage of CFSE-positive cells was determined.
Results
Nonsupervised hierarchical clustering of gene expression data reveals two major subsets of metastatic melanoma tumors that segregate based on the presence or absence of lymphocytes
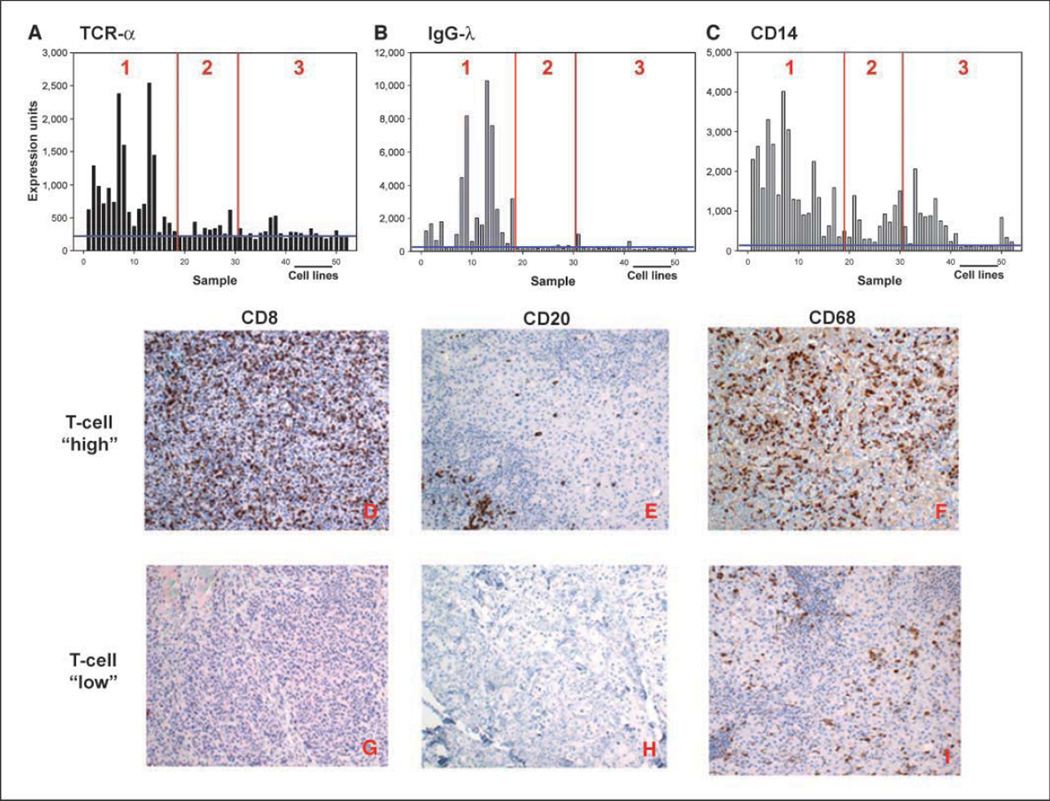
To begin gaining an understanding of the metastatic melanoma solid tumor microenvironment, Affymetrix gene expression profiling was done on a panel of 44 biopsies of melanoma metastases that satisfied defined quality control criteria. To minimize sampling error and to represent a balance of the cellular elements contained in these metastases, only core biopsies or excisional biopsies were used. As an approximation to infer expression of genes by the melanoma cells themselves versus by stromal cell components, a set of five melanoma cell lines and three primary melanocyte cell lines were included in the analysis. Nonsupervised hierarchical clustering analysis revealed two major clusters of samples, with a second major split in the dendrogram in the second cluster (Supplementary Fig. S1). These were designated groups 1, 2, and 3. The most striking transcriptional differences that distinguished the samples were those that suggested the presence of lymphocytes. T-cell receptor (TCR)-α, TCRβ, and TCRγ transcripts were expressed in group 1, as were the T-cell-specific adaptor proteins Slp76 and Fyb (Supplementary Table SI). Immunoglobulin genes were also expressed, including κ and λ light-chain genes and IgG and IgM heavy-chain genes. Of the 58 unique genes that characterize group 1 tumors, 41 (71%) are clearly immune related and 20 (34%) are thought to be predominantly or exclusively expressed by lymphocytes. Collectively, these data suggest the presence of both T- and B-lineage cells in these tumors. It is important to note that this pattern was not restricted to lymph node metastases but was also seen in melanoma biopsies obtained from brain, lung, skin, and small bowel sites. In addition, cutaneous metastases were the most commonly analyzed metastatic site, and there were cutaneous metastases that either contained or lacked lymphocyte transcripts. Thus, it is unlikely that the segregation of these samples was dominantly dictated by transcripts coming from contaminating normal cells of the tissues from which the metastases were obtained.
Regarding the other hematopoietic cells, CD14 transcripts were expressed at higher levels in the lymphocyte-rich group but were present at variable levels in all solid tumor samples above the background level seen in melanoma cell lines. This observation suggests that macrophages were likely present to varying degrees in all tumors examined. The magnitude of expression of selected genes indicating the presence of inflammatory cells in individual classified tumors is shown in Fig. 1A–C.
Figure 1.
Distribution of selected inflammatory cells’ transcripts in melanoma biopsies. A to C, gene array data. Expression levels of TCRα (indicating T-lineage cells), IgGλ (indicating B-lineage cells), and CD14 (indicating monocyte-lineage cells) transcripts represented as normalized hybridization intensity data are shown for individual samples. The vertical lines indicate separations between groups 1, 2, and 3, and the horizontal line indicates the data with melanoma cell lines. D to I, immunohistochemical confirmation of inflammatory cell infiltrates. Representative tissue samples from tumors that contained (top) or lacked (bottom) T-cell transcripts were stained with antibodies specific for CD8 (D and G), CD20 (E and H), and CD68 (F and I). Similar results were observed with two additional tumors from each group.
To confirm whether the expression of these transcripts indeed indicated the presence of T cells, B cells, and macrophages, and to determine whether these cell types were infiltrating within the tumors, immunohistochemical staining was done on a subset of samples. As represented in Fig. 1D–I, the tumors containing lymphocyte transcripts showed the presence of abundant infiltrating CD8+ cells and scattered CD20+ cells, whereas tumors that lacked lymphocyte transcripts did not. CD4+ cells were also seen in tumors that contained T cells, but in fewer numbers (data not shown). Scoring of four “T-cell-high” tumors revealed that all of them showed 2+ level of staining for CD8, whereas of the three “T-cell-low” tumors analyzed, two were scored as 0 and one as 1+ (indicating rare staining). In addition, CD68+ cells (indicating the presence of macrophages) were seen both in tumors that contained lymphocytes and in those that did not.
In addition to lymphocyte-specific transcripts, expression of other genes encoding putative positive regulators of antitumor immune responses was observed. The lymphocyte-rich tumors also contained transcripts for granzyme A, class II MHC, complement Clq, and multiple IFN-inducible proteins. Expression of these transcripts suggests a more extensive degree of inflammation in that subset of tumors that includes both innate and adaptive immune components.
Differential expression of key chemokines in individual melanoma tumors and confirmation by protein array
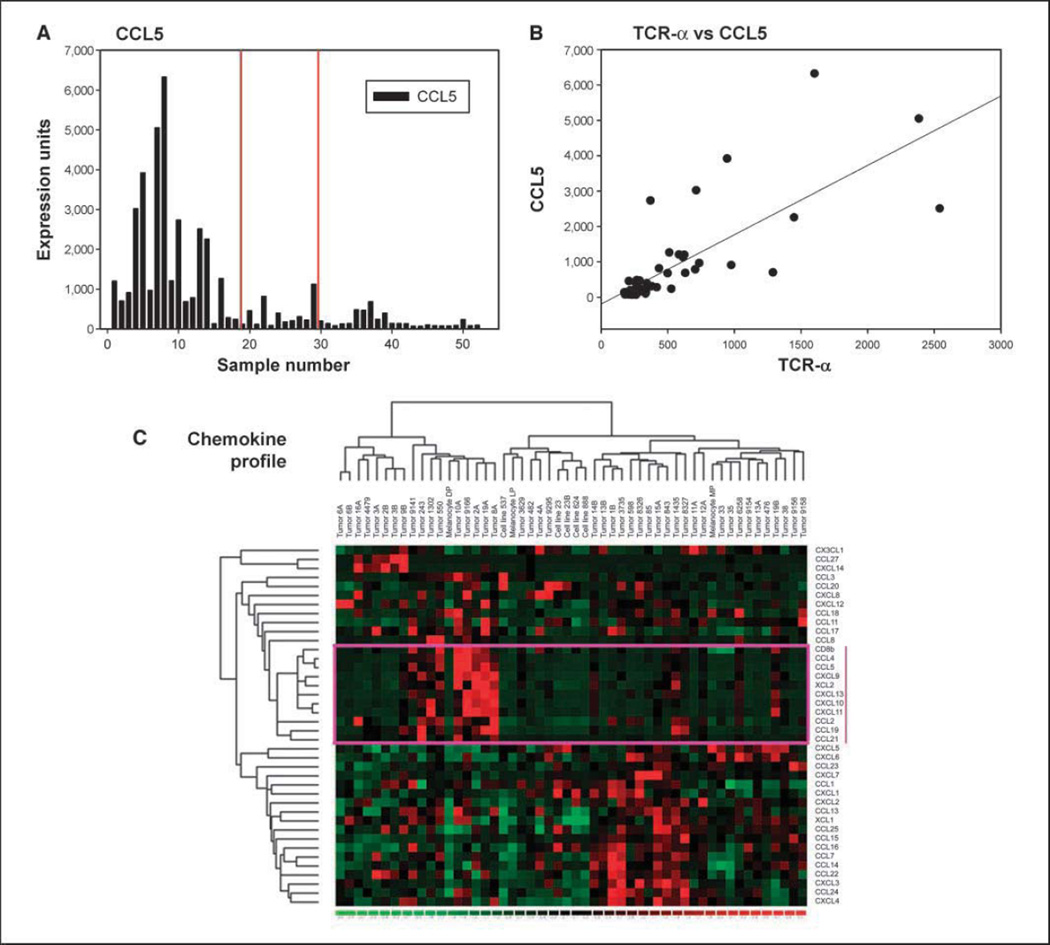
As the major distinguishing feature observed was differential expression of lymphocyte signatures, it was of interest to gain insight into the potential mechanism by which T cells seemed to be recruited into some melanoma lesions but not into others. Interestingly, these samples were also distinguished by expression of specific chemokine transcripts. Tumors from the lymphocyte-rich group uniquely expressed CCL2, CCL3, CCL4, CCL5, CCL19, CCL21, CXCL9, CXCL10, CXCL11, and CXCL13 (Supplementary Table S1). Also of note is that the majority of tumors, whether they contained lymphocytes or not, expressed detectable levels of IL-8 and CXCL12 transcripts. The magnitude of expression of CCL5 in individual tumors is represented as an example in Fig. 2A, which showed a positive correlation with the expression of TCRα transcripts (R2 = 0.6; Fig. 2B). As an alternative approach for examining correlation between specific chemokines and T-cell-specific transcripts, the data for all chemokines as well as CD8β were extracted and analyzed by nonsupervised hierarchical clustering. By this analysis, samples with high CD8β transcripts clustered tightly with those expressing high levels of CCL2, CCL4, CCL5, CCL19, CCL21, CXCL9, CXCL10, CXCL11, CXCL13, and XCL2 (Fig. 2C). Together, these data suggest that T-cell infiltration was associated with the presence of a broad array of chemokine transcripts, implying a potential mechanism for recruitment of activated lymphocytes into the tumor microenvironment.
Figure 2.
Correlation between chemokines and T-cell transcripts in individual tumors. A, expression levels of CCL5 transcripts represented as normalized hybridization intensity data are shown for individual samples. The vertical lines indicate separations between groups 1, 2, and 3. B, plot of CCL5 transcript levels versus TCRα levels in individual tumors. A positive correlation was observed (R2 = 0.6). C, nonsupervised hierarchical clustering of chemokine and CD8β transcript data. To examine associations between CD8 expression and a diverse panel of chemokines, nonsupervised hierarchical clustering analysis was done on the subset of transcripts that encode chemokines and CD8β. CD8β was found to spontaneously cluster with a subset of chemokine transcripts (indicated by the box).
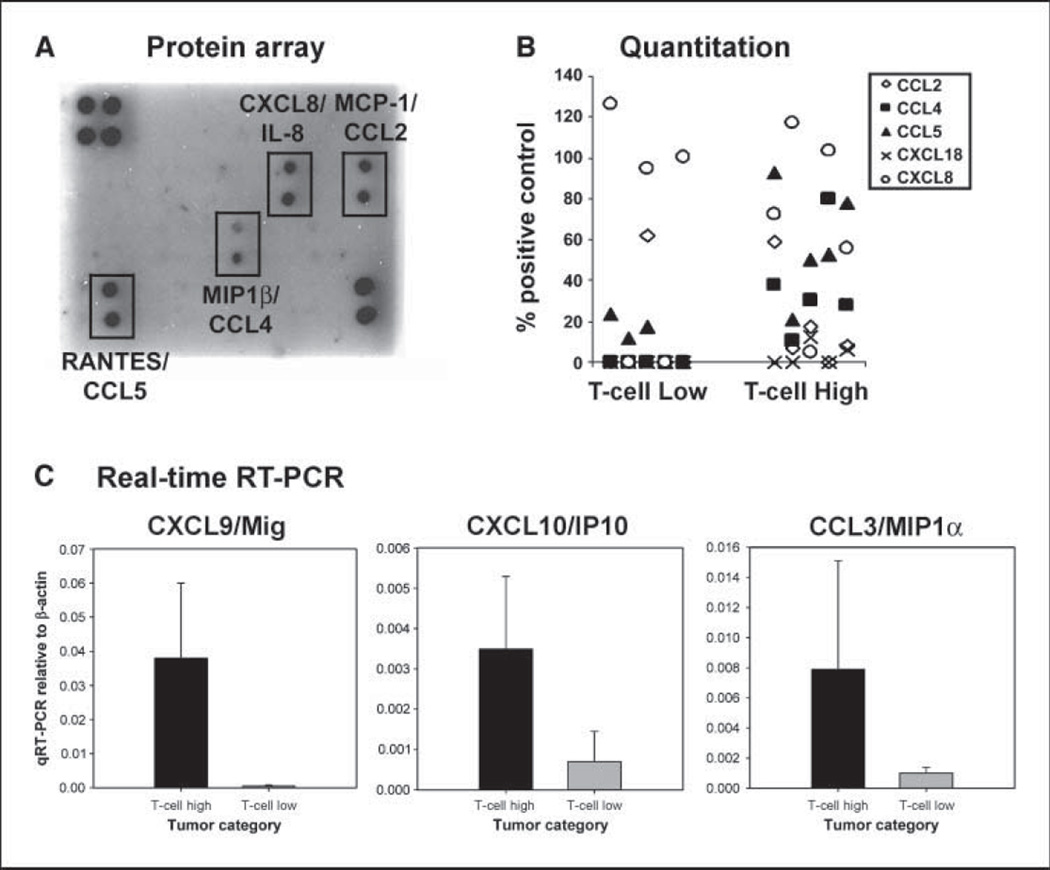
To determine whether differential chemokine expression could be confirmed at the protein level, antibody-based protein arrays that allow detection of 38 individual chemokines were used. Protein lysates generated from five tumors that contained high T-cell transcripts and five that contained low T-cell transcripts were used for analysis. A representative blot from each of these categories is shown in Fig. 3A, with quantitative data from individual tumors shown in Fig. 3B. Three chemokine proteins that were reproducibly detected at abundant levels in the tumors with T cells were CCL2/MCP-1, CCL4/MIP-1β, and CCL5/RANTES. In contrast, the T-cell-poor tumors showed expression predominantly of CXCL8/IL-8. IL-8 is often produced by melanoma cells themselves and has been reported to exert a proangiogenic effect in these tumors (12). Quantitatively, CCL4 expression was significantly greater in the T-cell-containing tumors (mean intensity 37 ± 25 compared with 0 ± 0; P = 0.005) as was CCL5 (mean intensity, 59 ± 27 versus 10 ± 10; P = 0.003). CCL2 showed a trend but was not statistically significantly different (18 ± 23 versus 12 ± 27; P = 0.35). Because it seemed likely that the protein array approach might not be sensitive enough to detect chemokines in low abundance, real-time RT-PCR was done for expression of CXCL9/Mig, CXCL10/IP-10, and CCL3/MIP-lα. Each of these also was confirmed to be expressed at higher levels in the T-cell-containing tumors (Fig. 3C).
Figure 3.
Quantitative expression of chemokine proteins in melanoma metastases that contain or lack T cells. Antibody-based protein arrays were used to assess the presence of 38 chemokines in tumor lysates. A, representative blots from a tumor rich in T-cell transcripts versus a tumor that lacked T-cell transcripts. B, quantitative data from scanned blots from five tumors that contained T-cell transcripts and five tumors that lacked T-cell transcripts. C, real-time RT-PCR was done for CXCL9, CXCL10, and CCL3 on five tumors that contained or lacked T-cell transcripts. Columns, mean; bars, SD.
Tumor-expressed chemokines support migration of human CD8+ effector T cells
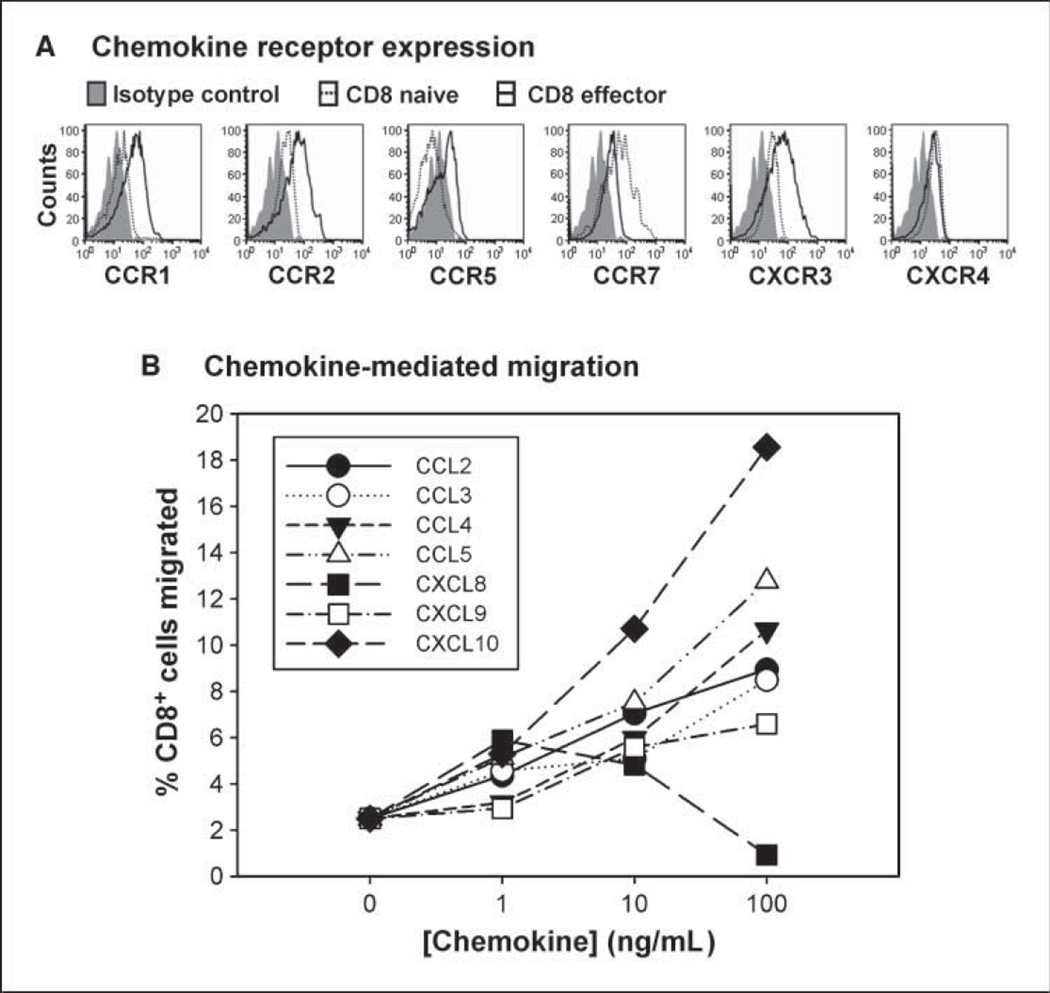
To narrow the candidate chemokines that may be most relevant for antitumor T-cell responses, the chemokine receptors expressed on naive versus effector CD8+ T cells obtained from normal donors were analyzed by flow cytometry. As shown in Fig. 4A, expression of CCR1, CCR2, CCR5, and CXCR3 was observed to be up-regulated in effector cells relative to the naive state. In addition, CXCR4 and CCR7 were expressed at low yet detectable levels. Similar relative levels were seen comparing naive and 7-day in vitro primed CD8+ effector cells at the mRNA level (data not shown). Based on well-defined specificities of binding of specific chemokines to specific chemokine receptors (10), the up-regulated receptors narrowed our focus to CCL2, CCL3, CCL4, CCL5, CXCL9, and CXCL10 as those that could be most relevant for effector CD8+ T-cell migration. Transwell migration assays were done and revealed that each of these chemokines was sufficient to recruit CD8+ effector T cells primed from normal donors in vitro (Fig. 4B). In contrast, CXCL8/IL-8 was not effective, correlating with lack of expression of the relevant chemokine receptors CXCR1/CXCR2. These results support the hypothesis that this set of six chemokines may be the most important for recruitment of activated CD8+ T cells into melanoma metastases and may have partially redundant roles.
Figure 4.
Chemokines relevant for recruitment of CD8+ effector T cells. A, chemokine receptor expression. Flow cytometric analysis was done comparing naive versus effector CD8+ T cells obtained from normal donors for expression of CCR1, CCR2, CCR5, CXCR3, CCR7, and CXCR4. Similar results were seen with at least two independent donors, and with effector cells generated by in vitro priming with anti-CD3/anti-CD28 mAb–coated beads. B, chemokine-mediated migration. In vitro transwell migration was done using in vitro primed CD8+ effector cells in the upper chamber and the indicated chemokines in the lower chamber. The percentages of T cells in the lower chamber were determined by flow cytometry at 2 h and are representative of at least two experiments.
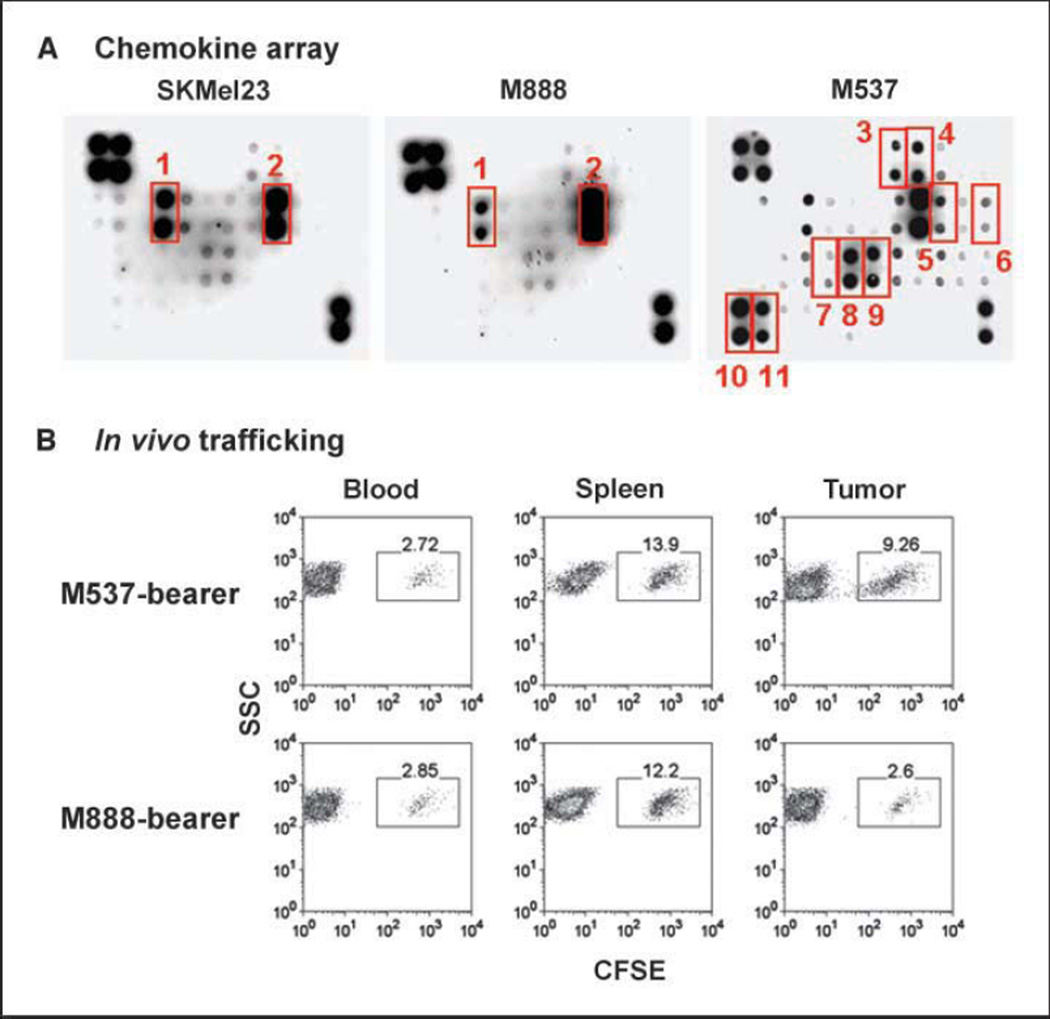
To investigate the potential relevance of these chemokines for functional recruitment of CD8+ effector T cells into melanoma tumors, a panel of human melanoma cell lines was screened for chemokine production using protein arrays. Most melanoma cell lines (e.g., SKMel23, SKMel 28, and 888) produced a restricted set of chemokines (predominantly IL-8 with variable levels of GRO; Fig. 5A) that showed similarity to the lymphocyte-poor melanoma metastases analyzed ex vivo from patients. However, rare melanoma cell lines produced an expanded set of chemokines. For example, melanoma cell line M537 produced detectable levels of CCL2, CCL3, CCL4, CCL5, CXCL9, and CXCL10, as well as a few additional chemokines (Fig. 5A). Thus, it is possible for some melanoma tumor cells themselves to produce a set of chemokine proteins capable of attracting lymphocytes.
Figure 5.
Chemokines produced by a subset of melanoma cell lines can attract human CD8+ effector T cells in a xenograft setting in vivo. A, supernatants from a series of melanoma cell lines were examined for chemokine content using a protein array. Melanoma cell line M537 showed a more diverse chemokine production profile. The indicated chemokines are GRO (1), IL-8 (2), CTACK (3), CXCL16 (4), IP-10 (5), MCP-1 (6), MIG (7), MIP-1α (8), MIP-1β (9), PARC (10), and RANTES (11). Each sample is represented in duplicate. The four top left corner spots and two bottom right corner spots represent loading controls. B, human melanoma cell lines were implanted s.c. into NOD/scid mice. Once they grew to a solid tumor, CD8+ effector T cells were prepared from normal human donors, labeled with CFSE as a fluorescent indicator, and injected i.v. The indicated tissues were then harvested and analyzed by flow cytometry for the presence of labeled T cells versus side scatter. Similar results were seen in at least two independent experiments.
To investigate whether the melanoma cells expressing a broader chemokine array were actually more effective at recruiting human CD8+ effector T cells, a mouse xenograft model was established. Tumors were established s.c. by injecting melanoma cell suspensions in NOD/scid mice. Polyclonal CD8+ effector T cells were generated from normal donors by stimulation with anti-CD3/anti-CD28–coated beads and IL-2, labeled with CFSE as a fluorescent marker, and injected i.v. Twenty-four hours later, the tumors and lymphoid organs were harvested and analyzed by flow cytometry. As shown in Fig. 5B, melanoma cell line M537 was substantially more effective at recruiting CD8+ effector cells than was melanoma cell line 888. This difference was not observed in the blood or spleen, arguing that the specific enrichment of T cells in the M537 tumors is a feature of the local tumor microenvironment. Similar results of superior recruitment of CD8+ effector cells by M537 tumor cells compared with 888 tumor cells were obtained by analyzing tumor-derived supernatants in a transwell migration assay in vitro (data not shown).
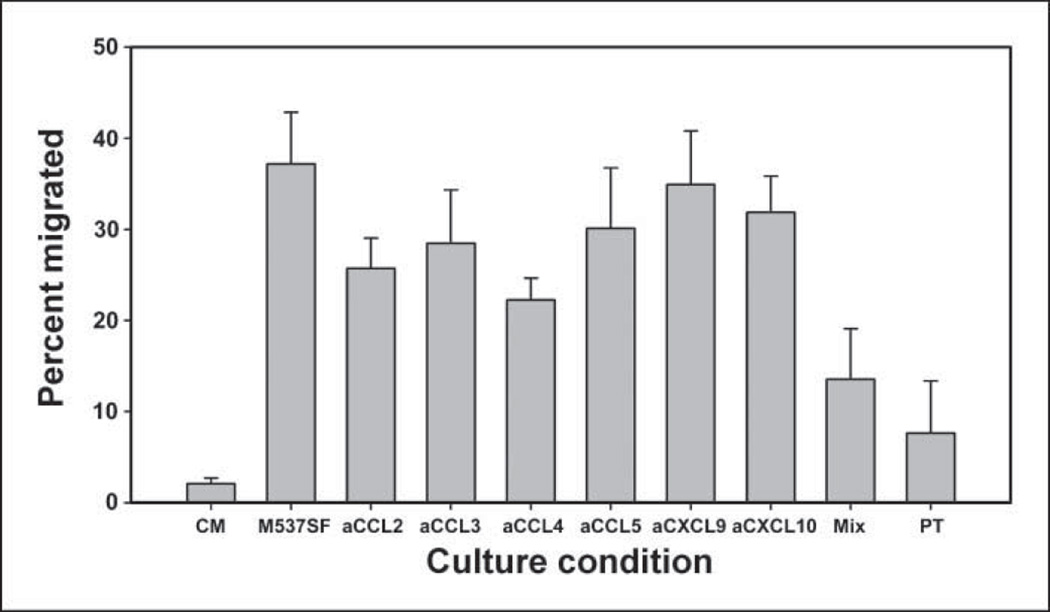
It was of interest to determine which chemokines produced by M537 tumor cells were necessary for promoting migration of CD8+ effector T cells. This was evaluated in vitro using specific neutralizing antibodies. Supernatants from M537 cells were generated and used in a transwell assay to measure recruitment of human primed CD8+ effector T cells. Migration was observed to be very vigorous, the majority of which was blocked by pertussis toxin, supporting dependency on chemokine receptor signaling. The addition of individual antibodies targeting CCL2, CCL3, or CCL4 each had a modest effect, showing partial inhibition of migration. However, a cocktail of antibodies against those chemokines plus those against CCL5, CXCL9, and CXCL10 was required to effectively inhibit T-cell recruitment near to the level seen with pertussis toxin poisoning (Fig. 6). These results suggest that multiple chemokines produced by melanoma cells contribute to achieving superior recruitment of CD8+ effector T cells.
Figure 6.
Chemokine blockade inhibits recruitment of CD8+ effector T cells by M537 tumor-derived supernatants. Supernatants were generated from M537 tumor cells and assessed for the ability to attract human CD8+ effector T cells from normal donors. Migration was analyzed in the presence of the indicated chemokine-specific antibodies or with pertussis toxin pretreatment of the T cells (PT). Culture medium alone (CM) was used as a negative control. Similar results were seen in at least two experiments.
Discussion
The relatively low clinical activity of melanoma vaccines despite induction of specific T-cell responses detected in the blood has suggested the possibility of downstream resistance mechanisms at the level of the tumor microenvironment. Our current results indicate that some tumors lack key chemokines that can be critical for recruitment of activated T cells into metastatic sites, which could represent an important barrier for effective T-cell–mediated rejection of tumors in vivo.
Several cell types within the tumor microenvironment could potentially produce the chemokines important for recruitment of effector CTL. Our analysis of melanoma cell lines indicates that a subset of such lines is capable of producing an expanded array of chemokines themselves. However, additional cell types may contribute to chemokine synthesis within the complexity of the tumor microenvironment in vivo. Macrophages, endothelial cells, and even recruited T cells could theoretically secrete relevant chemokines and positively reinforce recruitment of additional specific T-cell subsets (13). Further work will be required to identify the cell types producing each chemokine within the metastatic melanoma microenvironment in individual patients using in situ assays.
It is of interest that the majority of melanoma tumors expressed stromal cell–derived factor-1 (SDF-1)/CXCL12 transcripts, even those tumors that lack T cells. There is some controversy in the literature on whether this chemokine attracts or repulses T cells (14, 15). The fact that some tumors seem to express SDF-1 without a T-cell infiltrate argues at least that expression of this factor is not sufficient for effective recruitment of activated CTL, and it is interesting to speculate that fugetaxis may be the dominant effect of SDF-1 in vivo. Chemokines may have additional properties that extend beyond cell trafficking that could be functionally relevant here. Recent work has suggested that CXCR4 and CCR5 may cooperatively provide a costimulatory signal for T-cell activation (16) and that CCR5 on CD8+ T cells may be important for the generation of a memory phenotype (17). Thus, expression of CCR5 ligands, in particular, may help to maintain functional properties of activated T cells in vivo. In addition to chemokines, our current work has not interrogated the likely contribution of adhesion molecules and homing receptors in the trafficking of T cells into tumor sites, a question that is worth pursuing in future studies.
Why some melanoma metastases seem to be capable of generating an inflammatory microenvironment and recruit T cells whereas others do not is currently unknown. Several different oncogenic pathways are thought to be capable of contributing to a malignant phenotype from the melanocyte lineage [e.g., Ras, B-Raf, Akt, Notch, and signal transducer and activator of transcription 3 (Stat3); refs. 18, 19], and it is plausible to consider that differential involvement of individual signaling pathways within the tumor cells themselves may lead to differential expression of chemokine genes or other factors that influence the tumor cell/immune system interface. Manipulation of Stat3 expression has indicated that down-regulation of Stat3 can result in increased expression of chemokine genes by tumor cells (20), suggesting that differential activity of a single oncogenic pathway may be capable of controlling the recruitment of inflammatory cells into the tumor vicinity. It will be of interest to investigate a potential association between Stat3 or other signaling events in the tumor cells and the presence of activated T cells in tumor sites through future studies.
The question arises about how some melanoma metastases can express high levels of chemokines and recruit activated CD8+ T cells and yet still not be spontaneously rejected by the immune system. Other studies have shown that the tumor-infiltrating lymphocyte population includes tumor antigen–specific T cells (21, 22), suggesting that this recruitment does include relevant cells. However, the function of those T cells seems to be suppressed. Multiple mechanisms of immune suppression that act within the tumor microenvironment have been proposed, and several of these have been validated to be functionally important in mouse preclinical studies (7). Preliminary data have suggested that melanoma metastases that contain CD8+ T cells show greater presence of FoxP3+ regulatory T cells, as well as higher expression of PD-L1 and indoleamine-2,3-dioxygenase (data not shown), supporting the notion that the tumor microenvironment can be rich in immune suppressive mechanisms that could contribute to T-cell dysfunction.
Our results have implications for the use of autologous tumor-infiltrating lymphocytes (TIL) as an adoptive transfer immunotherapeutic strategy (23). By definition, those clinical trials are limited to patients from whom a substantial number of T cells are present in the resected tumor specimen for T-cell harvest. That patient population is thus enriched for individuals with tumors that are capable of recruiting activated T cells, and clinical results could theoretically lead to a superior clinical outcome based on this property. It will thus be critical to determine in prospective clinical studies whether a specific gene expression profile of the melanoma tumor microenvironment can predict clinical outcome from melanoma vaccines and other immunotherapeutic interventions.
Opportunities exist for manipulating chemokine expression in the tumor microenvironment with therapeutic intent. Transfection of tumor cells to express several chemokines before implantation in mice has shown improved tumor control in several models (24). One attractive alternative is the tumor necrosis factor superfamily member LIGHT, which elicits chemokine expression from infiltrating stromal cells through engagement of the lymphotoxin β-receptor (25). Transfection of tumor cells to express LIGHT results in potent tumor rejection (26), and introduction of an adenoviral vector encoding LIGHT can improve the control of preestablished tumors and promote rejection of micrometastases (27). Clinical data have suggested an association between CXCR3 expression by peripheral T cells and favorable clinical outcome in stage III melanoma patients (28), arguing that the chemokine receptors expressed by T cells may be important in patients. Because most melanomas express SDF-1 and IL-8, it is conceivable to transfer expression of the corresponding receptors into CD8+ effector T cells before adoptive transfer. Such an approach has been tested preclinically with GRO-α (29), and the active clinical efforts ongoing with adoptive T-cell transfer in cancer patients make this approach attractive to consider in the future.
Supplementary Material
Acknowledgments
Grant support: Translational research award from the Burroughs Wellcome Fund, P01 CA97296, R01 CA90575, and a grant from the Ludwig Trust.
We thank Todd Kuna, Alpana Sahu, and Terry Li for technical assistance; Xinmin Li in the University of Chicago Functional Genomics Facility for the gene array analysis; and Marisa Alegre for careful reading of the manuscript.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Rosenberg SA. Cancer vaccines based on the identification of genes encoding cancer regression antigens. Immunol Today. 1997;18:175–182. doi: 10.1016/s0167-5699(97)84664-6. [DOI] [PubMed] [Google Scholar]
- 2.Van den Eynde B, Gaugler B, van der Bruggen P, Coulie P, Brichard V, Boon T. Human tumour antigens recognized by T-cells: perspectives for new cancer vaccines. Biochem Soc Trans. 1995;23:681–686. doi: 10.1042/bst0230681. [DOI] [PubMed] [Google Scholar]
- 3.Peterson AC, Harlin H, Gajewski TF. Immunization with Melan-A peptide-pulsed peripheral blood mononuclear cells plus recombinant human interleukin-12 induces clinical activity and T-cell responses in advanced melanoma. J Clin Oncol. 2003;21:2342–2348. doi: 10.1200/JCO.2003.12.144. [DOI] [PubMed] [Google Scholar]
- 4.Cormier JN, Salgaller ML, Prevette T, et al. Enhancement of cellular immunity in melanoma patients immunized with a peptide from MART-1/Melan A. Cancer J Sci Am. 1997;3:37–44. [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Sherry RM, Morton KE, et al. Tumor progression can occur despite the induction of very high levels of self/tumor antigen-specific CD8+ T cells in patients with melanoma. J Immunol. 2005;175:6169–6176. doi: 10.4049/jimmunol.175.9.6169. [DOI] [PubMed] [Google Scholar]
- 6.Schadendorf D, Ugurel S, Schuler-Thurner B, et al. Dacarbazine (DTIC) versus vaccination with autologous peptide-pulsed dendritic cells (DC) in first-line treatment of patients with metastatic melanoma: a randomized phase III trial of the DC study group of the DeCOG. Ann Oncol. 2006;17:563–570. doi: 10.1093/annonc/mdj138. [DOI] [PubMed] [Google Scholar]
- 7.Gajewski TF, Meng Y, Blank C, et al. Immune resistance orchestrated by the tumor microenvironment. Immunol Rev. 2006;213:131–145. doi: 10.1111/j.1600-065X.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 8.Kedl RM, Mescher MF. Migration and activation of antigen-specific CD8+ T cells upon in vivo stimulation with allogeneic tumor. J Immunol. 1997;159:650–663. [PubMed] [Google Scholar]
- 9.Ogawa M, Tsutsui T, Zou JP, et al. Enhanced induction of very late antigen 4Aymphocyte function-associated antigen 1-dependent T-cell migration to tumor sites following administration of interleukin 12. Cancer Res. 1997;57:2216–2222. [PubMed] [Google Scholar]
- 10.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 11.Monteagudo C, Martin JM, Jorda E. Llombart-Bosch A CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathologic prognostic factors. J Clin Pathol. 2007;60:596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rofstad EK, Halsor EF. Vascular endothelial growth factor, interleukin 8, platelet-derived endothelial cell growth factor, and basic fibroblast growth factor promote angiogenesis and metastasis in human melanoma xenografts. Cancer Res. 2000;60:4932–4938. [PubMed] [Google Scholar]
- 13.Zlotnik A, Morales J, Hedrick JA. Recent advances in chemokines and chemokine receptors. Crit Rev Immunol. 1999;19:1–47. [PubMed] [Google Scholar]
- 14.Zhang T, Somasundaram R, Berencsi K, et al. CXC chemokine ligand 12 (stromal cell-derived factor la) and CXCR4-dependent migration of CTLs toward melanoma cells in organotypic culture. J Immunol. 2005;174:5856–5863. doi: 10.4049/jimmunol.174.9.5856. [DOI] [PubMed] [Google Scholar]
- 15.Vianello F, Papeta N, Chen T, et al. Murine B16 melanomas expressing high levels of the chemokine stromal-derived factor-l/CXCL12 induce tumor-specific T-cell chemorepulsion and escape from immune control. J Immunol. 2006;176:2902–2914. doi: 10.4049/jimmunol.176.5.2902. [DOI] [PubMed] [Google Scholar]
- 16.Molon B, Gri G, Bettella M, et al. T-cell costimulation by chemokine receptors. Nat Immunol. 2005;6:465–471. doi: 10.1038/ni1191. [DOI] [PubMed] [Google Scholar]
- 17.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Massi D, Tarantini F, Franchi A, et al. Evidence for differential expression of Notch receptors and their ligands in melanocytic nevi and cutaneous malignant melanoma. Mod Pathol. 2006;19:246–254. doi: 10.1038/modpathol.3800526. [DOI] [PubMed] [Google Scholar]
- 20.Burdelya L, Kujawski M, Niu G, et al. Stat3 activity in melanoma cells affects migration of immune effector cells and nitric oxide-mediated antitumor effects. 1 Immunol. 2005;174:3925–3931. doi: 10.4049/jimmunol.174.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harlin H, Kuna TV, Peterson AC, Meng Y, Gajewski TF. Tumor progression despite massive influx of activated CD8+ T cells in a patient with malignant melanoma ascites. Cancer Immunol Immunother. 2006;55:1185–1197. doi: 10.1007/s00262-005-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortarini R, Piris A, Maurichi A, et al. Lack of terminally differentiated tumor-specific CD8+ T cells at tumor site in spite of antitumor immunity to self-antigens in human metastatic melanoma. Cancer Res. 2003;63:2535–2545. [PubMed] [Google Scholar]
- 23.Rosenberg SA, Dudley ME. Cancer regression in patients with metastatic melanoma after the transfer of autologous antitumor lymphocytes. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14639–14645. doi: 10.1073/pnas.0405730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mule JJ, Custer M, Averbook B, et al. RANTES secretion by gene-modified tumor cells results in loss of tumorigenicity in vivo: role of immune cell sub-populations. Hum Gene Ther. 1996;7:1545–1553. doi: 10.1089/hum.1996.7.13-1545. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Fu YX. The role of LIGHT in T cell-mediated immunity. Immunol Res. 2004;30:201–214. doi: 10.1385/IR:30:2:201. [DOI] [PubMed] [Google Scholar]
- 26.Yu P, Lee Y, Liu W, et al. Priming of naive T cells inside tumors leads to eradication of established tumors. Nat Immunol. 2004;5:141–149. doi: 10.1038/ni1029. [DOI] [PubMed] [Google Scholar]
- 27.Yu P, Lee Y, Wang Y, et al. Targeting the primary tumor to generate CTL for the effective eradication of spontaneous metastases. J Immunol. 2007;179:1960–1968. doi: 10.4049/jimmunol.179.3.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins IM, Slingluff CL, Lee JK, et al. CXC chemokine receptor 3 expression by activated CD8+ T cells is associated with survival in melanoma patients with stage III disease. Cancer Res. 2004;64:7697–7701. doi: 10.1158/0008-5472.CAN-04-2059. [DOI] [PubMed] [Google Scholar]
- 29.Kershaw MH, Wang G, Westwood JA, et al. Redirecting migration of T cells to chemokine secreted from tumors by genetic modification with CXCR2. Hum Gene Ther. 2002;13:1971–80. doi: 10.1089/10430340260355374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.