Sir,
Isolates from a collection of 177 antibiotic-resistant Acinetobacter recovered at Westmead Hospital, Sydney over the period 1995–20021 were further characterized. The relationships among these isolates had previously been examined by PFGE of ApaI-digested DNA, determination of MICs of several antibiotics and PCR screening to determine whether the blaOXA-23 gene or class 1 integrons were present.1 Eight PFGE pulsotypes were detected. The majority of isolates were A. baumannii but five isolates (PFGE pulsotypes III and IV) were not, and these were not examined here. One or two isolates from each year for each remaining pulsotype were tested using triplex PCRs targeting the oxa-Ab, csuE and ompA genes,2 to determine whether they belong to global clone 1 (GC1) or global clone 2 (GC2). Pulsotype I isolates were GC1 and isolates from pulsotypes VI, VII and VIII, which first appeared in 1999, were GC2. The 54 GC2 isolates were the only isolates resistant to imipenem and carried the blaOXA-23 gene.1 Types II (n = 64) and V (n = 2) did not belong to either global clone. This is consistent with a previous report using a smaller number of isolates.3
Pulsotype I included 52 isolates and was present in the hospital intensive care unit from 1995 to 1999, with a single isolate recovered in 2000. Isolates in this group are among the earliest multiply antibiotic-resistant A. baumannii reported in Australia. They were gentamicin resistant and carried the aacC1-orfP-orfP-orfQ-aadA1 cassette array in a class 1 integron.1 They were also resistant to ciprofloxacin but susceptible to third-generation cephalosporins and carbapenems.1 Consistent with the GC1 designation, WM98, one of two representatives examined previously, was found here to belong to sequence type ST109 [Oxford multilocus sequence typing (MLST) scheme]. Ten representative isolates, including WM98, covering each of the years 1995–99, were examined in more detail, and, in contrast to the previous report,1 WM98 and other members of this group did not include a copy of ISAba1.
The resistance profile was expanded using disc diffusion, and the resistance genes present were determined by PCR.4 The comM gene was interrupted, and they were found to also carry the sul1 sulphonamide resistance gene, the tet(A) tetracycline resistance determinant and transposon Tn6020 carrying the aphA1b kanamycin and neomycin resistance gene. The catA1 and blaTEM genes were also present. PCR mapping5 of WM98 together with sequencing of all IS26 junctions revealed the presence of a genomic resistance island very closely resembling AbaR3 in AB00576 (GenBank accession number CP001182).
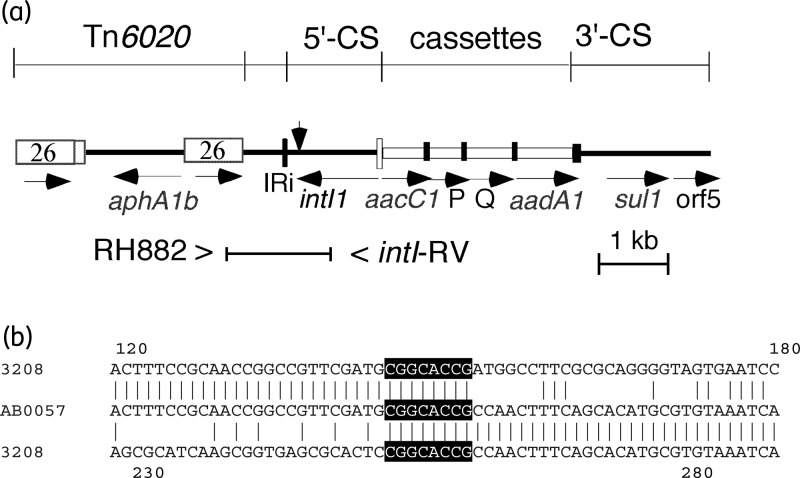
We had noticed that AbaR3 contained a deletion of 108 bp in the 5′-conserved segment (5′-CS) of the class 1 integron, and that this deletion was not present in AbaR5,4 which now has been completely sequenced (GenBank accession number FJ172370), or in AbaR1 in AYE (GenBank accession number CU459141) or in AbaR2 in the GC2 isolate ACICU (GenBank accession number CP000863). The location of this deletion is indicated in Figure 1(a). It removes the last 52 bp of the intI1 gene, and the loss of conserved amino acids in the 16 amino acids replaced at the C-terminus of IntI17 is likely to inactivate it. The deletion appears to have arisen via a rare recombination event or a replication slippage event involving a very short, 8 bp duplication present in the 5′-CS (Figure 1b). Whether the deletion was present in the AbaR of the WM98 group was examined using PCR with primers RH882 (5′-GATGCGTGCACTACGCAAAG-3′) and intI1-RV (5′-GGGCATGGTGGCTGAAGGACC-3′), which generate an amplicon of 1222 bp when the 5′-CS is complete and 1114 bp when the deletion is present and fragments of 659 + 563 or 659 + 455 bp after digestion with BamHI. The 5′-CS is intact in WM98 and related isolates, and this was confirmed by sequencing the PCR amplicon from WM98. This configuration with a complete 5′-CS is likely to pre-date the lineage that includes the deletion, making it the most likely ancestor of AbaR3 and the various AbaR configurations that have arisen subsequently. Therefore, we propose that the AbaR island of WM98 should be named AbaR0. The complete sequence of AbaR0 was assembled8 from the whole genome sequence of WM98 determined using Illumina HiSeq, generating 100 bp paired-end reads, and has been deposited in the GenBank DNA database under accession number KF483599.
Figure 1.
(a) Structure of the part of AbaR islands containing the integron and adjacent Tn6020. The positions of PCR primers used to detect the deletion are indicated below, and the vertical arrow indicates the location of the deletion in the 5′-CS. (b) Alignment of the 3208 and AB0057 sequences in the vicinity of the deletion. Identical bases are indicated by vertical lines and bases that occur twice in 3208 are white on black.
As the deletion in intI1 results from a rare event, it has the potential to assist in tracking different AbaR (i.e. islands in comM with a Tn6019 backbone) and hence GC1 lineages. Therefore its presence in other Australian GC1 isolates was examined. Isolates with an intact 5′-CS and ones with the deletion were both present among these strains. In AbaR21 in the reference GC1 strain RUH875/A297 from 1984,9 as well as in AbaR6 and AbaR7 found in isolates from 2006 and 2005,5 the 5′-CS is intact. In contrast, AbaR8, found in isolates from 2008,10 includes the deletion. Three sporadic isolates from different Australian cities, A85, RBH3 and 6772166, recovered in 2002 or 2003 and described recently as carrying AbaR3,11 included the deletion. This deletion, which can be simply detected by PCR, will serve as an additional marker for diverged lineages within the GC1 clone.
Funding
This study was supported by NHMRC Project Grant APP1026189 and Wellcome Trust grant number 098051. M. H. was supported by a University of Sydney Postgraduate Research Award. K. E. H. was supported by an NHMRC PostDoctoral Fellowship (no. 628930).
Transparency declarations
None to declare.
Acknowledgements
We thank Dr Jon Iredell, Westmead Hospital, Sydney for kindly supplying the isolates used in this study.
References
- 1.Valenzuela JK, Thomas L, Partridge SR, et al. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J Clin Microbiol. 2007;45:453–60. doi: 10.1128/JCM.01971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turton JF, Gabriel SN, Valderrey C, et al. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13:807–15. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 3.Eijkelkamp BA, Stroeher UH, Hassan KA, et al. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett. 2011;323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 4.Post V, Hall RM. AbaR5, a large multiple antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:2667–71. doi: 10.1128/AAC.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;16:1162–70. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 6.Adams MD, Goglin K, Molyneaux N, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008;190:8053–64. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collis CM, Kim M-J, Stokes HW, et al. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol Microbiol. 2002;46:1415–27. doi: 10.1046/j.1365-2958.2002.03260.x. [DOI] [PubMed] [Google Scholar]
- 8.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigro SJ, Post V, Hall RM. The multiresistant Acinetobacter baumannii European clone I type strain RUH875 (A297) carries a genomic antibiotic resistance island AbaR21, plasmid pRAY and a cluster containing ISAba1-sul2-CR2-strB-strA. J Antimicrob Chemother. 2011;66:1928–30. doi: 10.1093/jac/dkr213. [DOI] [PubMed] [Google Scholar]
- 10.Post V, Hamidian M, Hall RM. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J Antimicrob Chemother. 2012;67:1039–40. doi: 10.1093/jac/dkr586. [DOI] [PubMed] [Google Scholar]
- 11.Hamidian M, Hall RM. Tn6168, a transposon carrying an ISAba1-activated ampC gene and conferring cephalosporin resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2014;69:77–80. doi: 10.1093/jac/dkt312. [DOI] [PubMed] [Google Scholar]