Abstract
The marine alkaloid chlorizidine A contains chlorinated pyrroloisoindolone and pyrrolizine rings that are rare chemical features in bacterial natural products. Herein, we report the biosynthetic logic of their construction in Streptomyces sp. CNH-287 based on the identification of the chlorizidine A biosynthetic gene cluster. Using whole pathway heterologous expression and genetic manipulations, we show that chlorizidine A is assembled by a polyketide synthase that uniquely incorporates a fatty acid synthase-derived dichloropyrrolyl extender unit into the pyrroloisoindolone enzymatic product. We further provide the first biochemical characterization of a flavoenzyme associated with the oxidative formation of chlorizidine A’s distinctive pyrrolizine ring. This work illuminates new enzymatic assembly line processes to rare nitrogen-containing rings in nature.
Polyketide synthase (PKS) enzymes construct their remarkable collection of chemical products chiefly from malonyl-CoA and methylmalonyl-CoA substrates.1,2 Recently, a new metabolic pathway has emerged for the elaboration of assorted PKS extender units derived from α,β-unsaturated acyl-CoA substrates.3 Their reductive carboxylation by enzymes belonging to the crotonyl-CoA reductase/carboxylase (CCR) family yields α-substituted malonyl-CoA molecules that transfer their unique side chain chemistry to polyketide backbones. Despite displaying diversity in carbon chain length, CCR-derived extender units thus far have consisted of linear and branched-chain alkylmalonyl-CoA substrates derived mostly from fatty acid metabolism.4,5 Herein, we report the unanticipated pathway for chlorizidine A (1) biosynthesis, which involves a dedicated fatty acid synthase (FAS) for the construction of a dichloropyrrole intermediate that is further oxidized, activated and incorporated into a PKS pathway to yield a bicyclic pyrrolizine ring.
Chlorizidine A is a tetrachlorinated alkaloid recently isolated from the marine bacterium Streptomyces sp. CNH-287.6 Its novel structure contains a 5H-pyrrolo[2,1-a]isoindol-5-one ring system not previously observed in a natural product connected through a single C–C bond to a dichlorinated 1H-pyrrolizine. Comparing the chemical structure of 1 to other known dichloropyrrole-containing microbial products such as pyrrolomycin C (pyr, 2)7 and marinopyrrole A (mpy, 3),8 we hypothesized that its biosynthetic assembly involves the fusion of two dichloropyrrole-based polyketide units. While the biosynthesis of 2 and 3 involves a C7–C12 aldol condensation of a linear polyketide intermediate, the construction of 1 was unclear, thereby motivating us to explore the molecular logic of its assembly. Hughes and co-workers6 proposed the participation of two distinct PKS systems that use dichloropyrrolyl-S-carrier protein (4) to generate polyketide intermediates terminating at C8 and C12 within 1 that combine intermolecularly to form the pyrroloisoindolone ring. We, on the other hand, suspected an alternative mechanism in which the tricyclic ring forms via a C6–C11 aldol cyclization of a single polyketide intermediate (Figure 1). Such a scenario, however, would necessitate the attachment of the pyrrolizine unit to C9 via an unknown reaction mechanism.
Figure 1.
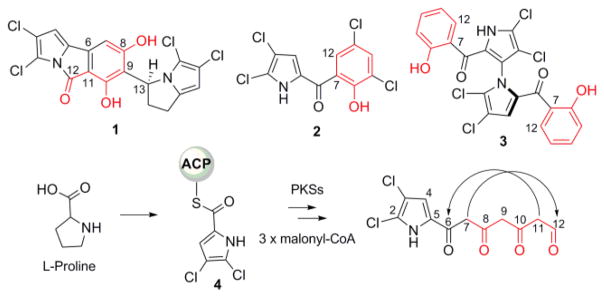
Structures of chlorizidine A (1) and other dichloropyrrole-containing natural products and their general biogeneses from a common intermediate. The full numbering scheme of 1 is shown in SI (page S11).
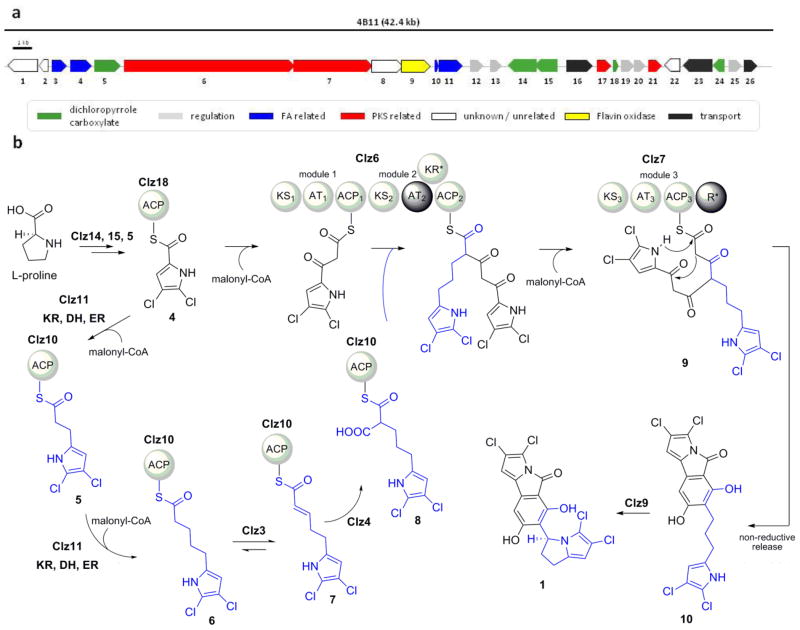
To clarify the strategy employed by nature in the structural diversification of dichloropyrrole-based natural products, we screened a genomic DNA library of Streptomyces sp. CNH-287 and identified a fosmid clone containing genes homologous to those associated with 4 biosynthesis.7,8 Next generation sequencing of the selected fosmid revealed a contiguous region of 42.4 kb containing 26 open reading frames (ORFs) (Figure 2). Targeted integration of the redesigned fosmid9 into the genome of Streptomyces coelicolor M512 imparted this surrogate strain the ability to produce 1, confirming that the identified gene cluster is sufficient for the biosynthesis of chlorizidine A. As such we designated the genes clz1–26 (Table S1).
Figure 2.
Molecular basis for chlorizidine A biosynthesis. (a) Gene organization of the clz biosynthetic cluster from Streptomyces sp. CNH-287. See Table S1 for proposed function assignments. (b) Proposed polyketide biosynthesis of chlorizidine A (1). The conversion of 4 to the Clz6 AT-2 substrate 8 is proposed to involve a divergent FAS pathway (blue) via the FabF ketosynthase Clz11 and primary FAS reducing enzymes encoded elsewhere in the genome. Compounds 5a and 7a correspond to the free carboxylic acid of intermediates 5 and 7, respectively. Abbreviations: ACP, acyl carrier protein; AT, acyl transferase; CoA, coenzyme A; DH, dehydratase; ER, enoyl reductase; KR, ketoreductase; KS, ketoacyl synthase; R, thioester reductase.
Bioinformatic analysis of the clz genes revealed that clz5, clz15, clz16 and clz19 are homologous to those involved with the similar construction of the dichloropyrrole intermediate 4 as previously observed in 2 and 3 biosynthesis. Further inspection of the clz locus identified a trimodular PKS encoded by clz6 and clz7 that is predicted to incorporate two malonate units by modules 1 and 3 and an α-branched malonate in module.4 No other PKS-encoding genes were present in the clz cluster, thereby suggesting that 1 is not assembled from the condensation of two distinct polyketide molecules.6 Rather we identified an acyl carrier protein (ACP), gene clz10, overlapping with a FAS gene clz11 that encode FabC and FabF homologues, respectively. FabF enzymes are known to catalyze iterative condensation of an acyl-ACP substrate with malonyl-ACPs to generate β-ketoacyl-ACP molecules that get reduced each cycle in three steps by ketoreductase, dehydratase and enoyl-reductase enzymes.10 Since none of the three Clz6/7 PKS modules contain reductive domains, we hypothesized that the pyrrolizine moiety may originate in a parallel FAS pathway involving two consecutive malonate extensions of 4 to yield the ACP-tethered intermediates 5 and 6 after full reductive cycles catalyzed by primary metabolic FAS enzymes (Figure 2). Additional type II FAS-based genes encoded in the clz locus include an enoyl-ACP-reductase (clz3) and a CCR (clz4) with low sequence identity with known enzymes. We thus envisaged that the unsaturated intermediate 7 is formed by Clz3 oxidation to facilitate its reductive carboxylation by the Clz4 CCR to give the dichloropyrrolo-substituted malonate 8 for assimilation into the main polyketide chain by Clz6 AT-2 (Figure 2).
To explore this sequence-based hypothesis, we generated a series of mutants in which we deleted targeted clz genes by λ-Red recombination.11 As expected, inactivation of the FAD-dependent halogenase clz5 and the PKS gene clz6 abolished the production of 1. In the case of the clz6 deletion mutant, we further observed the accumulation of a new dichlorinated molecule (5a). On the basis of high-resolution MS (m/z 205.9780, calculated 205.9781, C7H5NO2Cl2) and MS2 fragmentation data that clearly revealed decarboxylated (m/z 147) and dichloromethylpyrrole (m/z 162) fragments (Figure S5), 5a is the hydrolyzed product of protein-bound 5. This series of experiments not only confirmed the central importance of the Clz5 halogenase and the Clz6 PKS in chlorizidine biosynthesis, but it also supported our hypothesis that the dichloropyrrole intermediate 4 is additionally chain extended by a synthase other than the PKS.
We next turned our attention to the fatty acid biosynthesis genes suspected in the proposed pathway to 8. Gene inactivation of the FabF KS clz11 significantly decreased the production of 1 by approximately 90%, while deletion of clz11 and the coupled ACP clz10 further reduced production to just trace levels (Figure S2 and S3). We did not detect the accumulation of additional chlorinated compounds such as 5 in either case. These data confirmed the importance of the fatty acid pathway to 1 biosynthesis and suggested, to our surprise, that the S. coelicolor host can partially complement the native chlorizidine FAS pathway. Clz10 and Clz11 are homologous with the S. coelicolor genes SCO2389 (FabC, 49% identity) and SCO2390 (FabF, 53% identity). While the utilization of diverse starter units by PKSs is greatly appreciated,1,12 there are precious few examples of FASs primed with exotic precursors such as 4.13 The interaction of Clz11 with the ACPs Clz10 and Clz18 may have an important role in catalyzing the extension of 4.14
Inactivation of the enoyl-ACP-reductase clz3 and the putative CCR clz4 similarly abolished 1 while resulting in the accumulation of 5a (Figure S5). Further analysis of extracts of the Δclz4 mutant revealed the accumulation of a second dichlorinated species (7a, m/z 231.9937, calculated 231.9938, C9H7NO2Cl2). As with 5a, we measured decarboxylated (m/z 188) and dichloromethylpyrrole (m/z 147) MS fragments consistent with the α,β-unsaturated carboxylic acid derivative of protein-bound 7 (Figure S6). To exclude potential polar effects on expression of the downstream genes in the mutants, we reintroduced the deleted genes clz3 and clz4 back to the deficient mutants through conjugation. In each case, the production of 1 was restored with loss of 5a and 7a (Figure S4). These data further support our proposal that 4 primes both the type I PKS Clz6 and the type II FAS Clz11.
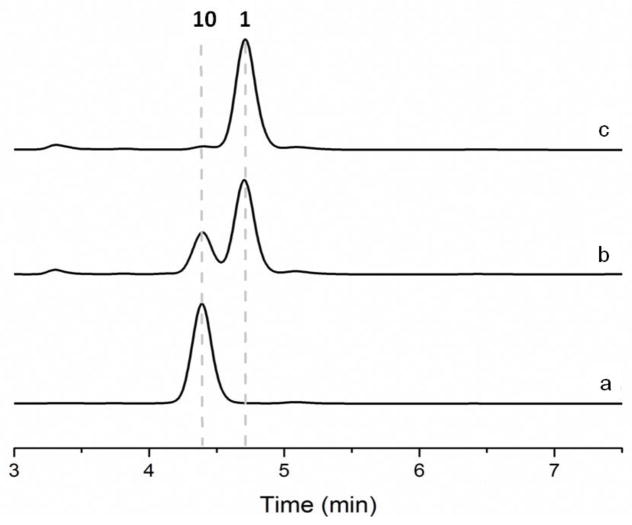
Further evidence for the proposed chlorizidine pathway was achieved with the characterization of the oxidoreductase gene clz9 and its flavoprotein product. Its genetic disruption led to the formation of a new metabolite, compound 10, which we isolated at ~5 mg/L. Inspection of its HR-MS supported a molecular formula of C18H12N2O3Cl4 (m/z 442.9532, calculated 442.9529) which differed by one degree of unsaturation compared to 1. NMR spectroscopic analysis clearly revealed that the dichlorinated 1H-pyrrolizine ring system of 1 was absent in 7 while the 5H-pyrrolo[2,1-a]isoindol-5-one ring system remained intact. We observed three contiguous methylene groups that connected a dichloropyrrole residue to the tricyclic alkaloid suggesting that the Clz9 flavoprotein catalyzes the final biosynthetic reaction to form the pyrrolizine ring.
Clz9 is annotated as a flavin adenine dinucleotide (FAD)-linked oxidase containing the berberine bridge enzyme (BBE) sequence motif by BLAST analysis. Such enzymes characteristically bind the flavin cofactor bicovalently in the enzyme active site. Sequence alignment of Clz9 with other well-characterized BBE family members confirmed the conserved active site histidine and cysteine residues at positions 99 and 157, respectively, necessary for bicovalent FAD attachment (Figure S6).15,16 We prepared recombinant Clz9 fused to a maltose binding protein to facilitate its purification. Upon incubation with 10, we showed the time-dependent conversion to 1, thereby confirming Clz9 as a new member of the BBE family with a novel enzymatic activity (Figure 3). The optical rotation of enzymatically produced 1 agreed with the value reported for natural (S)-1, thereby confirming Clz9’s functional role in catalyzing the stereoselective cyclization of 10 (prechlorizidine A) to 1.
Figure 3.
HPLC profiles at 254 nm of the in vitro conversion of 10 to 1 catalyzed by Clz9. (a) Control reaction using heat-inactivated enzyme. (b) 15 min reaction. (c) 16 hr reaction.
Enzymes containing bicovalently attached flavin cofactors have been reported in biosynthetic pathways of alkaloids,16 terpenes,17 and carbohydrates18 primarily of plant origin. These enzymes often function as oxidases that transfer hydride from the substrate to the flavin cofactor to generate reactive intermediates prone to nucleophilic chemistry. In the case of 1’s pyrrolizine ring, its formation is achieved by the creation of a new C–N bond between 10’s pyrrole nitrogen and the benzylic carbon C13. We propose a mechanism inspired by other BBE members16,17 in Scheme 1. The formation of the pyrrolizine residue in 1 is biosynthetically distinct from well-known plant pyrrolizine natural products that rather derive from homospermidine19. Structural and mechanistic details of Clz9’s biochemistry are presently underway and will be reported in due course.
Scheme 1.
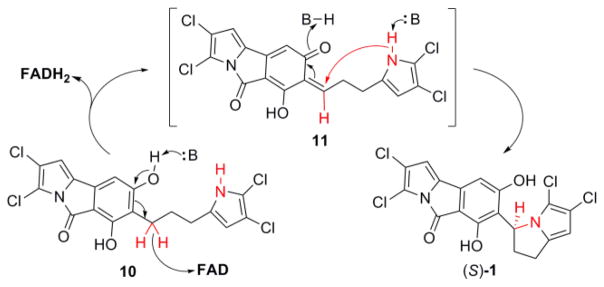
Proposed mechanisms for the stereoselective cyclization of 10 to (S)-1. Deprotonotation of the phenolic hydroxyl facilitates abstraction of the hydride by the FAD cofactor and generation of the intermediate 11. Further nucleophilic attack from the pyrrole-nitrogen yields (S)-1. The presence of molecular oxygen can restore the reduced FADH2 cofactor.
Based on the confirmed structure of 10 and its biosynthetic intermediacy, we propose that the pyrrolisoindolone core polyketide is formed upon release from Clz7 via an intramolecular condensation reaction involving the terminal thioester reductase domain (Figure 2). No other orphan oxidases are encoded in the clz cluster to alternatively support this function. Thioester reductase domains that catalyze intramolecular condensation reactions have been reported in several other microbial biosynthetic pathways20 and can explain the different cyclization pattern observed for 1 biosynthesis (Figure 1). Mutational studies regarding the active site residues of the reductive domain are undergoing.
In conclusion, we successfully characterized the genetic basis for chlorizidine A biosynthesis to reveal a series of unexpected biochemical reactions. Our proposed PKS incorporation of a dichloropyrrole-containing extender unit is unprecedented and extends CCR-based PKS building blocks into new chemical space that portends future polyketide bioengineering. Recently, the discovery of new pyrrolomycin derivatives21 containing linear fatty acid substituents suggests a similar CCR-based biogenesis in their construction. The further post-PKS tailoring of prechlorizidine A’s acyl pyrrole side chain to form chlorizidine A’s pyrrolizine heterocycle not only reveals a new function for a flavin-dependent BBE, but also uniquely expands post-PKS tailoring reactions to include alkaloid synthesis.
Supplementary Material
Acknowledgments
We kindly thank W. Fenical and P.R. Jensen (UCSD) for providing Streptomyces sp. CNH-287, C. C. Hughes (UCSD) for an authentic standard of chlorizidine A, J.P. Noel (Salk Institute for Biological Studies) for providing access to the Ion Torrent sequencing instrument, and B.M. Duggan and Y. Su (UCSD) for NMR and HR-MS assistance, respectively. We also greatly appreciate helpful guidance from K. Yamanaka, L. Kaysser and P. Jordan. Funding was generously provided by the NIH research grant R01-AI47818, NIH instrument grant S10-OD010640, and a postdoctoral fellowship from CNPQ to S.M.M.
Footnotes
The authors declare no competing financial interests.
Experimental section, Tables S1–S3, Figures S1–S16 containing protein sequence alignment and detailed compound characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hertweck C. Angew Chem Int Ed. 2009;48:4688. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 2.Ridley CP, Lee HY, Khosla C. Proc Natl Acad Sci USA. 2008;105:4595. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Hazzard LC, Eustaquio AS, Reynolds KA, Moore BS. J Am Chem Soc. 2009;131:10376. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale A, Kobayashi Y, Reynolds KA, Moore BS. Proc Natl Acad Sci U S A. 2009;106:12295. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mo S, Kim DH, Lee JH, Park JW, Basnet DB, Ban YH, Yoo YJ, Chen S-w, Park SR, Choi EA, Kim E, Jin Y-Y, Lee S-K, Park JY, Liu Y, Lee MO, Lee KS, Kim SJ, Kim D, Park BC, Lee S-g, Kwon HJ, Suh J-W, Moore BS, Lim S-K, Yoon YJ. J Am Chem Soc. 2011;133:976. doi: 10.1021/ja108399b. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu Z, Ding L, Hertweck C. Angew Chem Int Ed. 2011;50:4667. doi: 10.1002/anie.201008265. [DOI] [PubMed] [Google Scholar]; Quade N, Huo L, Rachid S, Heinz DW, Müller R. Nat Chem Biol. 2012;8:117. doi: 10.1038/nchembio.734. [DOI] [PubMed] [Google Scholar]; Lechner A, Wilson MC, Ban YH, Hwang J, Yoon YJ, Moore BS. ACS Synth Biol. 2013;2:379. doi: 10.1021/sb3001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson MC, Moore BS. Nat Prod Rep. 2012;29:72. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- 5.Koryakina I, McArthur JB, Draelos MM, Williams GJ. Org Biomol Chem. 2013;11:4449. doi: 10.1039/c3ob40633d. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez-Mico X, Jensen PR, Fenical W, Hughes CC. Org Lett. 2013;15:988. doi: 10.1021/ol303374e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Parry RJ. Antimicrob Agents Chemother. 2007;51:946. doi: 10.1128/AAC.01214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka K, Ryan KS, Gulder TAM, Hughes CC, Moore BS. J Am Chem Soc. 2012;134:12434. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaysser L, Bernhardt P, Nam SJ, Loesgen S, Ruby JG, Skewes-Cox P, Jensen PR, Fenical W, Moore BS. J Am Chem Soc. 2012;134:11988. doi: 10.1021/ja305665f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gago G, Diacovich L, Arabolaza A, Tsai SC, Gramajo H. FEMS Microbiol Rev. 2011;35:475. doi: 10.1111/j.1574-6976.2010.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc Natl Acad Sci U S A. 2003;100:1541. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore BS, Hertweck C. Nat Prod Rep. 2002;19:70. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 13.Moore BS, Poralla K, Floss HG. J Am Chem Soc. 1993;115:5267. [Google Scholar]; Mo S, Sydor PK, Corre C, Alhamadsheh MM, Stanley AE, Haynes SW, Song L, Reynolds KA, Challis GL. Chem Biol. 2008;15:137. doi: 10.1016/j.chembiol.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Poskon E, Arthur CJ, Kanari AMP, Wattana-amorn P, Williams C, Crosby J, Simpson TJ, Willis CL, Crump MP. Chem Biol. 2010;17:776. doi: 10.1016/j.chembiol.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Huang CH, Lai WE, Lee MH, Chen CJ, Vasella A, Tsai YC, Liaw SH. J Biol Chem. 2005;280:38831. doi: 10.1074/jbc.M506078200. [DOI] [PubMed] [Google Scholar]
- 16.Winkler A, Hartner F, Kutchan TM, Glieder A, Macheroux P. J Biol Chem. 2006;281:21276. doi: 10.1074/jbc.M603267200. [DOI] [PubMed] [Google Scholar]; Winkler A, Lyskowski A, Riedl S, Puhl M, Kutchan TM, Macheroux P, Gruber K. Nat Chem Biol. 2008;12:739. doi: 10.1038/nchembio.123. [DOI] [PubMed] [Google Scholar]
- 17.Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F. J Biol Chem. 2004;279:39767. doi: 10.1074/jbc.M403693200. [DOI] [PubMed] [Google Scholar]; Shoyama Y, Tamada T, Kurihara K, Takeuchi A, Taura F, Arai S, Blaber M, Shoyama Y, Morimoto S, Kuroki R. J Mol Biol. 2012;423:96. doi: 10.1016/j.jmb.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Alexeev I, Sultana A, Mantsala P, Niemi J, Schneider G. Proc Natl Acad Sci U S A. 2007;104:6170. doi: 10.1073/pnas.0700579104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carlson JC, Shengying L, Shamila S, Gunatilleke Y, Anzai DAB, Podust LM, Sherman DH. Nat Chem. 2011;8:628. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ober D, Hartmann T. Proc Natl Acad Sci USA. 1999;96:14777. doi: 10.1073/pnas.96.26.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims JW, Schmidt EW. J Am Chem Soc. 2008;130:11149. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]; Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ. Chem Bio Chem. 2008;9:585. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]; Liu X, Walsh CT. Biochemistry. 2009;48:8746. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Du L, Lou L. Nat Prod Rep. 2010;27:255. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 21.Dufour C, Wink J, Kurz M, Kogler H, Olivan H, Sable S, Heyse W, Gerlitz M, Toti L, Nuber L, Rey A, Couturier C, Bauer A, Bronstrup M. Chem Eur J. 2012;18:16123. doi: 10.1002/chem.201201635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.