Background: In bacterial transcription complexes, nascent RNA structures increase pause duration.
Results: Antisense RNA and to a lesser extent DNA oligonucleotides mimic effects of RNA structures synergistically with NusA and competitively with RfaH.
Conclusion: Exit channel duplexes interact specifically with RNA polymerase and NusA and compete with RfaH to control clamp position.
Significance: Insights into conformational dynamics of RNA polymerase improve mechanistic understanding of transcriptional regulation.
Keywords: Bacterial Transcription, Gene Expression, Gene Regulation, RNA Polymerase, Transcription, Transcription Factors, Transcription Regulation, NusA, RfaH, Transcriptional Pausing
Abstract
Transcript elongation by bacterial RNA polymerase (RNAP) is thought to be regulated at pause sites by open versus closed positions of the RNAP clamp domain, pause-suppressing regulators like NusG and RfaH that stabilize the closed-clamp RNAP conformation, and pause-enhancing regulators like NusA and exit channel nascent RNA structures that stabilize the open clamp RNAP conformation. However, the mutual effects of these protein and RNA regulators on RNAP conformation are incompletely understood. For example, it is unknown whether NusA directly interacts with exit channel duplexes and whether formation of exit channel duplexes and RfaH binding compete by favoring the open and closed RNAP conformations. We report new insights into these mechanisms using antisense oligonucleotide mimics of a pause RNA hairpin from the leader region of the his biosynthetic operon of enteric bacteria like Escherichia coli. By systematically varying the structure and length of the oligonucleotide mimic, we determined that full pause stabilization requires an RNA-RNA duplex of at least 8 bp or a DNA-RNA duplex of at least 11 bp; RNA-RNA duplexes were more effective than DNA-RNA. NusA stimulation of pausing was optimal with 10-bp RNA-RNA duplexes and was aided by single-stranded RNA upstream of the duplex but was significantly reduced with DNA-RNA duplexes. Our results favor direct NusA stabilization of exit channel duplexes, which consequently affect RNAP clamp conformation. Effects of RfaH, which suppresses oligo-stabilization of pausing, were competitive with antisense oligonucleotide concentration, suggesting that RfaH and exit channel duplexes compete via opposing effects on RNAP clamp conformation.
Introduction
Transcriptional pausing plays multiple roles in regulating RNA synthesis and gene expression in all domains of life (1). In bacteria, pauses help to maintain coupling of transcription and translation, aid proper folding of nascent RNA, and provide time for recruitment of regulators and are the first step in both Rho-dependent and intrinsic (Rho-independent) termination of transcription (1–9).
Multiple lines of evidence suggest that bacterial RNA polymerase (RNAP)3 initially enters pause states by a sequence-induced structural isomerization that alters the active site and disrupts the nucleotide addition cycle. This initially formed pause state, termed the elemental pause, forms in competition with nucleotide addition; thus, some elongating RNAPs may bypass entry into the elemental paused state with a probability that varies among pauses (10, 11). At a subset of elemental pauses, additional rearrangements of the elongating transcription complex (EC) or interactions of regulators prolong the pause (1, 12). These rearrangements include (i) reverse translocation (backtracking) of the RNA and DNA chains through RNAP, which removes the RNA 3′-OH from the active site, and (ii) formation of RNA secondary structures (“pause hairpins”) in the RNA exit channel of RNAP, which is thought to inhibit nucleotide addition indirectly by stabilizing a conformation of RNAP in which the clamp domain is opened (Fig. 1).
FIGURE 1.
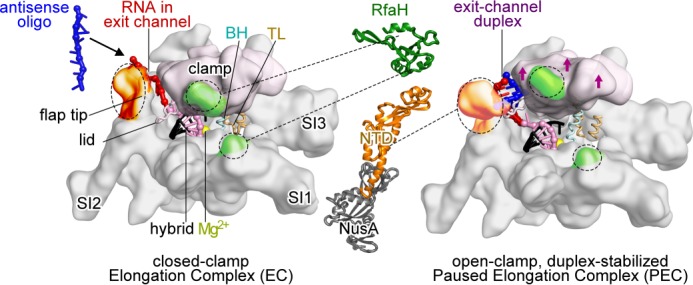
Models of E. coli RNAP EC and hairpin-stabilized paused EC. Left, closed clamp, active EC model (54) showing clamp (light pink), bridge helix (BH), trigger loop (TL), locations of major sequence insertions (SI1, SI2, and SI3) (55), flap tip, lid, active site Mg2+ (yellow), and binding sites for NusA-NTD (orange) and RfaH-NTD (green) (23, 30, 31). Most EC nucleic acids are omitted for clarity. The RNA-DNA hybrid (pink and black), exiting RNA (red), and 8-nt antisense RNA (blue) are shown. Middle, schematic representations of RfaH-NTD (Protein Data Bank code 2oug) (31, 56) and NusA-NTD, S1, KH1, and KH2 (Protein Data Bank code 1hh2) (31, 56). Dotted lines, contacts to RNAP. Right, open clamp paused EC (Protein Data Bank code 4gzy) (17). Arrows indicate clamp movement, and an 8-bp exit channel duplex (blue and red) is modeled into the expanded RNA exit channel.
A model pause signal from the his operon leader region uses a 5-bp stem, 8-nt loop pause hairpin that forms 12 nt from the RNA 3′-end to synchronize transcription of the his transcriptional attenuator with translation of the leader peptide coding region; the translating ribosome disrupts the hairpin to release the paused EC (1). Stabilization of the paused EC by the his pause hairpin appears to involve multiple inhibitory effects on steps in the nucleotide addition cycle in the RNAP active site (13). Non-paused nucleotide addition (50–100 s−1 for Escherichia coli RNAP) requires four steps: (i) translocation of RNA and DNA through RNAP to position the RNA 3′-OH and template base in the so-called i and i + 1 subsites; (ii) NTP binding in the i + 1 subsite; (iii) catalysis; and (iv) pyrophosphate release (14). Translocation shifts the DNA bubble downstream by shortening the RNA-DNA hybrid from 10 to 9 bp, shifting ∼5 nt of ssRNA that lies in the RNA exit channel upstream, melting one bp of the downstream DNA duplex directly in front of the RNA 3′-end, and reannealing one bp of upstream DNA duplex (15, 16). Rapid catalysis requires folding of the trigger loop into an α-helical hairpin (trigger helices) that contacts the NTP substrate and forms a three-helix bundle with the bridge helix. The bridge helix, which spans the active site cleft, is deformable and may occlude the i + 1 subsite when NTP is absent and the trigger loop is unfolded. The elemental pause appears to form when contacts between the clamp domain, the RNA-DNA hybrid, and the downstream DNA duplex are loosened in a partially translocated EC, leading to a rearrangement in which a deformed bridge helix prevents template base entry into the i + 1 subsite (17). Subsequent formation of the pause RNA hairpin then appears to stabilize the paused EC in the pretranslocated register with the RNA 3′ nt frayed off the template base and the clamp domain opened, inhibiting both translocation and formation of the trigger helices even after translocation and NTP binding (12, 18, 19). Recent structural, single-molecule, and biochemical studies have provided evidence for the flexibility of the RNAP clamp domain (20, 21), its movement associated with transcriptional pausing (17), and the inhibition of trigger loop folding in the hairpin-stabilized paused elongation complex (PEC) (19).
Regulators of pausing influence the RNAP active site through contacts to the pause hairpin, exit channel, or RNAP clamp domain (Fig. 1). NusA increases the duration of hairpin-stabilized pausing through contacts to the hairpin and the tip of the flap domain, which forms the wall of the RNA exit channel opposite the clamp (22–28). An alternative model in which NusA acts indirectly to stabilize the hairpins by displacing ssRNA from a different NusA-binding site outside the RNA exit channel has also been proposed (29). In contrast, the NusG paralog RfaH suppresses hairpin effects on pausing through contacts to RNAP thought to inhibit clamp opening (30–32).
Both enhancement of pausing by the his pause hairpin and the hairpin contribution to NusA enhancement of pausing can be mimicked by formation of an 8-bp RNA duplex using an RNA oligo complementary to the exiting RNA (23). Although there is some evidence that changes in hairpin structure can influence pause or NusA effects (27), it remains unclear (i) if any duplex that favors clamp opening is sufficient for these effects; (ii) if specific features of the duplex are needed to enhance pausing; or (iii) if the essential features might differ among exit channel duplexes for the basic hairpin effect, NusA enhancement of pausing, and RfaH suppression of pausing. To investigate these questions, we systematically varied RNA exit channel duplexes using complementary RNA and DNA oligos of varying lengths and concentrations and assayed pause prolongation, NusA enhancement, and RfaH suppression. Our results yield new insights into the mechanisms by which nascent hairpins affect pausing, support a model for direct NusA-hairpin interaction, and establish that duplexes and RfaH compete in a concentration-dependent manner for effect on EC.
EXPERIMENTAL PROCEDURES
Materials
All DNA and RNA oligonucleotides (Table 1) were obtained from Integrated DNA Technologies (Coralville, IA) and purified by denaturing PAGE before use. [α-32P]CTP was from PerkinElmer Life Sciences, and NTPs were from GE Healthcare.
TABLE 1.
Oligonucleotides used in this study
| Stock number | Description | Sequence (5′ → 3′) |
|---|---|---|
| DNA | ||
| 5420 | Template DNA | CTCTGAATCTCTTCCAGCACACATCAGGACGTACTGACC |
| 5069 | Non-template DNA | GGTCAGTACGTCCATTCGATCTTCGGAAGAGATTCAGAG |
| 6601 | 8-bp antisense DNA oligo | CCGGATGA |
| 7579 | 12-bp antisense DNA oligo | CCGGATGAAGCA |
| 7660 | 11-bp antisense DNA oligo | CCGGATGAAGC |
| 7663 | 10-bp antisense DNA oligo | CCGGATGAAG |
| 7666 | 9-bp antisense DNA oligo | CCGGATGAA |
| 8048 | 22-bp antisense DNA oligo | CCGGATGAAGCTCTACAAATGC |
| RNA | ||
| 6593 | 8-bp PEC RNA | UCAUCCGGCGAUGUGUG |
| 6598 | 8-bp antisense RNA oligo | CCGGAUGA |
| 7576 | 12-bp PEC RNA | UGCUUCAUCCGGCGAUGUGUG |
| 7578 | 12-bp antisense RNA oligo | CCGGAUGAAGCA |
| 7658 | 11-bp PEC RNA | GCUUCAUCCGGCGAUGUGUG |
| 7659 | 11-bp antisense RNA oligo | CCGGAUGAAGC |
| 7661 | 10-bp PEC RNA | CUUCAUCCGGCGAUGUGUG |
| 7662 | 10-bp antisense RNA oligo | CCGGAUGAAG |
| 7664 | 9-bp PEC RNA | UUCAUCCGGCGAUGUGUG |
| 7665 | 9-bp antisense RNA oligo | CCGGAUGAA |
| 7694 | 7-bp PEC RNA | AGUCAGGCGAUGUGUG |
| 7695 | 7-bp antisense RNA oligo | CCUGACU |
| 7696 | 6-bp PEC RNA | GUCAGGCGAUGUGUG |
| 7697 | 6-bp antisense RNA oligo | CCUGAC |
| 7698 | 5-bp PEC RNA | UCAGGCGAUGUGUG |
| 7699 | 5-bp antisense RNA oligo | CCUGA |
| 8043 | 8-bp U-tail RNA oligo | UUUUUUUUCCGGAUGA |
| 8045 | 8-bp hairpin-tail RNA oligo | CGCUGCAUUGCUGCAGCGCCCGGAUGA |
| 8046 | 22-bp PEC RNA | GCAUUUGUAGAGCUUCAUCCGGCGAUGUGUG |
| 8047 | 22-bp antisense RNA oligo | CCGGAUGAAGCUCUACAAAUGC |
Proteins
Core E. coli RNAP was purified as described previously (13, 33). His-tagged full-length and N-terminal domain of NusA were purified from BL21 λDE3 cells containing a NusA overexpression plasmid by a two-step purification protocol using HisTrap HP affinity chromatography followed by gel filtration on Superdex 200, as described previously (23, 28). RfaH-NTD was obtained by overexpression of an RfaH derivative containing a tobacco etch virus protease cleavage site between the NTD and CTD and containing a C-terminal hexahistidine tag from plasmid pIA777 (kindly provided by I. Artsimovitch). E. coli cells (BL21 λDE3) harboring pIA777 were grown at 37 °C in LB medium containing kanamycin (25 μg/ml) until apparent A600 reached 0.6. The temperature was then lowered to 20 °C, and isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.2 mm. Cells were harvested after overnight induction and pelleted at 2800 × g for 15 min at 4 °C. Cell pellets were resuspended in buffer A (50 mm Tris-HCl, pH 7.9, 0.05 mm EDTA, 8% glycerol, 500 mm NaCl, 2 mm β-mercaptoethanol) supplemented with 0.1 mg of PMSF/ml and a protease inhibitor mixture (final concentrations of 0.0125 mg of benzamide/ml, 2 × 10−4 mg of chymostatin/ml, 2 × 10−4 mg of leupeptin/ml, 4 × 10−5 mg of pepstatin/ml, 4 × 10−4 mg of aprotonin/ml, and 4 × 10−4 mg of antipain/ml) and lysed by sonication. After centrifugation at 27,000 × g at 4 °C for 15 min, the supernatant was applied to a 5-ml HisTrap HP affinity column (GE Healthcare). The eluted fractions containing the full-length RfaH were combined and dialyzed in buffer A overnight at 4 °C. The full-length RfaH was then cleaved with ∼500 units of His6-tobacco etch virus protease in buffer A for 2 h at 20 °C, as described previously (31). The cleavage products were then reapplied to the HisTrap HP column, and RfaH-NTD was recovered in the flow-through. The recovered fractions containing RfaH-NTD were pooled, dialyzed against storage buffer (10 mm Tris-HCl, pH 7.9, 50% glycerol, 250 mm NaCl, 0.1 mm EDTA, 0.1 mm DTT), and analyzed by SDS-PAGE.
In Vitro EC Reconstitution
Nucleic acid scaffolds for reconstituting ECs were assembled in reconstitution buffer (10 mm Tris·HCl, pH 7.9, 40 mm KCl, 5 mm MgCl2) by heating RNAs, tDNAs, and ntDNAs (0.5, 0.6, and 0.6 μm, respectively; Table 1) to 95 °C for 2 min, rapidly cooling to 45 °C, and then cooling to room temperature in 2 °C steps of 2 min each, as described previously (28). ECs reconstituted by incubating core E. coli RNAPs with the nucleic acid scaffold (3:1 RNAP/scaffold) in elongation buffer (25 mm HEPES-KOH, pH 8.0, 130 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.15 mm EDTA, 5% glycerol, and 25 μg of acetylated bovine serum albumin/ml) for 15 min at 37 °C.
In Vitro Transcriptional Pause Assays
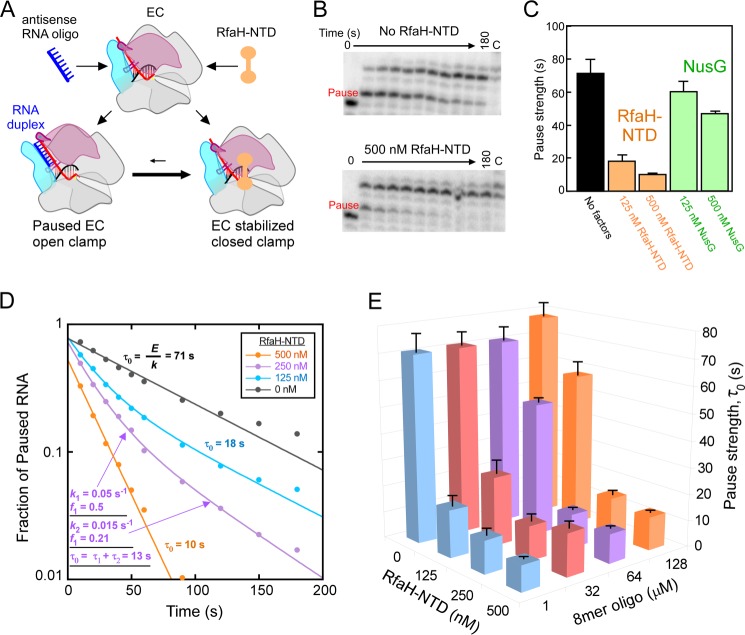
The nascent RNA in the reconstituted ECs was first radiolabeled by incubation with [α-32P]CTP (2 μm; 5 Ci/mmol) in elongation buffer at 50 nm EC for 1 min at 37 °C to give C18 ECs (28). Reconstitution buffer (as control) or antisense DNA or RNA oligos (1 μm final concentration if not indicated otherwise) were then added to the solution, and incubation was continued at 37 °C for 10 min. The C18 ECs were then incubated with 10 μm GTP and 100 μm UTP. Reaction samples were removed at the indicated times and quenched with an equal volume of 2× stop buffer (8 m urea, 50 mm EDTA, 90 mm Tris borate buffer, pH 8.3, 0.02% bromphenol blue, and 0.02% xylene cyanol). After the final time point, high concentrations of GTP and UTP (500 μm each) were added to chase all active ECs out of the paused state. The radiolabeled RNA reaction products were then separated by electrophoresis using a denaturing 20% polyacrylamide gel in 0.5× TBE buffer (34). Experiments were conducted with three variations: (i) with no oligonucleotide (reconstitution buffer solution); (ii) with RNA oligos ranging from 5 to 12 or 22 nt in length; or (iii) with DNA oligos ranging from 8 to 12 or 22 nt in length. The experiments were repeated with full-length NusA (∼2.5 μm), NusA-NTD (4 μm), or RfaH-NTD (125, 250, or 500 nm), which were added immediately after [α-32P]CTP radiolabeling and allowed to bind for 10 min at 37 °C.
Data Quantitation and Analysis
Gels were exposed to PhosphorImager screens, scanned using a Typhoon PhosphorImager, and quantitated using ImageQuant software (GE Healthcare). The RNA present in each lane was quantitated as a fraction of the total RNA in each lane (Fig. 2) and corrected for the unreacted fraction remaining in the chase lane. The rates of escape from the his pause site (k; reported as half-lives, t½) and the fractions of EC in the one or two pause states (E) were obtained by fitting the data to either a single- or double-exponential decay equation. In most experiments, the slow pause fraction, which may represent backtracked C18 or U19 ECs (28), was negligible and was not included in the calculation of pause half-life. For experiments with RfaH-NTD, two significant pause fractions were present in some conditions. Thus, for the experiments with RfaH-NTD (Fig. 7), we calculated and reported pause strength, τ0, as E/k, where E is the pause efficiency when only one paused state existed, or as (f1/k1) + (f2/k2) when two paused states existed (35). f1 and f2 are the fractions of paused RNAPs that enter the paused states with escape rates k1 and k2 (see Fig. 7D). All measurements of pause half-lives or pause strengths are reported as means ± S.D. of experimental triplicates.
FIGURE 2.
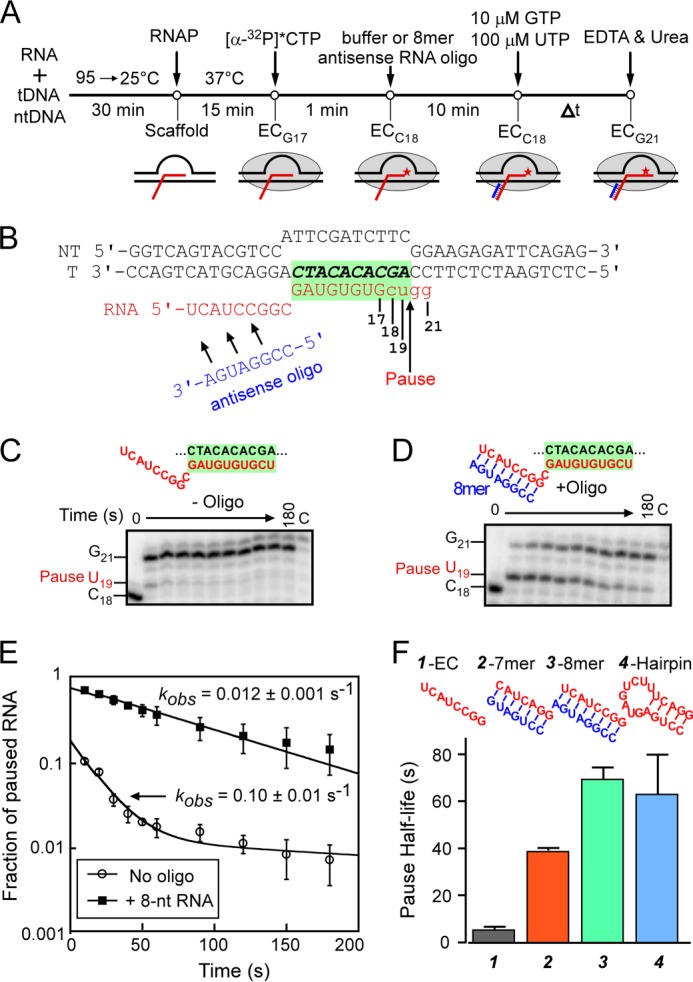
An antisense RNA oligo anneals to the nascent RNA and mimics the his pause hairpin. A, a schematic of the transcriptional pause assay. ECs were first preformed by assembling E. coli RNAP on the nucleic acid scaffold (RNA 6593, tDNA 5420, and ntDNA 5069; Table 1) shown in B (see “Experimental Procedures”). Preformed ECs (G17) were elongated to the C18 position by the addition of [α-32P]CTP. The complexes were then incubated with or without an 8-nt antisense RNA oligo. The pause assay was initiated by the addition of 10 μm GTP and 100 μm UTP; aliquots were removed at the indicated times and mixed with 2× loading dye, and RNA products were separated on 20% denaturing acrylamide gel. B, a representative example of a nucleic acid scaffold used in this study (for 8-mer antisense oligo; yields EC G17). The RNA-DNA hybrid region is highlighted in green, and the antisense RNA oligonucleotide (blue) anneals to C18 RNA upstream from the RNA-DNA hybrid (a 1-nt spacer separates the hybrid and the exit channel duplex). Elongation of RNA17 by RNAP is shown in lowercase type. C and D, two representative gels of transcription pause assay in the absence (C) or in the presence (D) of 1 μm 8-nt antisense RNA oligo. The corresponding nucleic acid scaffolds are depicted above the gels with the RNA-DNA hybrid highlighted in green. The lane marked C is a chase lane containing reaction products after ECs were incubated for 3 min with 0.5 mm GTP and UTP after the 3-min time course. E, the 19-mer RNA (U19) present in each lane was quantitated as a fraction of the total RNA in each lane, normalized for the fraction remaining in the chase lane and plotted as a function of reaction time. The rate of escape, kobs, is obtained by fitting the disappearance of U19 RNA to a single exponential (for 8-nt antisense RNA) or to a double exponential equation (for no oligo control). The rates of escape in the presence or absence of 8-nt RNA oligo are indicated on the plot. F, pause half-lives of ECs containing no oligo (gray), 32 μm 7-nt RNA oligo (red), 32 μm 8-nt RNA oligo (green), or hairpin RNA (blue). Error bars, S.D.
FIGURE 7.
RfaH-NTD and the 8-mer RNA oligo compete for binding to ECs. A, a model of competition between RfaH and oligo binding through the movement of the RNAP clamp domain (pink). Red, RNA; blue, antisense RNA oligo; orange, RfaH. Formation of RNA duplex requires opening of the clamp domain, which is inhibited when RfaH is bound to EC and holds the clamp in the closed position. B, representative pause assay gel panels for ECs assembled on the nucleic acid shown in Fig. 2B. ECs (C18; 50 nm) were elongated through the his pause site in the presence of 1 μm 8-mer antisense RNA oligo with or without 500 nm RfaH-NTD at 100 μm UTP and 10 μm GTP (see “Experimental Procedures”). C, pause strengths derived from the experiment shown in B and similar experiments with NusG (antisense RNA at 1 μm). Pause strengths were calculated as shown in D. D, derivation of pause strengths. The fractions of pause RNA for the experiment shown in B and similar experiments with 125 and 250 nm RfaH-NTD were plotted as a function of reaction time. Apparent fraction paused (E) and pause escape rate (k) were determined by non-linear regression. Pause strength (τ0) was calculated from E and k (τ0 = E/k). Two pause rate components were evident at 125 and 250 nm RfaH-NTD, necessitating use of a double-exponential fit. In these cases, pause strengths were calculated from the two rate components and two fractions present (τ0 = f1/k1 + f2/k2; τ0 = τ2 + τ2) as illustrated for 250 nm RfaH-NTD in the figure (see “Experimental Procedures”). E, a three-dimensional plot showing pause strengths as a function of concentrations of antisense 8-mer and RfaH-NTD present in the pause assays. Error bars, S.D.
RESULTS
The Length of RNAP Exit Channel RNA-RNA Duplexes Affects PEC Lifetime
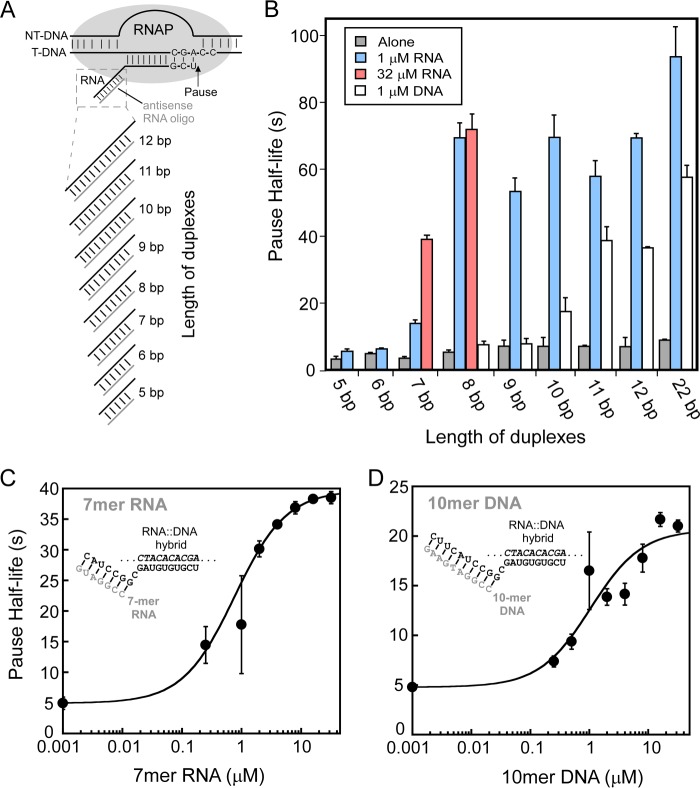
An 8-bp RNA-RNA duplex formed by pairing an RNA oligo to the nascent RNA at the location of the his pause hairpin (transcript 3′-proximal end of duplex at −12) mimics the pause lifetime-increasing effect of the hairpin (23). To test whether the length of the RNA duplex influences this effect, we first asked if shortening the duplex to 7 bp without changing its location altered the duplex effect on pause lifetime. We performed this experiment using reconstituted ECs formed with E. coli RNAP on partially complementary nucleic acid scaffolds known to recapitulate the properties of the his paused EC, as described previously (28) (Fig. 2, A and B; see “Experimental Procedures”). C18 complexes were labeled with 32P by incubating G17 ECs formed 2 nt upstream from the pause site with [α-32P]CTP. After annealing an 8-mer RNA oligo to the exiting RNA, the lifetimes of paused ECs formed at U19 were measured by tracking the 32P-U19 RNA as a function of time after the addition of GTP and UTP (to 10 and 100 μm, respectively) using denaturing polyacrylamide gel electrophoresis (Fig. 2, C and D). Consistent with previous findings, formation of the 8-bp exit channel duplex strongly stimulated pausing, and the magnitude of this effect (∼10-fold on pause duration) was equivalent to the difference in pause duration between an EC lacking an exit channel duplex and one containing the natural his pause RNA hairpin (Fig. 2F). However, shortening the exit channel duplex to 7 bp decreased pause duration by a factor of ∼2 even at saturating concentrations of complementary 7-mer oligo (Fig. 2, E and F; see below). (Note that the nascent RNA used to test the 7 mer antisense oligo is 1 nt shorter than the RNA used to test the 8-mer oligo; thus, C17 and U18 ECs for the 7-mer are equivalent to C18 and U19 ECs for the 8-mer.) Thus, an 8-bp but not a 7-bp RNA duplex is sufficient to recapitulate the full hairpin effect. This result confirms our previous report (23) that the hairpin stem, but not the hairpin loop, generates the hairpin-stabilizing effect and suggests that the length of the stem determines the magnitude of the effect.
Although we allowed ample time for equilibration of antisense oligo binding to C17 ECs used to test the 7-mer oligo before assaying pausing at position U18, we were concerned that oligos might bind the U18 ECs more tightly and that our results could be compromised by incomplete equilibration with the transient U18 ECs. In principle, the posttranslocated C17 EC, which is thought to predominate at equilibrium (15), should present the same nascent RNA target as the pretranslocated U18 ECs that would form upon nucleotide addition. Nonetheless, to test for possible complications, we compared effects of the 7-mer oligo when bound to ECs halted at C17 or at U18 (Fig. 3A). We found that 7-mer oligo binding to C17 ECs actually had stronger effects on pausing, especially at saturating 32 μm concentrations (Fig. 3B). We suspect that this result reflects backtracking of ECs halted at U18, which is known to occur (28) and which would shorten the RNA in the exit channel available for hybridization. These results validated our use of C17 complexes as preferred substrates for the oligo-simulated pause assay.
FIGURE 3.
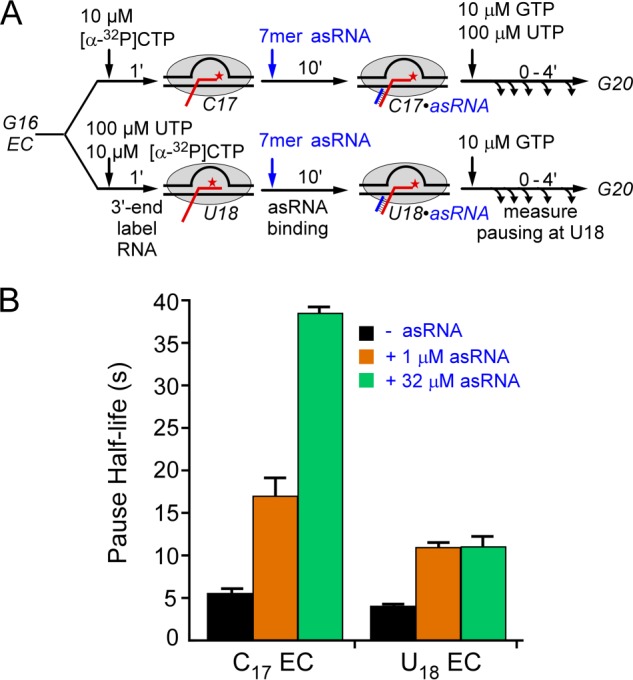
Comparison of antisense oligo annealing 1 nt before the pause site or at the pause site. A, reaction scheme to test the effects of EC position on oligo-stimulated pausing. ECs were halted at either C17 (1 nt before the pause) or U18 (at the pause) and then incubated with 1 μm or 32 μm antisense oligo (7-mer asRNA) prior to assay of pausing. The nascent RNA is 1 nt shorter than that shown in Fig. 2 to allow formation of a 7-bp exit channel duplex. B, pause half-lives for assays in which no oligo (black), 1 μm 7-mer antisense oligo (orange), or 32 μm 7-mer antisense oligo (green) was annealed to ECs halted at C17 or U18. Error bars, S.D.
The Maximal Increase in PEC Lifetime Requires an Exit Channel RNA-RNA Duplex ≥8 bp
In principle, differences in either the strength of oligo binding to the nascent RNA in halted ECs or a requirement for a longer duplex for maximum effect on RNAP could explain the reduced effect of the 7-bp exit channel RNA duplex on pause duration. Therefore, we next tested a range of possible exit channel RNA duplexes from 5 to 22 bp using 1 μm oligo during the annealing step (Fig. 4A). RNA oligos generating duplexes longer than 8 bp did not increase enhancement of pause duration significantly, giving ∼14-fold longer pause durations than the scaffold lacking an exit channel RNA duplex (Fig. 4B). Although 1 and 32 μm 8-mer RNA oligos gave the same pause durations, showing that oligo binding saturated at 1 μm, much less stimulation of pause duration was detected at 1 μm 7-mer relative to 32 μm (Fig. 4B). This result established that the 7-mer oligo binds the G17 EC more weakly than the 8-mer oligo. By testing various concentrations of 7-mer oligo, we found that the ∼7-fold stimulation of pause duration at 32 μm was saturating and that half-maximal stimulation occurred near 1 μm 7-mer (Fig. 4C). Consistent with observations from Ha and colleagues (36) that at least 7 complementary nt are necessary to seed oligo binding, we detected little if any increase in pause duration with 5-mer and 6-mer oligos at 1 μm (1.75-fold maximum; Fig. 4B), and increasing 5-mer or 6-mer concentrations did not change this result (data not shown). These results establish (i) that an 8-bp or longer RNA duplex maximally stimulates pause duration; (ii) that a 7-bp RNA duplex is both more difficult to form and, once formed, stimulates pause duration less than an 8-bp duplex; and (iii) that exit channel RNA duplexes less than 7 bp either are unable to form or have little to no effect on PEC lifetime.
FIGURE 4.
Duplexes of sufficient length in the RNA exit channel of RNAP increase pause duration. A, ECs with nascent RNA annealed to oligonucleotides of varying lengths (Table 1). B, pause half-life of ECs incubated without oligos (alone) or with different lengths of 1 μm RNA oligos or 1 μm DNA oligos. The maximum pause half-lives of 7- and 10-nt antisense RNA oligos at saturating concentration (32 μm) are shown in red. C, 7-mer RNA oligo concentration dependence of pause half-life. D, 10-mer DNA oligo concentration dependence of pause half-life. Error bars, S.D.
Exit Channel DNA-RNA Duplexes Increase PEC Lifetime Less than RNA-RNA Duplexes
We next tested whether DNA oligos targeting the exiting RNA at positions identical to the RNA oligos could also stimulate pause duration and found that DNA oligos exhibited significantly less effect on PEC lifetime than RNA duplexes (Fig. 4B). Further, at 1 μm, DNA oligos ≥11 nt in length were required for maximal effect (in contrast to ≥8-mer for RNA oligos). Further, the maximal effect of DNA oligos on pause duration was about half that observed for the comparable length RNA oligo (Fig. 4B). We conclude that exit channel DNA-RNA duplexes increase PEC lifetime less than RNA-RNA duplexes. Because a 10-mer DNA oligo at 1 μm gave about the same partial effect as a 7-mer RNA oligo, we next tested whether its binding also was submaximal by measuring pause duration at varying 10-mer DNA oligo concentrations. We found that pause duration saturated at ∼4-fold stimulation (less than the ∼7-fold seen for the 7-mer RNA oligo) but that half-maximal binding again occurred at ∼1 μm oligo (Fig. 4D). These results establish (i) that DNA-RNA exit channel duplexes are both more difficult to form than RNA-RNA duplexes (e.g. a 10-mer DNA oligo gives only half-maximal effect at 1 μm, whereas 1 μm 8-mer RNA is saturating); (ii) that once formed, DNA-RNA exit channel duplexes stimulate pause duration significantly less than a RNA-RNA duplex of comparable length; and (iii) that DNA-RNA exit channel duplexes must be ≥11 bp to give the maximum possible effect.
The approximate equivalence of 10–11-bp DNA-RNA duplexes with 7–8-bp RNA-RNA duplexes for pause stimulation (even if not to similar levels) is consistent with calculations of DNA-RNA versus RNA-RNA stabilities in our assay conditions (Table 2). Thus, these differences in length requirements probably reflect basic properties of the nucleic acid duplexes rather than differences in duplex-RNAP contacts. However, the reduced maximal stimulation of pause duration of all length DNA-RNA versus RNA-RNA duplexes cannot be similarly explained and must reflect differences in the way these two types of duplexes affect the enzyme (see “Discussion”).
TABLE 2.
Predicted thermodynamics of RNA-RNA or RNA-DNA duplex formation
| Sequence | Length | ΔG037 (1 m NaCl)a | ΔG037 (100 mm NaCl)b | Tmc |
|---|---|---|---|---|
| nt | kcal mol−1 | kcal mol−1 | °C | |
| RNA-RNA duplex | ||||
| r(CCUGA)/r(GGACU) | 5 | −6.14 ± 0.26 | −5.53 ± 0.56 | 23.6 |
| r(CCUGAC)/r(GGACUG) | 6 | −7.95 ± 0.26 | −6.67 ± 0.68 | 35.9 |
| r(CCUGACU)/r(GGACUGA) | 7 | −10.46 ± 0.27 | −8.25 ± 0.84 | 45.3 |
| r(CCGGAUGA)/r(GGCCUACU) | 8 | −12.70 ± 0.30 | −9.67 ± 0.98 | 54.5 |
| r(CCGGAUGAA)/r(GGCCUACUU) | 9 | −14.06 ± 0.29 | −10.52 ± 1.1 | 55.8 |
| r(CCGGAUGAAG)/r(GGCCUACUUC) | 10 | −15.71 ± 0.30 | −11.56 ± 1.20 | 59.3 |
| r(CCGGAUGAAGC)/r(GGCCUACUUCG) | 11 | −19.13 ± 0.31 | −13.71 ± 1.40 | 66.2 |
| r(CCGGAUGAAGCA)/r(GGCCUACUUCGU) | 12 | −21.67 ± 0.32 | −15.31 ± 1.56 | 70.4 |
| r(CCGGAUGAAGCUCUACAAAUGC)/r(GGCCUACUUCGAGAUGUUUACG) | 22 | −39.82 ± 0.38 | −26.75 ± 2.73 | 80.2 |
| RNA-DNA duplex | ||||
| d(CCGGATGA)/r(GGCCUACU) | 8 | −9.1 ± 0.52 | −7.4 ± 0.75 | 35.5 |
| d(CCGGATGAA)/r(GGCCUACUU) | 9 | −10.1 ± 0.58 | −8.0 ± 0.82 | 35.6 |
| d(CCGGATGAAG)/r(GGCCUACUUC) | 10 | −11.9 ± 0.68 | −9.2 ± 0.93 | 37.8 |
| d(CCGGATGAAGC)/r(GGCCUACUUCG) | 11 | −14.6 ± 0.83 | −10.8 ± 1.11 | 45.7 |
| d(CCGGATGAAGCA)/r(GGCCUACUUCGU) | 12 | −15.5 ± 0.88 | −11.4 ± 1.17 | 49 |
| d(CCGGATGAAGCTCTACAAATGC)/r(GGCCUACUUCGAGAUGUUUACG) | 22 | −31 ± 1.76 | −21 ± 2.15 | 59.2 |
A Maximal NusA Effect on PEC Lifetime Requires RNA-RNA Duplexes ≥10 bp
NusA stimulates the duration of some pauses by factors of 3 or more, and this effect of NusA depends on both the formation of an RNA structure in the exit channel and the NusA N-terminal domain that contacts RNAP at the flap tip, near the RNA exit point (23). Thus, we next asked how the effect of NusA varied as a function of different exit channel duplexes using 1 μm RNA oligos (Fig. 5). NusA increased the duration of oligo-stabilized pausing, and this effect depended on the length and structure of the exit channel duplex but with patterns that differed from those observed for oligos in the absence of NusA (Fig. 5A). The maximal effect of NusA (∼10-fold stimulation of pause duration relative to oligo alone) required longer RNA duplexes (≥10 bp instead of ≥8 bp). As a control, we also tested the effect of NusA on PECs lacking an exit channel duplex (ssRNA in exit channel; Fig. 5C). We found no significant NusA effect on pause duration for PECs lacking exit channel duplexes (Fig. 5C), consistent with the idea that NusA acts through structured RNA in the RNAP exit channel to affect pausing. Consistent with previous results (23), NusA required only an extended duplex and not an ssRNA hairpin loop to stimulate pausing to extents even greater than observed for the native his pause hairpin (Fig. 2, D and F).
FIGURE 5.
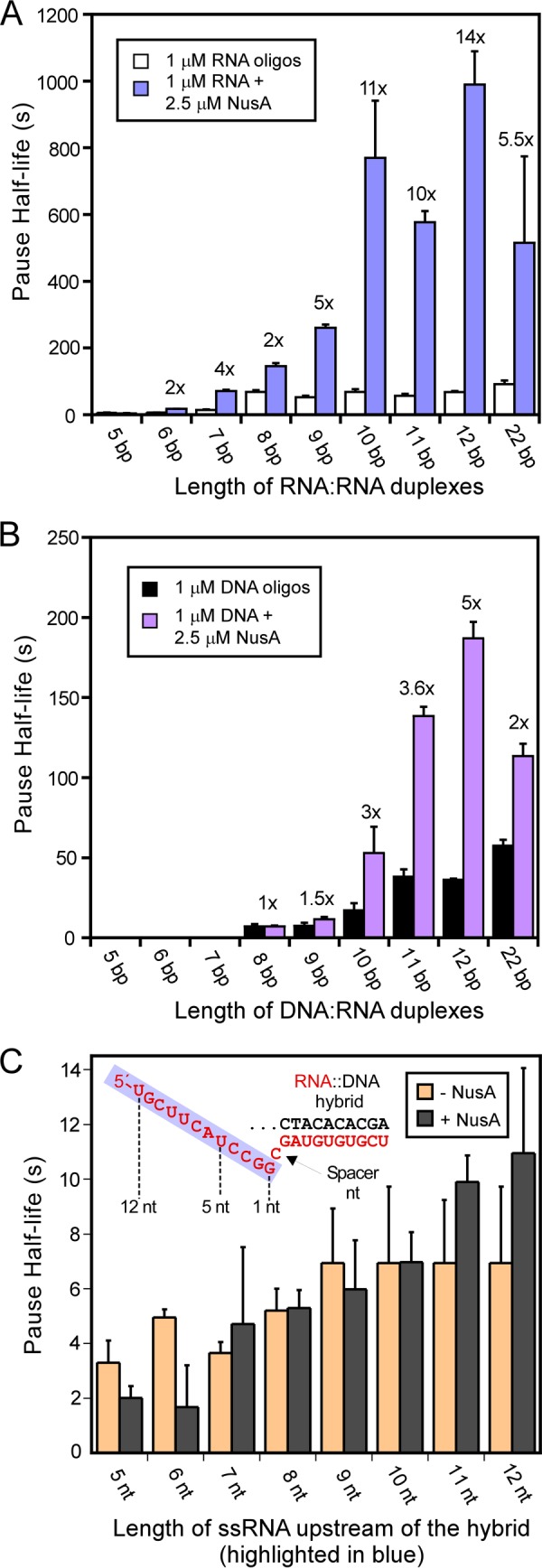
NusA increases pausing stimulated by oligos of varying length. A, NusA effect on RNA oligo stimulation of pause lifetime was measured using ECs reconstituted with DNAs 5069 and 5420 and different sizes of RNAs (Table 1; see “Experimental Procedures”). Antisense RNA oligos were added at 1 μm to 50 nm ECs as illustrated in Fig. 2A. For samples with NusA, 1 μm antisense oligo and 2.5 μm full-length NusA protein were added at the same time. The enhancement of pause half-life by NusA was calculated, and the -fold effect is indicated above the bars. B, bar graph of pause half-life for ECs with 1 μm antisense DNA oligos and in the presence or absence of 2.5 μm full-length NusA. The experiment was performed identically to that shown in A except that DNA oligos with various lengths were added to generate a range of ECs containing exit channel RNA-DNA heteroduplexes. C, NusA has no effect on the pause half-life of ECs when antisense RNA oligos are absent. ECs were assembled on nucleic acid scaffolds (shown in the inset) with different RNAs by changing the length of RNA highlighted in the blue box located upstream from the CMP spacer nucleotide. Pause half-lives of ECs with 2.5 μm full-length NusA (gray) or without NusA (orange) were determined as described under “Experimental Procedures.” Error bars, S.D.
Given that the 7-mer RNA oligo required significantly higher than 1 μm concentration of oligo to achieve maximal effect in the absence of NusA, we also tested whether NusA affected the oligo concentration dependence of pause stimulation by comparing the effects of 1 and 32 μm oligo (Table 3). Although high oligo concentration gave the expected increase in pause duration in the absence of NusA, 1 μm 7-mer RNA oligo proved to be a saturating concentration in the presence of NusA. This result demonstrates that NusA stabilizes RNA oligo binding to the exiting RNA. The reduced stimulation of pause lifetime by 7-bp relative to 8-bp exit channel RNA duplexes at saturating concentrations in both the absence and presence of NusA suggests that contacts of the exit channel duplex to both RNAP and to NusA can extend to positions >18 nt from the transcript 3′-end. Further, stabilization of exit channel duplexes by NusA when no ssRNA is present upstream from the duplex argues strongly against the indirect NusA action model in which the duplex-stabilizing effect is proposed to result from NusA displacement of the upstream hairpin arm from a sequestering ssRNA binding site on RNAP (29) (see “Discussion”).
TABLE 3.
Pause enhancement by NusA for different types of duplexes and oligo concentrations
The pause half-life was calculated from the rate of pause escape on the scaffolds containing different exit channel duplexes using an in vitro transcriptional pause assay as described under “Experimental Procedures.” 2.5 μm full-length NusA was used in all experiments.
| Types of duplex | [Oligo] | Pause half-life |
||
|---|---|---|---|---|
| −NusA | +NusA | +NusA/−NusA | ||
| μm | s | |||
| 7-bp RNA-RNA duplex | 1 | 14 ± 1 | 62 ± 5 | 4.4 ± 0.5 |
| 32 | 38 ± 1 | 59 ± 5 | 5.2 ± 0.3 | |
| 10-bp DNA-RNA duplex | 1 | 15 ± 0.6 | 48 ± 2 | 3.2 ± 0.2 |
| 32 | 26 ± 2 | 88 ± 7 | 3.4 ± .04 | |
| 12-bp DNA-RNA duplex | 1 | 35 ± 3 | 170 ± 15 | 4.8 ± 0.6 |
| 32 | 36 ± 3 | 186 ± 17 | 5 ± 0.6 | |
Exit Channel DNA-RNA Duplexes Reduce NusA Stimulation of PEC Lifetime
We next tested whether NusA would have comparable effects on pause duration for PECs containing exit DNA-RNA duplexes. Using the same set of exit channel DNA-RNA duplexes that elicited maximal NusA effect with a ≥10-bp RNA-RNA duplex (Fig. 4, A and B), we found that NusA had no significant effect on pause duration for DNA-RNA <10 bp and that the maximal NusA effect of a 3.5–5-fold increase in pause duration occurred with DNA-RNA duplexes of 11–12 bp (in contrast to ≥10-fold stimulation for 11- and 12-bp RNA-RNA duplexes; Fig. 5B). To test whether DNA oligos simply bind more weakly even in the presence of NusA, we compared the effect of 10-mer and 12-mer DNA oligos at 1 μm and 32 μm with and without NusA (Table 3). In contrast to the result with a 7-mer RNA oligo, 1 μm 10-mer DNA oligo proved to be subsaturating even in the presence of NusA, whereas a 12-mer DNA oligo gave saturating effects at 1 μm in the absence or presence of NusA. Thus, not only are RNA-RNA duplexes more effective at stimulating pausing than DNA-RNA duplexes even at saturating oligo concentrations, but the formation of RNA-RNA duplexes also appears to be stabilized by NusA in ways that DNA-RNA duplexes are not.
Single- or Double-stranded RNA Upstream of the Exit Channel Duplex Does Not Contribute to the Direct Effect of Exit Channel Duplexes on Pause Duration
When the his pause hairpin or other exit channel structures form during natural transcription, the exit channel must accommodate three RNA strands: the two strands of the RNA duplex and the third RNA strand that connects the bottom of the hairpin stem near the lid domain to the upstream RNA transcript. This third RNA strand is thought to exit under the RNAP flap domain and is not required for effects on pause duration of the his pause hairpin or of artificial exit channel duplexes (23, 28). However, it remains unclear whether RNA upstream from the exit channel duplex, in single- or double-stranded form, may positively or negatively affect the formation of exit channel duplexes, for instance by affecting clamp or flap positioning, because the his pause hairpin may form robustly and obscure such effects. To determine the impact of upstream RNA on duplex formation, we designed RNA oligos that could form 8-bp exit channel duplexes identical to those formed by our 8-mer RNA oligo but that contained additional non-complementary nt upstream of the duplex (Fig. 6). In one case, 8 non-complementary Us were present upstream (U-tail RNA), and in the other, a short hairpin structure was present (hairpin-tail RNA; 7-bp stem, 4-nt loop). At high oligo concentration (32 μm), we found that the U-tail RNA and hairpin-tail RNA stimulated pause duration equivalently to the 8-mer RNA oligo (Fig. 6). Thus, neither the U-tail nor hairpin-tail altered the effect of the exit channel duplex per se. However, at 1 μm, the hairpin-tail RNA, but not the U-tail RNA, increased pausing much less effectively than the 8-mer RNA previously found to be saturating at 1 μm (see above). Thus, the hairpin-tail RNA, but not the single-stranded upstream RNA, appears to inhibit formation of the exit channel duplex.
FIGURE 6.
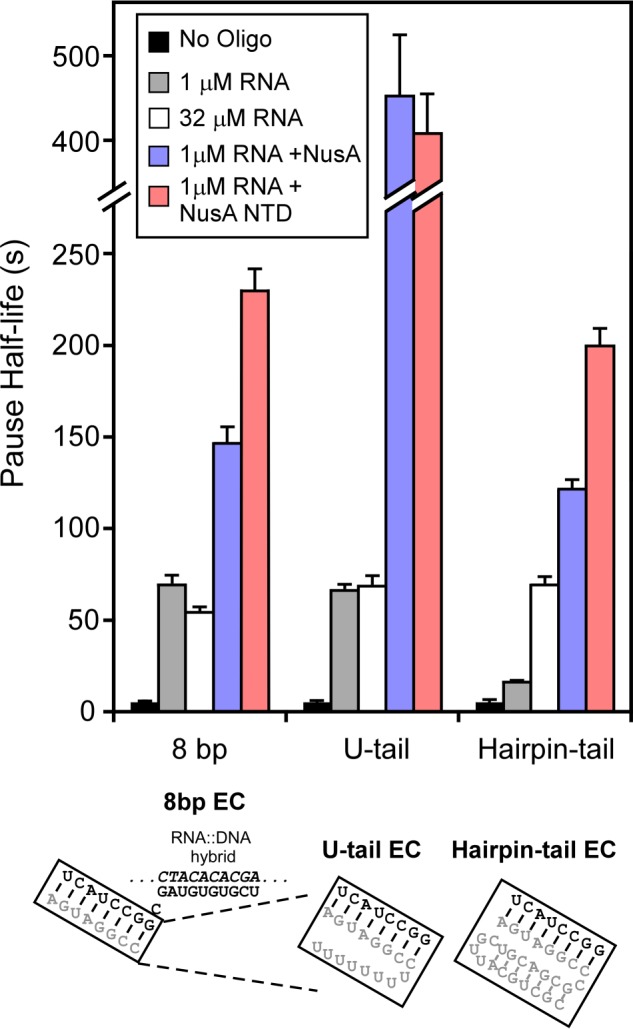
Full-length NusA favors but does not require exit channel duplexes with upstream ssRNA for enhancement of PEC lifetime. Pause stimulation by the antisense 8-mer RNA oligo, RNA oligos with single strand tails (U-tail EC), and RNA oligos with double strand tails (Hairpin-tail EC). ECs were shown in Fig. 2B (see “Experimental Procedures”). Antisense 8-mer RNA oligo, U-tail antisense RNA oligo, and hairpin-tail antisense RNA oligo were added to 1 μm or 32 μm to generate the 8-bp EC, U-tail EC, and hairpin-tail EC, respectively. Pause half-lives were measured as in the preceding figures (see “Experimental Procedures”) in the absence of NusA, with full-length NusA (2.5 μm) or NusA-NTD (residues 1–137; 4 μm). Error bars, S.D.
ssRNA Upstream from an Exit Channel Duplex Enhances the Effect on Pause Duration of Full-length NusA but Not of NusA-NTD
Ha et al. (23) found that full-length NusA required about 2-fold higher concentration to stimulate pausing on a his pause scaffold relative to on a promoter-initiated template for which RNA is present upstream of the pause hairpin, but that this difference was insignificant for the NusA-NTD alone, which binds PECs more weakly (20–40-fold higher apparent Ki). Thus, we wondered if ssRNA upstream from an exit channel duplex could contribute to NusA stimulation of pause duration. To address this question, we tested NusA enhancement of pause duration using the U-tail and hairpin-tail RNAs oligos. Although the effects of 8-mer RNAs with or without the U-tail and the NusA effect on an 8-bp exit channel duplex saturate at 1 μm oligo, pause duration increased for the U-tail RNA relative to the 8-mer RNA in the presence of NusA (Fig. 6). Consistent with findings of Ha et al. (23), some of this effect disappeared when using the NusA-NTD. Although upstream RNA is not needed for NusA action, single-stranded, upstream RNA can contribute to full-length NusA action. In contrast, PECs containing the hairpin-tail RNA reached about the same pause duration in the presence of NusA as observed for the 8-mer oligo. Hence, duplex RNA upstream of the exit channel duplex does not appear to aid NusA action. These results are consistent with an interaction of the NusA KH1 and KH2 domains with single-stranded RNA upstream of the exit channel duplex that contributes to NusA enhancement of pausing, as proposed by Ha et al. (23).
RfaH Pause Suppression Can Be Overcome by High Concentrations of Antisense RNA
RfaH, a NusG paralog found in some enterobacteria, was found previously to inhibit pausing at the his pause site (5, 31). This effect of RfaH appears to be mediated by RfaH contacts to the RNAP clamp and lobe domains that prevent clamp opening required for exit channel duplex formation (Figs. 1 and 7A) (30). If this model is correct, then binding of antisense oligos in the RNAP exit channel and binding of RfaH to RNAP may compete indirectly; bound RfaH would be expected to inhibit oligo binding by locking the clamp shut, whereas bound oligo would be expected to inhibit RfaH binding by favoring an open clamp position that precludes simultaneous RfaH contacts to the clamp and lobe. Alternatively, RfaH may act through effects on the active site even when an exit channel duplex is formed; in this latter case, high concentrations of antisense oligo should not outcompete pause suppression by RfaH.
To test whether RfaH competes with antisense oligo binding or acts on ECs even in the presence of an exit channel duplex, we tested pause enhancement by the 8-mer antisense RNA and pause suppression by the isolated N-terminal domain of RfaH (RfaH-NTD). RfaH-NTD lacks the autoinhibitory C-terminal RfaH domain and is capable of RfaH action even in the absence of an ops (operon polarity suppressor) sequence on the EC non-template strand (31, 37) (see “Experimental Procedures”). RfaH-NTD decreased pause dwell time at the his pause by a factor of ∼5 at 1 μm antisense 8-mer (pause half-life ∼69 s without RfaH-NTD versus 14 s with 500 nm RfaH-NTD and required only 125 nm RfaH-NTD for a large effect; Fig. 7, B and C, and Table 4). Before testing for competition between RfaH and antisense RNA, we next asked if NusG might exhibit a similar effect. Although the NusG CTD does not inhibit NusG binding to RNAP (38) and full-length NusG has little effect on hairpin-stabilized pausing (12), we reasoned that high concentrations of NusG might compete for antisense RNA binding. Indeed, we found that 500 nm NusG partially suppressed pause stimulation by 1 μm antisense RNA (Fig. 7C), but the weak effect and absence of any effect at lower concentration precluded using NusG for our planned experiment. We concluded that NusG probably binds the closed clamp EC more weakly than RfaH-NTD and therefore used RfaH-NTD for subsequent experiments.
TABLE 4.
Antagonistic effects of RfaH and antisense 8-mer RNA oligo on pausing
Pause half-life and fraction paused species were calculated from pause assays using the 8-bp scaffold (Fig. 2B), as described under “Experimental Procedures.” Errors are S.D. from experimental triplicates. ND, not determined.
| RfaH-NTD | 1 μm 8-mer |
32 μm 8-mer |
64 μm 8-mer |
128 μm 8-mer |
||||
|---|---|---|---|---|---|---|---|---|
| Half-life | Fraction | Half-life | Fraction | Half-life | Fraction | Half-life | Fraction | |
| nm | s | s | s | s | ||||
| 500 | 14 ± 0.3 | 0.48 ± 0.01 | 17 ± 3 | 0.67 ± 0.07 | 14 ± 1 | 0.55 ± 0.01 | 17 ± 1 | 0.52 ± 0.01 |
| 250 | 14 ± 2 | 0.51 ± 0.07 | 14 ± 2 | 0.53 ± 0.08 | 16 ± 1 | 0.55 ± 0.01 | 20 ± 2 | 0.51 ± 0.01 |
| 46 ± 9 | 0.22 ± 0.09 | 46 ± 9 | 0.23 ± 0.09 | |||||
| 125 | 14 ± 3 | 0.47 ± 0.07 | 16 ± 7 | 0.35 ± 0.1 | 44 ± 1 | 0.84 ± 0.02 | 62 ± 6 | 0.64 ± 0.01 |
| 58 ± 9 | 0.32 ± 0.09 | 69 ± 14 | 0.43 ± 0.1 | |||||
| 0 | 69 ± 5 | 0.71 ± 0.07 | 71 ± 6 | 0.69 ± 0.01 | 72 ± 6 | 0.68 ± 0.004 | 79 ± 7 | 0.68 ± 0.01 |
| 125a | 14 ± 5 | 0.3 ± 0.1 | ND | ND | ND | ND | ND | ND |
| 60 ± 15 | 0.38 ± 0.1 | ND | ND | ND | ND | ND | ND | |
a 127 μm non-complementary oligo was also added.
To test whether the 8-mer antisense RNA and RfaH-NTD compete as a function of their relative concentrations, we conducted the oligo-stimulated pause assay at various RfaH-NTD and 8-mer concentrations. In some conditions, the rate of pause RNA disappearance was biphasic (e.g. at 250 nm RfaH-NTD and 1 μm 8-mer; Fig. 7D and Table 4). When present, the faster pause escape rate was relatively constant with a t½ of ∼14 s (Table 4). In contrast, the t½ of the slow species, when evident, was inversely related to the concentration of RfaH-NTD (Table 4). To simplify analysis given the presence of the two pause species, we calculated pause strengths (τ0; fraction paused/escape rate; Fig. 7D; see “Experimental Procedures”) because τ0 of multiple pause species at one position can be added. We found that high concentrations of RfaH-NTD could outcompete the effect of even high concentrations of 8-mer antisense oligo. However, at intermediate concentrations of RfaH (125 nm RfaH-NTD), high concentrations of 8-mer could overcome pause suppression by RfaH-NTD, although 1 μm 8-mer was sufficient to give the full effect of the 8-mer in the absence of RfaH-NTD. To verify that this effect was specific to formation of an exit channel duplex, we also tested high concentrations of a non-complementary 8-mer RNA in the presence of 1 μm antisense 8-mer and 125 nm RfaH-NTD (Table 4). We found that high concentrations of non-complementary 8-mer did not compete for pause suppression by RfaH, verifying that competition occurs through formation of an exit channel duplex. In our experiments, we added RfaH-NTD and the oligo at the same time. To test whether the order of addition is important, we added RfaH-NTD followed by the addition of oligo or vice versa. We found that the competition between RfaH and the oligo was unaffected by the order of RfaH and oligo addition. We conclude that RfaH-NTD and 8-mer antisense oligo compete in a concentration-dependent manner consistent with the model in which RfaH stabilizes the closed clamp, which inhibits 8-mer binding, whereas exit channel duplexes stabilize the open clamp, which inhibits RfaH binding. Our results suggest that E. coli NusG acts similarly but with reduced ability to favor clamp closing (see “Discussion”).
DISCUSSION
Our study provides several new insights into the mechanism of pause enhancement by exit channel duplexes and the mechanisms of the elongation factors NusA and RfaH. We found that the length and type of the exit channel duplexes affect duplex-stabilized pausing and NusA enhancement of pausing differently. Beyond simply being harder to form, DNA-RNA exit channel duplexes are less effective at stabilizing the paused state even in the absence of NusA. The maximal NusA stimulation of pausing requires duplexes 2 bp longer than the maximal exit channel duplex effect without NusA (≥10-bp versus ≥8-bp RNA-RNA duplexes, respectively). Our results suggest that NusA functions through direct NusA-duplex interaction rather than indirect displacement of ssRNA binding to RNAP and that RfaH and exit channel duplexes compete for opposite effects on RNAP clamp conformation.
The Bacterial RNAP Exit Channel Appears to Recognize RNA-RNA Duplexes Preferentially
Although many studies provide evidence for direct RNA hairpin-RNAP interaction in the stimulation of transcriptional pausing by E. coli RNAP (22, 25–27, 39, 40), little is known about the structural requirements for these effects. Chan and Landick (26) found that a UUCG tetraloop could replace the 8-nt loop normally present on the 5-bp stem of the his pause hairpin without loss of pause stabilization but that the tetraloop his pause hairpin reduced the effect of NusA. Toulokhonov et al. (27) found that increasing the length of the his pause hairpin stem to 8-bp gave a 50% increase in pause duration, about the same effect as increasing the distance of the hairpin from the RNA 3′-end from 11 to 12 nt. Our results here suggest that the 8-bp RNA-RNA stem is sufficient for the maximal effect of an exit channel duplex, whereas a 7-bp stem is less effective, and a ≤6-bp stem has little effect. The requirement for ≥7 bp for pause stimulation is consistent with the recent report of a “rule of seven” for rapid pairing of oligonucleotides (36); for both RNA and DNA, and both in vivo and in vitro, seven contiguous bp greatly increase kon for pairing. We infer that the same rule applies to pairing of nascent RNA in the RNAP exit channel to oligos in solution.
Interestingly, the concentration dependences of pause stimulation by 7-mer antisense RNA oligo and 10-mer antisense DNA oligo (Fig. 4, C and D) are not much different than those predicted for pairing of the pure nucleic acids at 37 °C and the buffer conditions used in our assay (50% of 50 nm transcript predicted to be paired by ∼2 μm 7-mer RNA or ∼0.7 μm 10-mer DNA based on data in Table 2). Thus, the RNAP exit channel does not appear to enhance or inhibit oligo binding dramatically, although direct measures of oligo binding will be required to verify this conclusion.
However, the natural 5-bp stem of the his pause hairpin is more effective than our 7-bp artificial duplex. This result suggests that the hairpin loop, which is likely to assume some degree of secondary structure, contributes to the effect of the his pause hairpin although a single-stranded loop is not required for the effect of artificial duplexes. Thus, the natural his pause hairpin may resemble the artificial 8-bp exit channel duplex in overall shape, evident in a model of the his pause hairpin loop (23), and this duplex length apparently is required for the full effect of the exit channel duplex.
Interestingly, DNA-RNA duplexes of any size are unable to mimic the full effect of the his pause hairpin (Fig. 4). Although DNA-RNA duplexes are expected to be less stable than RNA-RNA duplexes (41–43), DNA-RNA duplexes predicted to be as stable as somewhat shorter RNA-RNA duplexes (44, 45) (Table 3) still fail to increase pause duration to the same extent as those RNA-RNA duplexes (Fig. 4). We suggest that this reduced effect of DNA-RNA exit channel duplexes is likely to reflect some extent of specificity in the interactions of pause-enhancing duplexes with the RNAP exit channel. In crystal structures, a 10-bp DNA-RNA duplex is at least as large in both helix diameter and length as an 8-bp RNA-RNA duplex (Fig. 8). Hence, based simply on steric consequences for the exit channel, the DNA-RNA duplex should be as effective as the RNA-RNA duplex in altering RNAP clamp conformation. The small differences evident are in the positions of phosphate charges relative to the minor groove. Although differences in interactions within the grooves are possible, we favor the view that differences in locations of the phosphates on the RNA-RNA duplex increase interactions with positive charges on RNAP that either slow translocation or alter interactions with charges on RNAP that result in a greater degree of clamp opening. Distinguishing these two possible effects remains an important research question. A PEC crystal structure that includes an exit channel duplex would be highly informative, but all efforts to date to obtain such a structure have resulted either in insufficient resolution to detect the duplex or, more likely, degradation of the duplex during formation of the crystal lattice (17, 21).
FIGURE 8.
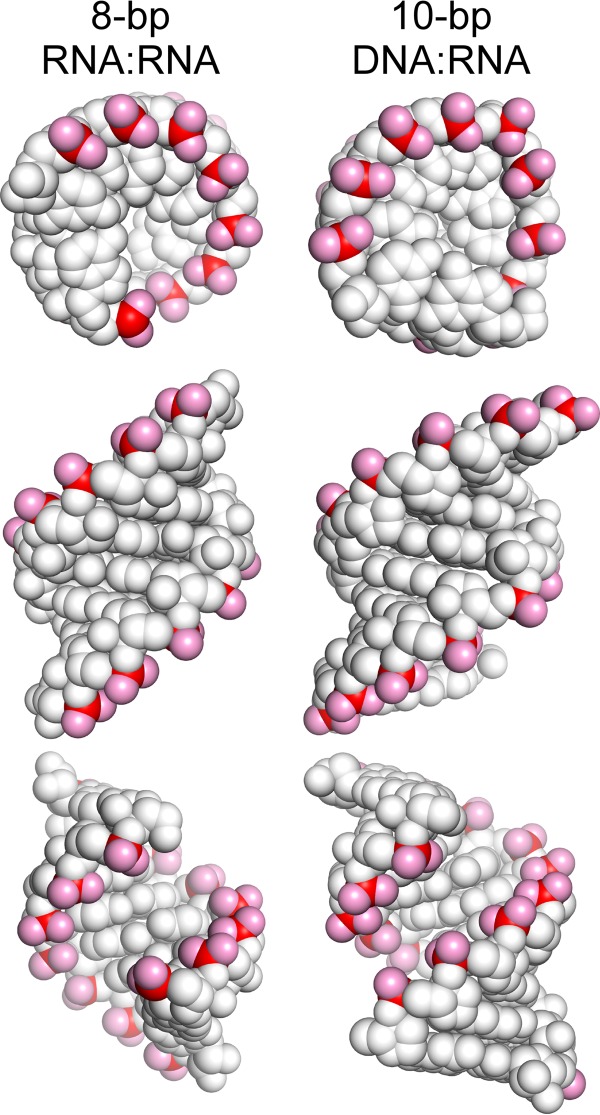
Structures of 8-bp RNA-RNA and 10-bp DNA-RNA duplexes. Representations of duplex structures (Protein Data Bank entries 1rna and 1fix) (57, 58) are shown top down (top), from the major groove side (middle), and from the minor groove side (bottom) with phosphates in red and phosphate oxygens in pink.
NusA Remodels RNAP Exit Channel Interaction Specificity
The E. coli hexadomain NusA regulator (NTD-S1-KH1-KH2-AR1-AR2) is known to interact with the RNAP exit channel via an essential NusA NTD-RNAP-flap tip contact (23, 24, 27), alone to promote hairpin-stabilized pausing and intrinsic termination (46, 47) and in concert with antiterminator proteins like λN or λQ to suppress termination by inhibiting formation of exit channel duplexes (29, 48). Based on nascent RNA-NusA cross-linking, Gusarov and Nudler (29) argue that alone NusA stimulates the exit channel duplex formation indirectly by binding to and displacing nascent RNA from a ssRNA-specific binding site near the exit channel. In a variant of this model, Yang and Lewis propose that the NusA AR1-2 interacts with the upstream arm of the exit channel hairpin to displace it from the ssRNA-binding site (49). Our results contradict the simple versions of either model. Pause enhancement by NusA requires the potential to form an exit channel duplex but remains strong even at high concentrations of antisense oligos that could both fully saturate an ssRNA-binding site and form the exit channel duplex. NusA-NTD alone can achieve this enhancement, but it requires a 10-bp rather than 8-bp duplex. These observations are most consistent with a direct interaction of NusA-NTD with the exit channel duplex that is optimal with a 10-bp duplex and either inhibits translocation or stabilizes clamp opening. This direct model of NusA interaction also explains the observations that either single-stranded nascent RNA or nascent hairpin RNA cross-links to NusA-NTD (23) and that a replacement of the 8-nt loop of the his pause hairpin with a UUCG tetraloop sequence (which stabilizes the hairpin but reduces the apparent duplex length) decreases the effect of NusA (26).
Thus, we favor the view that NusA remodels the RNA binding specificity of the RNAP exit channel to include additional duplex contacts extending up to −21 from the RNA 3′-end (a 10-bp duplex from −12 to −21). However, greater insight into the nature of direct NusA-nascent RNA interactions awaits PEC crystal structures that include both exit channel duplexes and NusA.
RfaH and Exit Channel Duplexes May Compete Indirectly by Tightest Binding to Different RNAP Conformations
Both RfaH and its general paralog NusG, the only elongation factor conserved in all three domains of life, are thought to stabilize the closed clamp, elongation-efficient conformation of RNAP (50–52). RfaH requires interaction of its CTD with a specific non-template strand DNA sequence called ops to undergo a structural rearrangement and liberate the RNAP-binding activity of the RfaH-NTD (5, 31), but both RfaH-NTD and NusG-NTD alone bind ECs through multiple contacts and exhibit antipausing activity (31, 38). However, RfaH-NTD, which contacts the EC on the clamp helices, the gate loop, and potentially the NT DNA strand (Fig. 1), is more potent and can override the pausing-enhancing effects of exit channel duplexes (30, 31, 37). Our findings clarify the action mechanisms of RfaH/NusG class regulators with the key insight that RfaH and exit channel duplex-generating oligos compete in a concentration-dependent manner for their opposite effects on pausing by RNAP (Fig. 7). This concentration dependence necessitates mutually exclusive binding of RfaH-NTD versus oligo to the EC states in which they exert their largest effects on pausing. In principle, mutually exclusive binding could occur because RfaH-NTD directly blocks exit channel duplex formation through contacts to nascent RNA or steric exclusion in the exit channel. However, such a direct effect of RfaH-NTD on exit channel duplexes is highly unlikely because structural models of RfaH binding, which are well constrained by mapped contacts to the clamp helices, gate loop, and NT DNA, place all of RfaH-NTD ≥25 Å from the exiting RNA with the bulky and negatively charged NT DNA strand and upstream DNA duplex between RfaH-NTD and RNA (30, 31). Thus, the simplest interpretation of the concentration-dependent competition of RfaH-NTD and antisense oligo for effects on pausing is that RfaH binds tightest to the closed clamp RNAP conformation and that duplexes form more readily in the open clamp conformation. We note that the results do not preclude RfaH binding weakly to open clamp RNAP (e.g. through a single contact to the clamp helices) or antisense oligo binding to the closed clamp conformation, as long as such binding results in weaker effects on pausing. Indeed, NusG, which binds ECs avidly (38) but competes with duplex formation much less effectively than RfaH (Fig. 7C), may primarily bind the clamp helices and make only weak clamp-closing contacts with other parts of RNAP. Interestingly, Bacillus subtilis NusG (53), like RfaH (5), can even enhance pausing at some sites, suggesting that significant regulatory plasticity exists for this class of regulators.
The concentration-dependent competition of RfaH-NTD and antisense oligos also has an important implication for antitermination, regardless of the precise mechanism. In addition to antipausing, RfaH, λN, and λQ can suppress transcription termination for at least a subset of intrinsic terminators; λN and λQ appear to achieve suppression by blocking formation of the terminator hairpin through contacts also involving NusA (29, 32, 48). For λN and λQ, a number of mechanisms for inhibiting hairpin formation have been considered, including direct antiterminator contacts to nascent RNA. However, our finding that the closed clamp may inhibit exit channel duplex formation suggests that indirect suppression of terminator hairpin formation could be achieved simply by RfaH, λN, or λQ stabilization of a closed clamp conformation. Such a view is also consistent with crystal structures of open and closed clamp RNAP conformations because only the open clamp conformation appears able to accommodate RNA secondary structures in the nascent RNA exit channel (17, 21).
The idea that different effects of regulators and nascent RNA structures on transcript elongation by RNAP may be mediated by competing effects on clamp position is attractive because it would allow synergy or antagonism between different regulators and structures interacting at different locations of the clamp. Depending on the strengths of interactions with open or closed clamp conformations, possible direct contacts among regulators and RNA structures, and the consequences of clamp conformations for RNAP activity, multiple regulatory inputs to elongation could be integrated. Much work remains, however, to understand specific interactions of regulators with the clamp and the consequences of different clamp conformations for different steps in the nucleotide addition cycle, such as translocation or catalysis mediated by trigger loop folding.
Acknowledgments
We thank members of the Landick laboratory for many helpful discussions and for comments on the manuscript, Rachel Mooney for providing plasmids, and Irina Artsimovitch for providing pIA777 plasmid and the RfaH purification protocol.
This work was supported, in whole or in part, by National Institutes of Health Grant GM39660 (to R. L.).
- RNAP
- RNA polymerase
- EC
- elongation complex
- PEC
- paused elongation complex
- nt
- nucleotide
- oligo
- oligonucleotide
- NTD and CTD
- N- and C-terminal domain, respectively.
REFERENCES
- 1. Landick R. (2006) The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 34, 1062–1066 [DOI] [PubMed] [Google Scholar]
- 2. Pan T., Artsimovitch I., Fang X. W., Landick R., Sosnick T. R. (1999) Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. U.S.A. 96, 9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan T., Sosnick T. (2006) RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 35, 161–175 [DOI] [PubMed] [Google Scholar]
- 4. Landick R., Carey J., Yanofsky C. (1985) Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. U.S.A. 82, 4663–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Artsimovitch I., Landick R. (2002) The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203 [DOI] [PubMed] [Google Scholar]
- 6. Gusarov I., Nudler E. (1999) The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 [DOI] [PubMed] [Google Scholar]
- 7. Peters J. M., Vangeloff A. D., Landick R. (2011) Bacterial transcription terminators. The RNA 3′-end chronicles. J. Mol. Biol. 412, 793–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wickiser J. K., Winkler W. C., Breaker R. R., Crothers D. M. (2005) The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 [DOI] [PubMed] [Google Scholar]
- 9. Proshkin S., Rahmouni A. R., Mironov A., Nudler E. (2010) Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kassavetis G. A., Chamberlin M. J. (1981) Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J. Biol. Chem. 256, 2777–2786 [PubMed] [Google Scholar]
- 11. Neuman K. C., Abbondanzieri E. A., Landick R., Gelles J., Block S. M. (2003) Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell 115, 437–447 [DOI] [PubMed] [Google Scholar]
- 12. Artsimovitch I., Landick R. (2000) Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 97, 7090–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toulokhonov I., Zhang J., Palangat M., Landick R. (2007) A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419 [DOI] [PubMed] [Google Scholar]
- 14. Zhang J., Landick R. (2009) Substrate loading, nucleotide addition, and translocation by RNA polymerase. in RNA Polymerase as Molecular Motors (Buc H., Strick T., eds) pp. 206–235, Royal Society of Chemistry, London [Google Scholar]
- 15. Malinen A. M., Turtola M., Parthiban M., Vainonen L., Johnson M. S., Belogurov G. A. (2012) Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 40, 7442–7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kireeva M., Kashlev M., Burton Z. F. (2010) Translocation by multi-subunit RNA polymerases. Biochim. Biophys. Acta 1799, 389–401 [DOI] [PubMed] [Google Scholar]
- 17. Weixlbaumer A., Leon K., Landick R., Darst S. A. (2013) Structural basis of transcriptional pausing in bacteria. Cell 152, 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J., Palangat M., Landick R. (2010) Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 17, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nayak D., Voss M., Windgassen T., Mooney R. A., Landick R. (2013) Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol. Cell 50, 882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakraborty A., Wang D., Ebright Y. W., Korlann Y., Kortkhonjia E., Kim T., Chowdhury S., Wigneshweraraj S., Irschik H., Jansen R., Nixon B. T., Knight J., Weiss S., Ebright R. H. (2012) Opening and closing of the bacterial RNA polymerase clamp. Science 337, 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tagami S., Sekine S., Kumarevel T., Hino N., Murayama Y., Kamegamori S., Yamamoto M., Sakamoto K., Yokoyama S. (2010) Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468, 978–982 [DOI] [PubMed] [Google Scholar]
- 22. Toulokhonov I., Landick R. (2003) The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell 12, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 23. Ha K. S., Toulokhonov I., Vassylyev D. G., Landick R. (2010) The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 401, 708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang X., Molimau S., Doherty G. P., Johnston E. B., Marles-Wright J., Rothnagel R., Hankamer B., Lewis R. J., Lewis P. J. (2009) The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 10, 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang D., Severinov K., Landick R. (1997) Preferential interaction of the his pause RNA hairpin with RNA polymerase β subunit residues 904–950 correlates with strong transcriptional pausing. Proc. Natl. Acad. Sci. U.S.A. 94, 8433–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan C. L., Landick R. (1993) Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 233, 25–42 [DOI] [PubMed] [Google Scholar]
- 27. Toulokhonov I., Artsimovitch I., Landick R. (2001) Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292, 730–733 [DOI] [PubMed] [Google Scholar]
- 28. Kyzer S., Ha K. S., Landick R., Palangat M. (2007) Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J. Biol. Chem. 282, 19020–19028 [DOI] [PubMed] [Google Scholar]
- 29. Gusarov I., Nudler E. (2001) Control of intrinsic transcription termination by N and NusA. The basic mechanisms. Cell 107, 437–449 [DOI] [PubMed] [Google Scholar]
- 30. Sevostyanova A., Belogurov G. A., Mooney R. A., Landick R., Artsimovitch I. (2011) The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol. Cell 43, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belogurov G. A., Vassylyeva M. N., Svetlov V., Klyuyev S., Grishin N. V., Vassylyev D. G., Artsimovitch I. (2007) Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 26, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Svetlov V., Belogurov G. A., Shabrova E., Vassylyev D. G., Artsimovitch I. (2007) Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35, 5694–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vassylyeva M. N., Lee J., Sekine S. I., Laptenko O., Kuramitsu S., Shibata T., Inoue Y., Borukhov S., Vassylyev D. G., Yokoyama S. (2002) Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta Crystallogr. D Biol. Crystallogr. 58, 1497–1500 [DOI] [PubMed] [Google Scholar]
- 34. Hein P. P., Palangat M., Landick R. (2011) RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry 50, 7002–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Landick R., Wang D., Chan C. (1996) Quantitative analysis of transcriptional pausing by RNA polymerase: the his leader pause site as a paradigm. Methods Enzymol. 274, 334–353 [DOI] [PubMed] [Google Scholar]
- 36. Cisse I. I., Kim H., Ha T. (2012) A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat. Struct. Mol. Biol. 19, 623–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Belogurov G. A., Sevostyanova A., Svetlov V., Artsimovitch I. (2010) Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol. 76, 286–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mooney R. A., Schweimer K., Rösch P., Gottesman M., Landick R. (2009) Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 391, 341–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Artsimovitch I., Landick R. (1998) Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 12, 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan C. L., Landick R. (1997) Effects of neutral salts on RNA chain elongation and pausing by Escherichia coli RNA polymerase. J. Mol. Biol. 268, 37–53 [DOI] [PubMed] [Google Scholar]
- 41. Nakano S., Fujimoto M., Hara H., Sugimoto N. (1999) Nucleic acid duplex stability. Influence of base composition on cation effects. Nucleic Acids Res. 27, 2957–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Owczarzy R., Moreira B. G., You Y., Behlke M. A., Walder J. A. (2008) Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry 47, 5336–5353 [DOI] [PubMed] [Google Scholar]
- 43. Owczarzy R., You Y., Moreira B. G., Manthey J. A., Huang L., Behlke M. A., Walder J. A. (2004) Effects of sodium ions on DNA duplex oligomers. Improved predictions of melting temperatures. Biochemistry 43, 3537–3554 [DOI] [PubMed] [Google Scholar]
- 44. Xia T., SantaLucia J., Jr., Burkard M. E., Kierzek R., Schroeder S. J., Jiao X., Cox C., Turner D. H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–14735 [DOI] [PubMed] [Google Scholar]
- 45. Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34, 11211–11216 [DOI] [PubMed] [Google Scholar]
- 46. Farnham P. J., Greenblatt J., Platt T. (1982) Effects of NusA protein on transcription termination of the tryptophan operon of Escherichia coli. Cell 29, 945–951 [DOI] [PubMed] [Google Scholar]
- 47. Landick R., Yanofsky C. (1984) Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J. Biol. Chem. 259, 11550–11555 [PubMed] [Google Scholar]
- 48. Shankar S., Hatoum A., Roberts J. W. (2007) A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol. Cell 27, 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang X., Lewis P. J. (2010) The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 7, 272–275 [DOI] [PubMed] [Google Scholar]
- 50. Belogurov G. A., Mooney R. A., Svetlov V., Landick R., Artsimovitch I. (2009) Functional specialization of transcription elongation factors. EMBO J. 28, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klein B. J., Bose D., Baker K. J., Yusoff Z. M., Zhang X., Murakami K. S. (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc. Natl. Acad. Sci. U.S.A. 108, 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martinez-Rucobo F. W., Sainsbury S., Cheung A. C., Cramer P. (2011) Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 30, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yakhnin A. V., Yakhnin H., Babitzke P. (2008) Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc. Natl. Acad. Sci. U.S.A. 105, 16131–16136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Opalka N., Brown J., Lane W. J., Twist K. A., Landick R., Asturias F. J., Darst S. A. (2010) Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 8, e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Artsimovitch I., Svetlov V., Murakami K. S., Landick R. (2003) Co-overexpression of E. coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 278, 12344–12355 [DOI] [PubMed] [Google Scholar]
- 56. Worbs M., Bourenkov G. P., Bartunik H. D., Huber R., Wahl M. C. (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell 7, 1177–1189 [DOI] [PubMed] [Google Scholar]
- 57. Dock-Bregeon A. C., Chevrier B., Podjarny A., Johnson J., de Bear J. S., Gough G. R., Gilham P. T., Moras D. (1989) Crystallographic structure of an RNA helix. [U(UA)6A]2. J. Mol. Biol. 209, 459–474 [DOI] [PubMed] [Google Scholar]
- 58. Horton N. C., Finzel B. C. (1996) The structure of an RNA/DNA hybrid. A substrate of the ribonuclease activity of HIV-1 reverse transcriptase. J. Mol. Biol. 264, 521–533 [DOI] [PubMed] [Google Scholar]