Abstract
DNA replication stress is an inefficient DNA synthesis process that leads replication forks to progress slowly or stall. Two main factors that cause replication stress are alterations in pools of deoxyribonucleotide (dNTP) precursors required for DNA synthesis and changes in the activity of proteins required for synthesis of dNTPs. Ribonucleotide reductase (RNR), containing regulatory hRRM1 and catalytic hRRM2 subunits, is the enzyme that catalyzes the conversion of ribonucleoside diphosphates (NDPs) to deoxyribonucleoside diphosphates (dNDPs) and thereby provides dNTP precursors needed for the synthesis of DNA. Here, we demonstrate that either endogenous or exogenous expression of Bcl2 results in decreases in RNR activity and intracellular dNTP, retardation of DNA replication fork progression and increased rate of fork asymmetry leading to DNA replication stress. Bcl2 co-localizes with hRRM1 and hRRM2 in the cytoplasm and directly interacts via its BH4 domain with hRRM2 but not hRRM1. Removal of the BH4 domain of Bcl2 abrogates its inhibitory effects on RNR activity, dNTP pool level and DNA replication. Intriguingly, Bcl2 directly inhibits RNR activity by disrupting the functional hRRM1/hRRM2 complex via its BH4 domain. Our findings argue that Bcl2 reduces intracellular dNTPs by inhibiting ribonucleotide reductase activity, thereby providing insight into how Bcl2 triggers DNA replication stress.
Keywords: Bcl2, Ribonucleotide reductase, Deoxyribonucleotide, DNA replication stress
Introduction
High-fidelity DNA replication and its accurate segregation to daughter cells are critical for maintaining genome stability and suppressing cancer (1). DNA replication is tightly coordinated with the increased cellular mass during the cell cycle. The kinetics of DNA replication depends on two fundamental parameters: the frequency of origin activation (initiation) and the rate of DNA synthesis at the replication forks (elongation). DNA synthesis is initiated at specific sites on the chromosome termed origins of replication, and proceeds bidirectionally to elongate and duplicate the chromosome. The processes of initiation and elongation determine the total rate of DNA synthesis in the cell. A multiprotein replication factory incorporates deoxyribonucleotides (dNTPs) into the growing DNA chain. The enzyme ribonucleotide reductase (RNR) is responsible for the reduction of nucleoside diphosphates (NDPs) to their corresponding deoxynucleoside diphosphates (dNDPs) and plays a key role in synthesizing the necessary dNTPs (2, 3), which are then used in the synthesis of DNA during replication or DNA repair (4). RNR is composed of identical large regulatory subunits (hRRM1) and identical small catalytic subunits (hRRM2) (4). The active complex is a mixture of α2β2 and α6βn (5, 6). Interestingly, it has also been reported that the radical is stored in the hRRM2 subunits and is relocated to the substrate, and that hRRM1 can also be regarded as the catalytic subunit with redox-active thiols and substrate binding (4). hRRM2 is expressed exclusively during the late G1/early S phase and is degraded in late S phase (7). RNR activity plays an important role not only in regulating the kinetics of DNA replication but also in maintaining the integrity of the genome. Synthesis of dNTPs by RNR allows for DNA replication to begin. Defective RNR activity often leads to inhibition of DNA replication fork progression, cell cycle arrest, growth retardation, high mutation rates and carcinogenesis (8, 9).
DNA replication stress results from slow or stalled DNA replication fork progression and/or asymmetrical fork elongation. There are numerous causes of DNA replication stress and replication stress-induced DNA damage and genetic instability (10). These include an insufficient level of dNTPs and enzymes required for DNA synthesis, decreased frequency with which initiation of DNA replication occurs at origins of replication (resulting in larger replicons), hyper-DNA replication caused by the repeated activation of origins more than once per S phase, DNA damage lesions that block replication forks, and inhibition of DNA replication by DNA damaging agents or RNR inhibitors. Replication stress also occurs in regions of DNA that are intrinsically difficult to replicate due to secondary structures or that are difficult to unwind during DNA replication. Proteins bound to DNA can also cause replication forks to pause, thus causing replication stress.
Bcl2, a major antiapoptotic and oncogenic protein, mediates inhibitory effects on the G1/S cell cycle transition and DNA repair leading to increased genetic instability(11, 12). Overexpression of Bcl2 results in lymphomagenesis in transgenic mice, suggesting that Bcl2, in addition to its pro-survival effect, may also promote oncogenic events (13). However, the mechanism(s) by which Bcl2 facilitates oncogenesis is not fully understood. The oncogenic effect of Bcl2 may result from its ability to influence multiple cell operational systems. Bcl2 was originally discovered as a gene product at the chromosomal breakpoint of t(14;18) (14), which may promote genetic instability and tumor development by impeding DNA repair or replication. DNA replication stress is a major mechanism through which spontaneous DNA damage and genetic instability (i.e. chromosome breaks) are generated in cells (10, 15). Our previous studies have demonstrated that Bcl2 strongly suppresses G1/S cell cycle progression and increases chromosomal/chromatid breaks (11, 12, 16). It is currently unknown whether and how Bcl2 affects DNA replication fork progression to induce DNA replication stress. In the present study, we demonstrate that expression of Bcl2 retards DNA replication fork progression with increased fork asymmetry, leading to DNA replication stress. The inhibitory effect of Bcl2 on DNA replication fork progression occurs through direct inhibition of RNR activity and subsequent lowering of intracellular dNTP levels.
Materials and Methods
Materials
Bcl2, hRRM1, hRRM2 and p53R2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-BrdU clone B44 was purchased from Becton Dickinson (San Jose, CA). Rat anti-BrdU BU1/75(ICR1) was purchased from Novus Biologicals (Littleton, CO). Purified, recombinant WT and BH deletion Bcl2 mutant proteins were obtained from ProteinX Lab (San Diego, CA). Purified hRRM2 was purchased from Abcam (Cambridge, MA). Purified hRRM1 was purchased from OriGene (Rockville, MD). Boronate resins were obtained from Thermo scientific (Rockford, IL). QD605 goat anti-rabbit IgG conjugate (red), QD705 goat anti-mouse IgG conjugate (green), Alexa Fluor 488 (green) goat anti-mouse and Alexa Fluor (red) 555 goat anti-rat were purchased from Invitrogen Life Technologies Inc (Carlsbad, CA). C14-CDP and dC14-CDP were obtained from Moravek Biochemicals, Inc (Brea, CA). AMP-PNP, CDP, CMP, dCMP and dUMP were purchased from Sigma Aldrich (St. Louis, MO). TLC Silica Plate was purchased from EMD Chemicals (Gibbstown, NJ). All of the reagents used were obtained from commercial sources unless otherwise stated.
Cell lines, plasmids and transfections
Normal human bronchial epithelial cell line (BEAS-2B), H1299 and H460 lung cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). WT and Bcl2 BH deletion mutants were created and stably expressed in H1299 or BEAS-2B cells as previously described (12). The expression levels of exogenous Bcl2 were analyzed by Western blot analysis, and three separate clones expressing similar amounts of exogenous Bcl2 were selected for further analysis.
DNA molecular combing
DNA fiber spreads were performed as described (17, 18). Briefly, cells were first labeled with 5’ chlorodeoxyuridine (CldU, 100 μM) 20 min. After washing, cells were then labeled with iododeoxyuridine (IdU, 100 μM) for another 20 min. 2 μl of cell suspension (106 cells/ml) in cold PBS were spotted onto a microscope slide and mixed with 12 μl lysis buffer (0.5% SDS, 50 mM EDTA, 200 mM Tris-Cl) for 10 min at room temperature. Samples were carefully tilted at a 15° angle to allow spreading of the genomic DNA into single molecule DNA fibers by gravity. Fibers were then fixed with methanol and acetic acid (3:1) buffer. Fibers were subsequently acid treated with HCl (2.5 M) to denature the DNA fibers. Slides were neutralized and washed four times with 1×PBS (pH 8.0 PBS one time, pH 7.4 PBS three times). Slides were blocked with 10% goat serum and 0.1% Triton-X in PBS for at least 1 hr, and then incubated with primary antibodies against IdU (mouse anti-BrdU clone B44) and CldU (rat anti-BrdU BU1/75(ICR1)) and secondary antibodies (Alexa Fluor 488 (green) goat anti-mouse and Alexa Fluor (red) 555 goat anti-rat) for 1 hr. Slides were mounted in Prolong with DAPI and scanned with a Zeiss Axioplan2 upright microscope (Axioplan, Zeiss). Images were analyzed by Zeiss AxioVision software. The lengths of CIdU (red) and IdU (green) labeled patches were analyzed using the ImageJ software (19). The fork rate (kb/min) was calculated from the length of fluorescent signal (kb) divided by the time of the pulse. The median replication rates, inter-origin distances and p-values derived from Mann-Whitney U test were computed with Prism v5.0. Representative images of DNA fibers were cropped from different fields of view and assembled with Adobe Photoshop, as described previously (20).
Nucleotide pool analysis
Levels of cellular deoxynucleotides were analyzed as described (21, 22). Briefly, cells were harvested, and cellular nucleotides were extracted with 0.4 N perchloric acid and neutralized with potassium hydroxide. Deoxynucleotides were separated from ribonucleotides using a boronate affinity column (21) Deoxynucleotides were analyzed by HPLC using UV absorbance at 254 and 281 nm for identification and quantification as previously described (22, 23). All data were plotted using the GraphPad Prism v 5.0 program (GraphPad software).
Ribonucleotide reductase activity assay
Ribonucleotide reductase activity was analyzed as described (24, 25). Briefly, cells were harvested and washed with 1×PBS. Low salt homogenization buffer (10 mM Hepes, 2 mM DTT, pH 7.2) was added to the cell pellets. After homogenization with a 27G1/2 syringe needle, cell debris was removed by centrifugation at 16,000 g at 4°C for 20 min. The supernatant was passed through a Sephadex G25 spin column. 600 μg of protein was added to a 40 μl reaction mixture (50 mM Hepes buffer, pH 7.2, 10 mM DTT, 4 mM AMP-PNP, 20 μM FeCl3, 2 mM magnesium acetate, 50 μM CDP and 100 μM C14-CDP) and incubated at 37°C for 1 h. Then, 4 μl of 10 M perchloric acid was added for 15 min on ice. After centrifugation, the supernatant was transferred to a new tube and boiled for 20 min. 4 μl of a marker solution (60 mM CMP, 60 mM dCMP, and 60 mM dUMP plus 12 μl 5 M KOH) was added and then incubated on ice for 15 min. Samples were centrifuged at 14,000 rpm for 5 min. The resulting supernatant containing nucleotides was spotted on a TLC plate and separated by thin-layer chromatography. TLC plates were analyzed with quantification using the variable scanner Typhoon 9210 (GE health) (26). All data were plotted using the GraphPad Prism v 5.0 program. RNR activity was calculated by 14C-dCDP/(14C -CDP+14C-dCDP).
Bcl2 silencing
Bcl2 shRNA and control shRNA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Hairpin sequence of Bcl2 shRNA: GAT CCG TGT GGA TGA CTG AGT ACC TGA TTC AAG AGA TCA GGG ACT CAG TCA TCC ACA TTT TTG. Hairpin sequence of control shRNA: GAT CCG GAA CGG CAT CAA GGT GAA CTT CAA GA GAG TTC ACC TTG ATG CCG TTC TTT TTG. For pseudovirus production, Bcl2 shRNA or control shRNA was cotransfected into 293FT cells with a lentivirus packaging plasmid mixture (System Biosciences, CA) using the Nanojuice transfection kit (EMD Chemical, Inc.) as described (27). After 48h, the virus-containing media were harvested by centrifugation at 20,000 × g. H460 cells were infected with the virus-containing media in the presence of polybrene (8 μg/ml) for 24h. Stable positive clones were selected using 1 μg/ml puromycin. Specific silencing of the targeted Bcl2 gene was confirmed by at least three independent experiments.
Statistical analysis
Significant differences between two groups were analyzed using Mann Whitney test or two-sided unpaired Student’s t-test. A p value < 0.05 was considered statistically significant.
Results
Expression of endogenous Bcl2 is associated with decreased levels of RNR activity and intracellular dNTPs
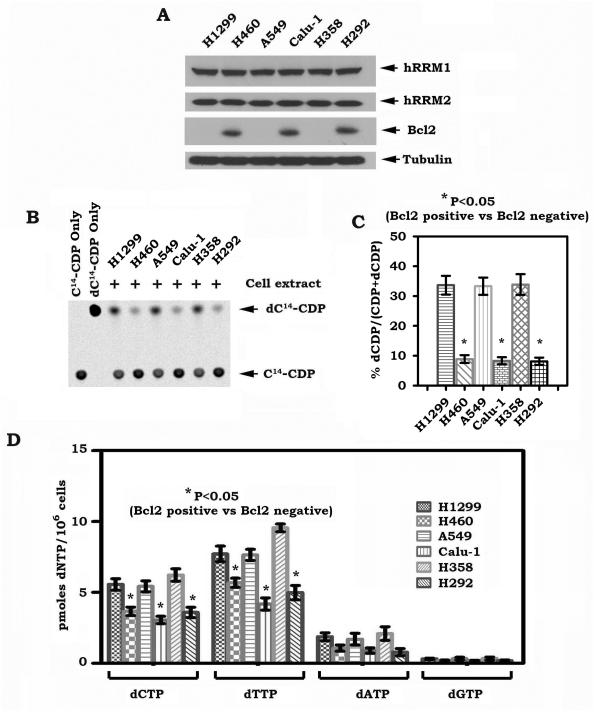
Bcl2 has been reported to delay DNA synthesis and DNA replication in vitro and in vivo (28). The mechanism of which is not fully understood. RNR is the “rate limiting” enzyme in the de novo dNTP synthesis pathway, which is critical for synthesizing the necessary dNTPs (2), which are required for normal DNA replication in mammalian cells (29, 30). Bcl2 may negatively regulate RNR to alter intracellular dNTPs levels. To test this possibility, RNR activity and cellular dNTP pools were measured in human lung cancer cells that express various levels of endogenous Bcl2 as described in “Methods”. Although expression of endogenous Bcl2 did not affect expression levels of hRRM1 and hRRM2 (Fig.1A), significant decreases in RNR activity and intracellular dNTPs were observed in H460, Calu-1 and H292 cells that express high levels of endogenous Bcl2 as compared to the other cell lines that express undetectable levels of Bcl2 (Fig. 1B, C and D). These findings suggest that expression of endogenous Bcl2 appears to be correlated with reduced levels of RNR activity and dNTPs in human lung cancer cells, which may negatively regulate DNA synthesis during DNA replication progression.
Figure 1.
Expression of endogenous Bcl2 is correlated with decreased levels of RNR activity and intracellular dNTPs. A, expression levels of Bcl2, hRRM1 and hRRM2 were analyzed by Western blot. B-C, extracts from various human lung cancer cells were incubated with 14C-CDP. The generation of 14C-dCDP was analyzed by a phosphorImager on TLC. The relative abundance of 14C-dCDP and 14C-CDP was quantified with imageQuant software. RNR activity was calculated by a formula as 14C-dCDP/(14C-CDP+14C-dCDP). Error bars represent ±SD of three repeated determinations. D, intracellular levels of dNTPs in various human lung cancer cells were measured. Error bars represent ± SD of three repeated determinations.
Bcl2 reduces intracellular dNTP levels by direct inhibition of RNR activity leading to retardation of DNA replication fork progression with increased fork asymmetry
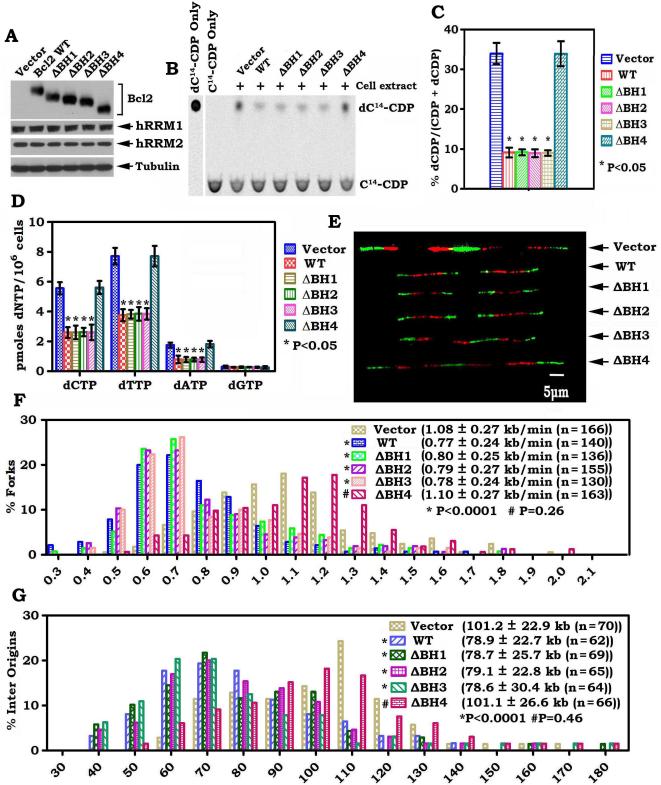
To further test the role of Bcl2 in regulating RNR activity and dNTP pool size, exogenous Bcl2 was stably overexpressed in H1299 cells (Fig. 2A). Overexpression of Bcl2 in H1299 resulted in a dramatic 4-fold decrease in RNR activity compared to vector-only control cells (Fig. S1A, B). To test whether Bcl2 directly inhibits RNR activity, purified recombinant Bcl2 protein was added directly to a reaction mixture containing RNR and substrates. Intriguingly, Bcl2 directly inhibited RNR activity in a dose-dependent manner (Fig. S1C, D). HPLC analysis of cellular dNTP pools revealed that exogenous overexpression of Bcl2 in H1299 cells resulted in decreased levels of all four dNTPs (Fig. S1E), indicating that the dNTP synthesis pathway is disturbed by Bcl2 overexpression. These findings proposed that Bcl2-mediated reduction of intracellular dNTPs may occur through direct suppression of RNR activity.
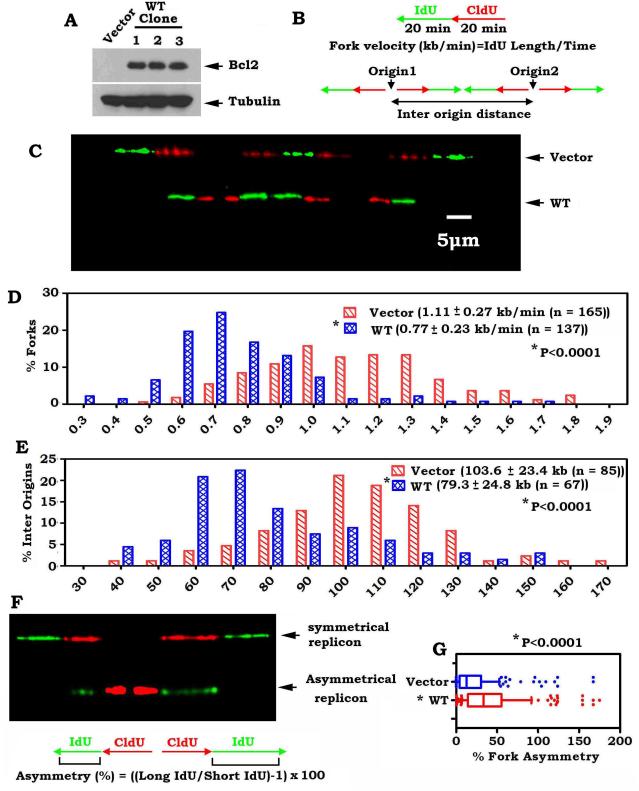
Figure 2.
Bcl2 suppresses DNA replication fork progression. A, Bcl2 was stably expressed in H1299 human lung cancer cells. B, schematic diagram for calculation of the fork rate (Top) and inter origin distance (Bottom) of replication fork progression. C, H1299 cells expressing Bcl2 or vector-only control were pulsed-labeled with 100 μM CldU for 20 min and 100 μM IdU for another 20 min. The labeled cells were processed for DNA combing. Representative pairs of sister replication forks are shown. Red, CldU. Green, IdU. Bar, 5 μm (1 μm = 2.59 kb). D, distribution of fork rate (kb/min) in H1299 cells expressing Bcl2 or vector-only control. The mean±SD for fork rate and number of scores are summarized. The p value was determined by a two-tailed Mann-Whitney test. E, distribution of inter origin distances (kb) were compared in H1299 cells expressing Bcl2 or vector-only control. The mean ±SD for distances and number of scores are summarized. F, representative images of symmetrical and asymmetrical replication. G, distribution of the degree of asymmetry of bidirectional replication forks was compared in H1299 cells expressing Bcl2 or vector-only control.
Since the dNTP pool size determines fork progression and origin usage under replication stress (22, 29), Bcl2-mediated reduction in RNR activity and dNTP pools may affect DNA replication fork progression. To test this possibility, we examined replication dynamics using single-molecule DNA fiber analysis in H1299 cells stably expressing Bcl2 or vector-only control as shown in Figure 2. This technique measures the speed of individual replication fork progression as well as the frequency of replication initiation events. The rate of individual replication fork progression was calculated by dividing the length of each fluorescent segment (1μm=2.59 kb) by the time of pulse (20 min), as previously described (31) (Fig. 2B). Newly synthesized DNA labeled with CldU (red) and IdU (green) was detected with fluorescent antibodies (Fig. 2C). A pronounced and reproducible decrease in the mean replication fork progression rate was observed in H1299 cells expressing Bcl2 (0.77 ± 0.23 kb/min, n=137) when compared to the vector-only control H1299 cells (1.11 ± 0.27 kb/min, n=165) (p< 0.0001; Fig. 2C, D). A dramatic increase in the percentage of slowly progressing forks was observed in the Bcl2-expressing cells (Figure 2D). These results reveal that expression of Bcl2 significantly reduces the rate of DNA-replication fork progression.
To test the effect of Bcl2 on the origin density, we measured the distance between two adjacent origins as described (22). These experiments revealed that the mean origin distance in vector-only control cells was 103.6 ± 23.4 Kb (n = 85), whereas in the Bcl2-expressing cells, it was significantly shorter, only 79.3 ± 24.8 Kb (n = 67) (p < 0.0001) (Fig. 2E). This decrease is expected from the observed decreased fork rate, as fork rate and inter-origin distances show a linear correlation (31, 32). A dramatically increased population with short origin distance was observed in the Bcl2-expressing cells (Fig. 2E). These results indicate that Bcl2 expression leads to activation of an increased number of origins.
It is known that the two replication forks emanating from the same origin should exhibit the same replication rate (31). However, under DNA replication stress, one fork may continue to replicate, whereas the other fork may slow down or stall. Thus, asymmetry between the left and right arms may occur. To test whether Bcl2 induces asymmetric progression of the outgoing forks, we compared the progression of the left and the right forks emerging from the same origin in the Bcl2-expressing cells and vector-only control cells (Fig. 2F). Only 22.8% of replication forks displayed asymmetric dynamics in the vector-only control cells. In contrast, 42.3% of the origins showed asymmetric replication progression in H1299 cells expressing Bcl2 (Fig. 2F, G). Collectively, these replication dynamics results reveal that DNA replication is significantly perturbed in the Bcl2-expressing cells. Similar results were also obtained using a normal human bronchial epithelial cell line (i.e. BEAS-2B) (Fig. S2), indicating that the role of Bcl2 in regulating RNR activity, dNTPs, DNA replication rate, origin distance and fork asymmetry is not a cell type-specific phenomenon.
Bcl2 co-localizes with hRRM1 and hRRM2, and directly interacts with hRRM2 via the BH4 domain
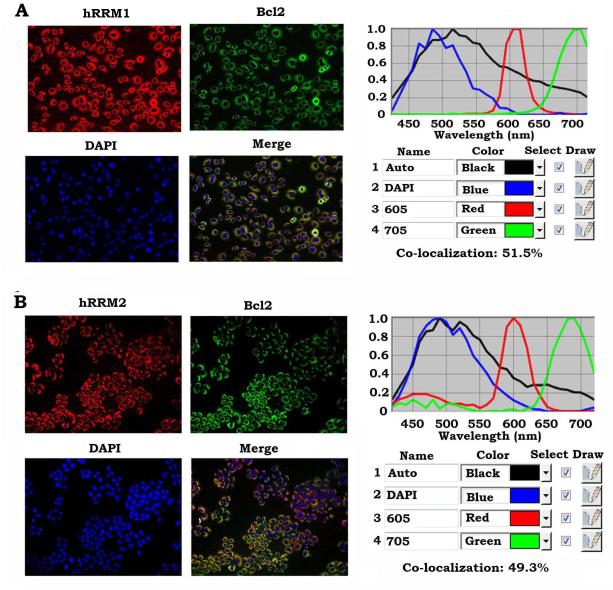
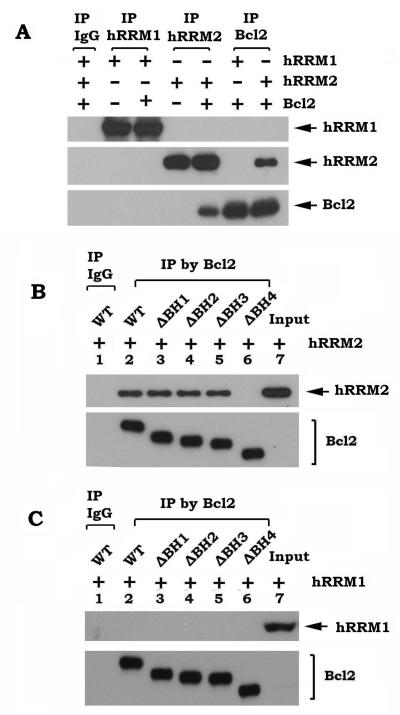
To elucidate the mechanism by which Bcl2 inhibits RNR activity, first, co-localization between Bcl2 and hRRM1 or hRRM2 was measured by quantum dot (QD)-based immunofluorescence (QD-IF) analysis in H460 cells that express high levels of endogenous Bcl2. QDs combined with an interrelated imaging and quantification system can be used to quantify the localization and/or co-localization of multiple proteins in the same sample. QD-based image analysis showed that Bcl2 is co-localized with hRRM1 (51.5%) and hRRM2 (49.3%) in the cytoplasm (Fig. 3A, B). To further test whether Bcl2 directly interacts with hRRM1 or hRRM2, purified recombinant Bcl2 protein was incubated with purified recombinant hRRM1 or hRRM2, respectively, in 1% CHAPS lysis buffer at 4°C for 2h. A co-immunoprecipitation (co-IP) was performed using Bcl2, hRRM1 or hRRM2 antibody, respectively. IgG was used as IP control. The co-IP results revealed that Bcl2 preferentially interacts with hRRM2 but not hRRM1 (Fig. 4A).
Figure 3.
Quantum dot-based immunofluorescence (QD-IF) analysis of co-localization of Bcl2 and RNR. Co-localization of Bcl2/hRRM1 (A) and Bcl2/hRRM2 (B) was measured by QD-IF in H460 cells expressing high levels of endogenous Bcl2, hRRM1 and hRRM2 as described in “Methods”. Quantification of co-localization was analyzed with Nuance imaging software (Caliper/PerkinElmer).
Figure 4.
Bcl2 directly interacts with hRRM2 via the BH4 domain. A, purified WT Bcl2 (20 ng/ml) was incubated with purified hRRM1 or hRRM2 (20 ng/ml) in 1% CHAPS lysis buffer at 4°C for 2h. Co-immunoprecipitation (co-IP) was performed using Bcl2, hRRM1 or hRRM2 antibody, respectively. Bcl2, hRRM1 and hRRM2 were analyzed by Western blot. B-C, purified WT or each of the BH deletion Bcl2 mutant proteins (20 ng/ml) was incubated with 20 ng/ml of purified hRRM2 (B) or hRRM1 (C) in 1% CHAPS lysis buffer at 4°C for 2h. Co-IP was performed using agarose-conjugated Bcl2 antibody. Bcl2, hRRM1 and hRRM2 were analyzed by Western blot.
Bcl2 family members share homology in the Bcl2 homology (BH) domains, including BH1, BH2, BH3, and BH4 (33). To determine the hRRM2 binding site on Bcl2 protein, co-IP experiments were performed using purified hRRM2 and purified recombinant WT Bcl2 or a series of Bcl2 deletion mutants lacking each of the BH domains (i.e. δBH1, δBH2, δBH3 and δBH4). Since hRRM2 but not hRRM1 was able to associate with WT, δBH1, δBH2 and δBH3 Bcl2, but not with the δBH4 Bcl2 mutant protein (Fig. 4B, C), this indicates that deletion of the BH4 domain ablated Bcl2’s ability to associate with hRRM2. Based on these findings, we conclude that the BH4 domain is the hRRM2 binding site on Bcl2.
p53R2 has been identified as a second radical-providing small subunit in mammalian cells (34). The major role of p53R2 is involved in regulating synthesis, replication and repair of mitochondrial DNA (mtDNA) in quiescent cells (35, 36). Interestingly, co-IP experiments show that Bcl2 can also interact with p53R2 in H1299 cells (Fig. S3).
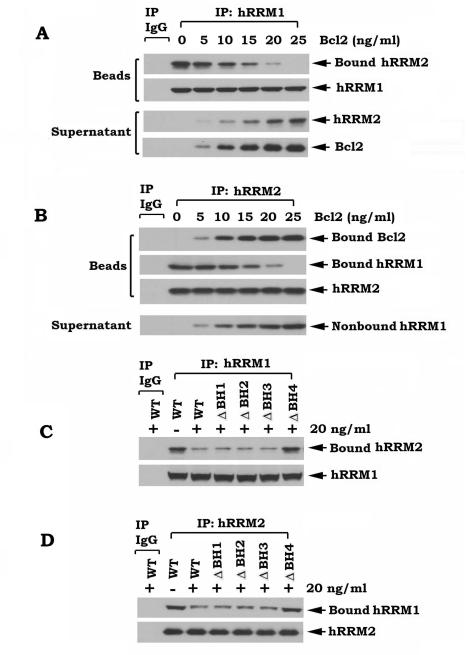
Bcl2 disrupts the functional hRRM1/ hRRM2 complex in vitro and in vivo, which requires the hRRM2 binding site on Bcl2
It is known that hRRM1 subunits are bound to hRRM2 subunits leading to formation of the functional RNR complex (24, 30). Since Bcl2 can directly interact with hRRM2 via its BH4 domain (Fig. 4), Bcl2 may affect the functional hRRM1/hRRM2 complex by binding to hRRM2. To directly test this, the hRRM1/hRRM2 complex was co-immunoprecipitated from H1299 parental cells. The immune complex was incubated with increasing concentrations of purified, recombinant Bcl2 at 4°C for 1–2 h, and proteins released from the complex were identified in the supernatant after centrifugation. Interestingly, addition of purified Bcl2 to the hRRM1 co-IP complex resulted in decreased levels of bound hRRM2 on beads and increased levels of nonbound hRRM2 in the supernatant in a dose-dependent manner (Fig. 5A). This indicates that Bcl2 is able to dissociate hRRM2 from the hRRM1 co-IP complex. Similarly, Bcl2 dissociates hRRM1 from the RRM2 co-IP complex (Fig. 5B). These findings provide insight into the mechanism by which Bcl2 inhibits RNR activity (Fig. S1A-D). To further test whether the hRRM2 binding site on Bcl2 is required for Bcl2 disruption of the hRRM1/hRRM2 complex, WT or each of the Bcl2 BH domain deletion mutant proteins were subjected to similar experiments. Intriguingly, WT, δBH1, δBH2 or δBH3, but not the δBH4 Bcl2 mutant protein could disrupt the hRRM1/hRRM2 complex (Fig. 5C, D). To test whether overexpression of Bcl2 also affects functional hRRM1/hRRM2 association in cells, co-IP experiments using hRRM1 or hRRM2 antibody, respectively, were performed in lysate isolated from H1299 cells expressing WT or each of the BH deletion Bcl2 mutants. Overexpression of Bcl2 did not affect expression levels of either hRRM1 or hRRM2 (Fig. S4A), but significantly inhibited functional hRRM1/hRRM2 association in cells (Fig. S4B, C). Similarly, the disruptive effect of Bcl2 on the hRRM1/hRRM2 complex in cells requires the BH4 domain (i.e. the hRRM2 binding site on Bcl2).
Figure 5.
Bcl2 directly disrupts the hRRM1/hRRM2 complex, which requires the BH4 domain. A-B, the hRRM1/hRRM2 complex was co-immunoprecipitated using hRRM1 or hRRM2 antibody, respectively, from H1299 vector-only cells, and incubated with increasing concentrations of purified WT Bcl2 in 1% CHAPS lysis buffer at 4°C for 2h. After centrifugation, the resulting supernatant or immunocomplexed beads were subjected to SDS-PAGE. Bcl2, hRRM1 and hRRM2 on beads or in supernatant were analyzed by Western blot. C-D, the hRRM1/hRRM2 complex was obtained as above and incubated with purified WT or each of the BH deletion Bcl2 mutant proteins in 1% CHAPS lysis buffer at 4°C for 2h. After centrifugation and washing, the immunocomplexes on beads were subjected to SDS-PAGE and analyzed by Western blot to measure hRRM1-associated hRRM2 and hRRM2-associated hRRM1.
The BH4 domain is essential for the inhibitory effects of Bcl2 on RNR activity, intracellular dNTPs and DNA replication dynamics
To test the functional role of the Bcl2/hRRM2 interaction, RNR activity, intracellular dNTP levels and DNA replication dynamics were measured in H1299 cells expressing similar levels of WT or each of the BH deletion Bcl2 mutants. These experiments revealed that removal of the BH4 domain from Bcl2 eliminates its inhibitory effect on RNR activity, intracellular dNTP levels and DNA replication fork progression (Fig. 6). In contrast, deletion of the BH1, BH2 or BH3 domain did not affect the ability of Bcl2 to suppress RNR activity, synthesis of dNTPs and DNA replication fork progression (Fig. 6). Additionally, deletion of the BH4 domain resulted in a decrease in the population of asymmetrical replication forks as compared to that with WT or other Bcl2 BH domain deletion mutants (Fig. S5). This supports the concept that the binding of Bcl2 to hRRM2 via the BH4 domain is required for the inhibitory effects of Bcl2 on RNR activity, dNTP levels and DNA replication dynamics.
Figure 6.
The BH4 domain is essential for Bcl2 to inhibit RNR activity, dNTPs synthesis and DNA replication fork progression. A, WT or each of the BH deletion Bcl2 mutants was stably expressed in H1299 cells. Expression of Bcl2, hRRM1 and hRRM2 were analyzed by Western blot. B-D, RNR activity and dNTP pools in H1299 cells expressing WT or each of the BH deletion Bcl2 mutants were measured and quantified as described in the legend of figure 1. Error bars represent ± SD of three repeated determinations. E, representative pairs of sister replication forks from H1299 cells expressing WT or each of the BH deletion Bcl2 mutants are shown. F-G, distributions of fork rate (kb/min) or inter origin distance in H1299 cells expressing WT or each of the BH deletion Bcl2 mutants were analyzed as described in the legend of figure 2.
To uncover a functional relationship between the antiapoptotic activity of Bcl2 and its role in inducing DNA replication stress, the effect of WT or each of the BH deletion Bcl2 mutants on survival was analyzed after VP-16 treatment. Interestingly, all H1299 cells expressing the δBH1, δBH2, δBH3, or δBH4 Bcl2 mutant displayed a lower level of viability than WT Bcl2 expressing cells (Fig. S6). These results indicate that all of the BH domains in Bcl2 are required for Bcl2’s full and potent antiapoptotic function, which is consistent with previous reports (12, 37, 38). However, only the BH4 domain is essential for the effect of Bcl2 on RNR activity and DNA replication (Fig. 6), suggesting that the antiapoptotic function of Bcl2 may not be required for Bcl2-mediated DNA replication stress.
Knockdown of endogenous Bcl2 leads to enhanced RNR activity and accelerated DNA replication fork progression
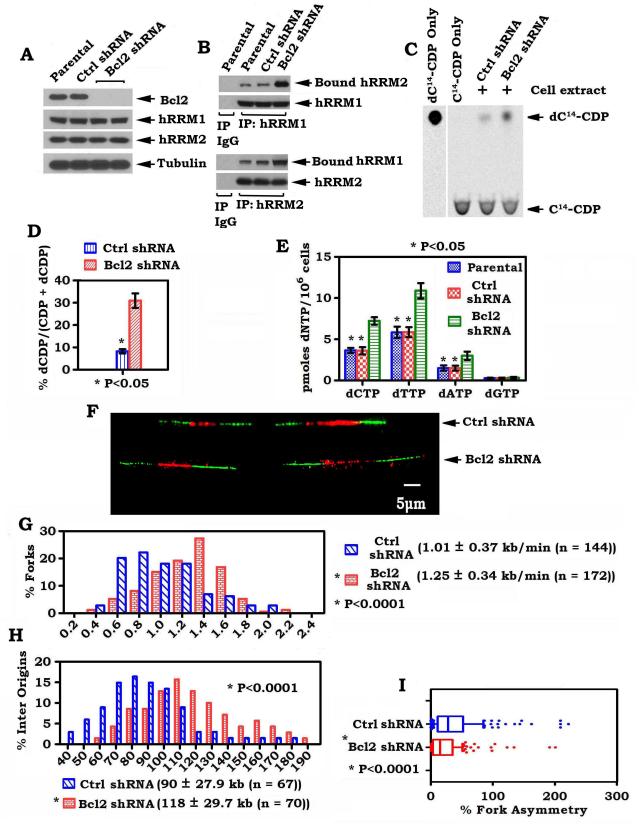
To test the role of endogenous Bcl2 in regulating RNR activity and DNA replication dynamics, Bcl2 was depleted by RNA interference (RNAi), using Bcl2 shRNA, from H460 cells that express elevated levels of endogenous Bcl2. Stable expression of Bcl2 shRNA significantly reduced the expression level of endogenous Bcl2 by more than 99% in H460 cells (Fig. 7A). This effect of shRNA on Bcl2 expression was highly specific because the control shRNA had no effect. Importantly, depletion of endogenous Bcl2 by RNAi from H460 cells enhanced the formation of functional hRRM1/hRRM2 complexes in association with increased RNR activity leading to elevated levels of intracellular dNTPs and accelerated DNA replication fork progression with decreased fork asymmetry (Fig. 7B-I). These results reveal a physiological role for Bcl2 in suppressing DNA replication fork progression through a mechanism involving RNR-mediated dNTP synthesis.
Figure 7.
Knockdown of endogenous Bcl2 upregulates RNR activity and intracellular levels of dNTPs, and accelerates DNA replication fork progression. A, Bcl2 shRNA or control shRNA was stably transfected in H460 cells. Expression levels of Bcl2, hRRM1and hRRM2 were analyzed by Western blot. B, H460 cells expressing Bcl2 shRNA or control shRNA were disrupted in 1% CHAPS lysis buffer. Co-IP was performed using hRRM1 or hRRM2 antibody, respectively. The hRRM1-associated hRRM2 and hRRM2-associated hRRM1 were analyzed by Western blot. C-E, RNR activity and dNTP pool in H460 cells expressing Bcl2 shRNA or control shRNA were analyzed as described in the legend of Figure 1. Error bars represent ±SD of three repeated determinations. F, representative pairs of sister replication forks from H460 cells expressing Bcl2 shRNA or control shRNA are shown. G-I, distributions of fork rate (kb/min), inter origin distance or asymmetry in H460 cells expressing Bcl2 shRNA or control shRNA were analyzed as described in the legend of figure 2.
Discussion
DNA replication stress can cause DNA damage and genetic instability. Insufficient levels of dNTPs and RNR activity required for DNA synthesis are major factors in the induction of DNA replication stress (10). RNR is the enzyme that catalyzes the conversion of NDPs to dNDPs, and phosphorylation of dNDPs by NDP kinase then yields dNTPs (2, 39, 40). The dNTPs, in turn, are used in the synthesis of DNA. The dynamics and fidelity of DNA replication are mainly dependent on the rate of RNR-mediated DNA synthesis at the replication forks (2). Here we demonstrate that Bcl2, a founding anti-apoptotic member of the Bcl2 family and an oncoprotein, strongly suppresses RNR activity resulting in reduction of intracellular dNTP levels, decreased DNA replication fork progression and increased rate of fork asymmetry leading to DNA replication stress (Figs. 1, 2, S1 and S2). These findings provide the basis for a model of oncoprotein-induced DNA replication stress. Because DNA replication stress can enhance DNA damage and genetic instability under certain conditions, the effect of Bcl2 on RNR and dNTP pool may be an early event of oncogenesis. Thus, our results reveal a new mechanism by which Bcl2 promotes early stage of cancer development (13).
It is known that a low nucleotide pool level in cells can result in DNA replication stress (22). RNR is the “rate limiting “enzyme in the de novo dNTP synthesis pathway, which is critical in regulating intracellular dNTP levels. Because expression of endogenous or exogenous Bcl2 in cells or addition of purified Bcl2 protein can directly inhibit RNR activity (Figs. 1, 2, S1 and S2), Bcl2 reduction of intracellular dNTP levels is likely to occur through inhibition of RNR activity. Endogenous Bcl2-induced reduction of RNR activity may slow down but not totally block DNA replication. A compensatory mechanism may also exist by regulating cytosolic deoxycytidine kinase or thymidine kinase 1 to ensure a basic rate of DNA replication in cells expressing endogenous Bcl2, as described (41). It is known that both hRRM1 and hRRM2 are localized in the cytoplasm (42). Ribonucleotide reduction by the hRRM1/hRRM2 complex takes place in the cytoplasm in mammalian cells. The resulting dNTPs diffuse into the nucleus or are transported into mitochondria for DNA synthesis to support DNA replication (42). Although Bcl2 was found to partially co-localize with hRRM1 (51.5%) and hRRM2 (49.3%) in the cytoplasm (Fig. 3), Bcl2 interacts selectively only with hRRM2 (Fig. 4). Co-IP experiments employing Bcl2 mutant proteins deleted in the BH domains revealed that Bcl2 directly interacts with hRRM2 via its BH4 domain but not the BH1, BH2 or BH3 domain (Fig. 4B, C), indicating that the BH4 domain is the hRRM2 binding site on the Bcl2 protein. It is known that formation of the hRRM1/hRRM2 complex is required for RNR activity in the reduction of NDPs to dNDPs (4). Our results further demonstrate that Bcl2 directly disrupts the hRRM1/hRRM2 complex in vitro (Fig. 5), and reduces the formation of the hRRM1/hRRM2 complex in cells (Fig. S4). Intriguingly, the hRRM2 binding site on Bcl2 (i.e. BH4) is required for Bcl2 to disrupt the functional hRRM1/hRRM2 complex both in vitro and in cells (Figs. 5 and S4). These results provide insight into the mechanism by which Bcl2 suppresses RNR activity and reduces dNTP pool size.
The BH4 domain is a known survival domain in Bcl2, as evidenced by the fact that its caspase-mediated cleavage or mutagenic removal abolishes the anti-apoptotic activity of Bcl2 (38, 43). The BH1, BH2 and BH3 domains form the surface binding pocket of Bcl2 (37), and are also necessary for the full and potent antiapoptotic function of Bcl2 (12, 37, 38). Because removal of the BH1, BH2 or BH3 domain does not affect the inhibitory effects of Bcl2 on RNR activity, intracellular dNTP levels and DNA replication fork progression (Fig. 6), but significantly attenuates its anti-apoptotic activity (Fig. S6), we propose that the anti-apoptotic function of Bcl2 is not required for its role in inducing DNA replication stress. The ability of Bcl2 to induce DNA replication stress is dependent on its hRRM2 binding, which requires the BH4 domain (Fig. 6). Bcl2 knockdown experiments further confirm a physiological role of Bcl2 in regulating RNR activity, dNTP pool size and DNA replication dynamics (Fig. 7).
The dNTP supply plays an essential role in maintaining genome integrity in normal eukaryotic cells (44). Appropriate levels of RNR activity are necessary to provide the appropriate amount of dNTPs for normal DNA replication and repair in cells (4). Optimization of intracellular concentrations of dNTPs is critical for the fidelity of DNA synthesis during DNA replication and repair because levels that are too high or too low can easily lead to increased rates of mutagenesis and promotion of cancer development (44). Consequently, overexpression of RNR is also associated with cancer progression (45). That reduced RNR activity can cause an insufficient dNTP pool leading to DNA replication stress (44), helps to explain our findings that Bcl2-mediated reduction of RNR activity causes decreased levels of intracellular dNTPs, disturbed DNA replication (Figs. 1-2 and S1-2). Thus, maintaining intracellular dNTPs at appropriate levels (not too high or too low) by manipulating RNR activity is the key to maintain high-fidelity DNA replication and prevent cancer formation.
Bcl2 is ubiquitously expressed in various types of cells, including normal and cancer cells, but its expression levels vary (46, 47). Overexpression of Bcl2 has been considered as a biomarker of resistance to chemotherapy or radiotherapy in various types of cancer (48, 49). The functional consequences of Bcl2 regulation of RNR activity should be different in resting and S-phase cells. In addition to hRRM2, p53R2 has been identified as a second radical-providing small subunit in mammalian cells (34). The main catalytically active enzyme is hRRM1/hRRM2 and not the hRRM1/p53R2 complex in cycling cells (35). However, p53R2 can substitute for hRRM2 as a subunit of ribonucleotide reductase in post-mitotic resting cells (35). p53R2 is primarily involved in regulating synthesis, replication and repair of mtDNA (35, 36). Our findings indicate that, in addition to hRRM2, Bcl2 can also interact with p53R2 (Fig. S3). It is possible that Bcl2 not only negatively regulates DNA replication via interaction with hRRM2 in cycling cells (i.e. the late G1–early S-phase) but may also affect synthesis, replication and repair of mtDNA by regulating RNR activity via its p53R2 binding in quiescent cells.
It has been recently reported that specific knockdown of hRRM2 by systemic delivery of a nanoparticle carrying hRRM2-specific siRNA resulted in down-regulation of Bcl2 in Tu212 and A549 cells (50). Knockdown of hRRM2 leads to Bcl2 degradation through the proteasome pathway (50). Thus, hRRM2 also contributes to stabilization of Bcl2 (50). Based on our findings and recent report, we propose that Bcl2 and hRRM2 regulate each other. It is possible that direct interaction between Bcl2 and hRRM2 not only benefits the stabilization of Bcl2 by hRRM2, but may also be the mechanism by which Bcl2 suppresses RNR activity.
In summary, our findings demonstrate that Bcl2 can induce DNA replication stress by retarding DNA replication fork progression with fork asymmetry via suppression of RNR activity and synthesis of dNTPs. This action of Bcl2 occurs through direct interaction of its BH4 domain with hRRM2 leading to disruption of the functional hRRM1/hRRM2 complex. The effect of Bcl2 on RNR and DNA replication dynamics is independent of its anti-apoptotic function, but depends on its hRRM2 binding capability. Based on our findings, we propose that the inhibitory effects of Bcl2 on RNR activity, intracellular dNTP pools and DNA replication fork progression may lead to DNA replication stress, enhance genetic instability and eventually contribute to the development of various types of cancer. Thus, Bcl2, in addition to its anti-apoptotic activity, may also function as a pro-oncogenic molecule that produces DNA replication stress through inhibition of RNR.
Supplementary Material
Acknowledgments
This work was supported by NCI, National Institutes of Health grants R01CA112183 and R01CA136534. We thank Anthea Hammond for editing of the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors disclose no potential conflicts of interest
References
- 1.Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J Mol Cell Biol. 2011;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 3.Xue L, Zhou B, Liu X, Heung Y, Chau J, Chu E, et al. Ribonucleotide reductase small subunit p53R2 facilitates p21 induction of G1 arrest under UV irradiation. Cancer Res. 2007;67:16–21. doi: 10.1158/0008-5472.CAN-06-3200. [DOI] [PubMed] [Google Scholar]
- 4.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 5.Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–22. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281:27705–11. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 7.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–6. [PubMed] [Google Scholar]
- 8.Lin ZP, Belcourt MF, Carbone R, Eaton JS, Penketh PG, Shadel GS, et al. Excess ribonucleotide reductase R2 subunits coordinate the S phase checkpoint to facilitate DNA damage repair and recovery from replication stress. Biochem Pharmacol. 2007;73:760–72. doi: 10.1016/j.bcp.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–31. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 10.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545–56. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X, Gao F, Flagg T, May WS., Jr. Mono- and multisite phosphorylation enhances Bcl2's antiapoptotic function and inhibition of cell cycle entry functions. Proc Natl Acad Sci U S A. 2004;101:153–8. doi: 10.1073/pnas.2533920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Gao F, May WS, Zhang Y, Flagg T, Deng X. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol Cell. 2008;29:488–98. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linette GP, Hess JL, Sentman CL, Korsmeyer SJ. Peripheral T-cell lymphoma in lckpr-bcl-2 transgenic mice. Blood. 1995;86:1255–60. [PubMed] [Google Scholar]
- 14.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce CM. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–3. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 15.Feng W, Di Rienzi SC, Raghuraman MK, Brewer BJ. Replication stress-induced chromosome breakage is correlated with replication fork progression and is preceded by single-stranded DNA formation. G3 (Bethesda) 2011;1:327–35. doi: 10.1534/g3.111.000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X, Gao F, May WS., Jr. Bcl2 retards G1/S cell cycle transition by regulating intracellular ROS. Blood. 2003;102:3179–85. doi: 10.1182/blood-2003-04-1027. [DOI] [PubMed] [Google Scholar]
- 17.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11:1109–17. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 20.Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & development. 2002;16:2479–84. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shewach DS. Quantitation of deoxyribonucleoside 5'-triphosphates by a sequential boronate and anion-exchange high-pressure liquid chromatographic procedure. Anal Biochem. 1992;206:178–82. doi: 10.1016/s0003-2697(05)80030-2. [DOI] [PubMed] [Google Scholar]
- 22.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: a new paradigm for antimetabolite radiosensitizers. Molecular cancer therapeutics. 2007;6:1858–68. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- 24.Zhou B, Liu X, Mo X, Xue L, Darwish D, Qiu W, et al. The human ribonucleotide reductase subunit hRRM2 complements p53R2 in response to UV-induced DNA repair in cells with mutant p53. Cancer Res. 2003;63:6583–94. [PubMed] [Google Scholar]
- 25.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature cell biology. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. The Journal of experimental medicine. 2009;206:2417–27. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Okumura K, Sinicrope FA. BH3 mimetic obatoclax enhances TRAIL-mediated apoptosis in human pancreatic cancer cells. Clin Cancer Res. 2009;15:150–9. doi: 10.1158/1078-0432.CCR-08-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vail ME, Chaisson ML, Thompson J, Fausto N. Bcl-2 expression delays hepatocyte cell cycle progression during liver regeneration. Oncogene. 2002;21:1548–55. doi: 10.1038/sj.onc.1205212. [DOI] [PubMed] [Google Scholar]
- 29.Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, et al. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–94. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–34. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18:3059–67. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–60. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 33.Kelekar A, Thompson CB. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–30. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 35.Pontarin G, Ferraro P, Bee L, Reichard P, Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc Natl Acad Sci U S A. 2012;109:13302–7. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 37.Castelli M, Reiners JJ, Kessel D. A mechanism for the proapoptotic activity of ursodeoxycholic acid: effects on Bcl-2 conformation. Cell Death Differ. 2004;11:906–14. doi: 10.1038/sj.cdd.4401433. [DOI] [PubMed] [Google Scholar]
- 38.Cheng EH, Kirsch DG, Clem RJ, Ravi R, Kastan MB, Bedi A, et al. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science. 1997;278:1966–8. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 39.Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci. 1992;17:119–23. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- 40.Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–14. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 41.Radivoyevitch T, Saunthararajah Y, Pink J, Ferris G, Lent I, Jackson M, et al. dNTP Supply Gene Expression Patterns after P53 Loss. Cancers (Basel) 2012;4:1212–24. doi: 10.3390/cancers4041212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci U S A. 2008;105:17801–6. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed JC, Miyashita T, Takayama S, Wang HG, Sato T, Krajewski S, et al. BCL-2 family proteins: regulators of cell death involved in the pathogenesis of cancer and resistance to therapy. J Cell Biochem. 1996;60:23–32. doi: 10.1002/(SICI)1097-4644(19960101)60:1%3C23::AID-JCB5%3E3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Niida H, Shimada M, Murakami H, Nakanishi M. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 2010;101:2505–9. doi: 10.1111/j.1349-7006.2010.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Page JL, Surtees JA, Liu H, Lagedrost S, Lu Y, et al. Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer Res. 2008;68:2652–60. doi: 10.1158/0008-5472.CAN-07-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xerri L, Devilard E, Bouabdallah R, Hassoun J, Chaperot L, Birg F, et al. Quantitative analysis detects ubiquitous expression of apoptotic regulators in B cell non-Hodgkin's lymphomas. Leukemia. 1999;13:1548–53. doi: 10.1038/sj.leu.2401533. [DOI] [PubMed] [Google Scholar]
- 47.Monni O, Franssila K, Joensuu H, Knuutila S. BCL2 overexpression in diffuse large B-cell lymphoma. Leuk Lymphoma. 1999;34:45–52. doi: 10.3109/10428199909083379. [DOI] [PubMed] [Google Scholar]
- 48.Condon LT, Ashman JN, Ell SR, Stafford ND, Greenman J, Cawkwell L. Overexpression of Bcl-2 in squamous cell carcinoma of the larynx: a marker of radioresistance. Int J Cancer. 2002;100:472–5. doi: 10.1002/ijc.10503. [DOI] [PubMed] [Google Scholar]
- 49.Stefanaki K, Rontogiannis D, Vamvouka C, Bolioti S, Chaniotis V, Sotsiou F, et al. Immunohistochemical detection of bcl2, p53, mdm2 and p21/waf1 proteins in small-cell lung carcinomas. Anticancer Res. 1998;18:1689–95. [PubMed] [Google Scholar]
- 50.Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19:3416–28. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.