Abstract
Our goal was to identify a unique gene expression signature for human colonic stem cells (SCs). Accordingly, we determined the gene expression pattern for a known SC-enriched region—the crypt bottom. Colonic crypts and isolated crypt subsections (top, middle, and bottom) were purified from fresh, normal, human, surgical specimens. We then used an innovative strategy that used two-color microarrays (∼18,500 genes) to compare gene expression in the crypt bottom with expression in the other crypt subsections (middle or top). Array results were validated by PCR and immunostaining. About 25% of genes analyzed were expressed in crypts: 88 preferentially in the bottom, 68 in the middle, and 131 in the top. Among genes upregulated in the bottom, ∼30% were classified as growth and/or developmental genes including several in the PI3 kinase pathway, a six-transmembrane protein STAMP1, and two homeobox (HOXA4, HOXD10) genes. qPCR and immunostaining validated that HOXA4 and HOXD10 are selectively expressed in the normal crypt bottom and are overexpressed in colon carcinomas (CRCs). Immunostaining showed that HOXA4 and HOXD10 are co-expressed with the SC markers CD166 and ALDH1 in cells at the normal crypt bottom, and the number of these co-expressing cells is increased in CRCs. Thus, our findings show that these two HOX genes are selectively expressed in colonic SCs and that HOX overexpression in CRCs parallels the SC overpopulation that occurs during CRC development. Our study suggests that developmental genes play key roles in the maintenance of normal SCs and crypt renewal, and contribute to the SC overpopulation that drives colon tumorigenesis.
Introduction
Identifying, isolating, and characterizing human colonic stem cells (SCs) has been challenging. First, there are a limited number of markers [1] that can unambiguously identify human gastrointestinal (GI) SCs, in large part because they are undifferentiated relative to other crypt cells, and lack typical tissue-specific differentiation markers. Second, human colonic SCs appear to be few in number, accounting for <5% of all crypt cells. Third, to purify them, human colonic SCs need to be isolated from their stromal support. Indeed, much of the research to date on human colonic SCs primarily has relied on anatomical and functional properties of SCs, including location at the crypt base, strong anchorage, extended proliferative potential over time, capacity for self renewal, and multipotency [2].
Accordingly, we devised an innovative strategy for the identification of human colonic SCs. Since colonic SCs have unique functional properties, the lower crypt, which contains most colonic SCs, should have a unique gene expression profile that should be discernable by microarray analysis. Therefore, in the current study, we (1) isolated pure crypts from surrounding stromal elements [3], (2) isolated crypt subsections (bottom, middle, and top), and then (3) used microarray-based gene expression profiling to compare the crypt bottom, a SC-enriched region, with the other crypt subsections (middle and top).
Identifying and characterizing colonic SCs also has clinical relevance because mutation-induced dysregulation of colonic SC dynamics is likely to contribute to the initiation and progression of colorectal cancer (CRC). Indeed, we showed [1] that APC mutations initiate and drive colon tumorigenesis by causing progressive colonic SC overpopulation.
Materials and Methods
Acquisition of tissue
We used freshly isolated colonic crypts from surgical specimens because we needed to obtain relatively large amounts of purified crypt subsections—bottom, middle, and top—that were free of contaminating stromal tissues such as fibroblasts, leukocytes, endothelial cells, and muscle cells. Specimens of fresh normal colonic epithelium were obtained from colectomy specimens through the Thomas Jefferson University Pathology Department. All tissues obtained and protocols used were done with the approval of the Institutional Review Board. Tissue samples (n=3) representing normal colon were taken from the distal margin of resection of tumors of surgical colectomy specimens (>10 cm away from the tumor) from colon cancer or colon adenoma patients who were otherwise healthy and who had no known clinical risk factors for hereditary colon cancer (ie, no other family members with CRC and none diagnosed under age 60). Normal-appearing, APC-mutant colon was obtained from prophylactic colectomy specimens from two familial adenomatous polyposis (FAP) patients with known germline APC mutations—one of whom was being treated with the chemopreventive agent Sulindac.
Isolation of whole colonic crypts and crypt subsections
Crypt subsections (top, middle, and bottom) were purified from tissue samples according to modification of a method that we developed [3] for isolation of whole human colonic crypts. Briefly, the mucosal layer was surgically dissected from underlying submucosal layers of the colon, placed in sterile phosphate-buffered saline (PBS), and washed 3×. The specimen was incubated in 0.04% Na hypochlorite for 15 min at room temp. To isolate crypt top subsections, each specimen was transferred to a PBS solution containing 0.05 mM EDTA, pH 8.0, incubated for 45 min at room temperature, and vigorously shaken by hand for 5 min. The supernatant was removed, the tissue washed thrice with sterile PBS, and the crypt top subsections that had been dislodged into solution were precipitated by centrifugation at 500 g at 4°C. To isolate crypt middle subsections, the same specimen was then transferred to media containing 0.5 mM EDTA in PBS and re-incubated for 30–60 min, and again the tube was shaken by hand for 5 min. The crypt middle subsections that were dislodged were precipitated as above. To isolate crypt bottom subsections the same specimen was then incubated in media containing 3 mM EDTA in PBS and re-incubated followed by shaking. Crypt bottom subsections were isolated and precipitated as above. Crypt subsection quality was checked by inverted phase microscopy (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
Two-color arrays
Two-color microarray profiling was used to characterize gene expression patterns in colonocytes from the three crypt subsections. Our primary objective was to compare the pattern of gene expression for cells at the crypt base with the patterns in the two other crypt regions to begin to determine which genes are preferentially expressed at the crypt base, the crypt subregion where SC reside. This involved isolating RNA from purified colonic crypt subsections that were obtained as described above and then doing microarray profiling for each crypt subsection.
Array-based mRNA expression profiling consisted of several steps: (1) preparation of labeled RNA targets, (2) preparation of the microarray, (3) hybridization and post-hybridization stringency washes, (4) detection and data acquisition of signal intensities, and (5) comparative data analysis and visualization. Note that according to standard array nomenclature the probes are bound to the array while the radio-labeled or dye-tagged target is in the solution.
Preparation of labeled RNA targets
This experiment was repeated twice. Isolated RNA was annealed to magnetic Dyna beads Oligo (dT)25 (Dynal Biotech, Lake Success, NY) on which first strand synthesis was initiated with reverse transcriptase. In brief, 20 μg of total RNA in 75 μL DEPC-treated water was heated at 65°C for 2 min to disrupt the secondary structures and was placed on ice. Then, 150 μL (750 μg) Dynabeads Oligo (dT)25 were added to a 1.5 mL Eppendorf® tube and the vial was placed on the Dynal MPC-S support. After 30 s, the supernatant was discarded, and the beads were washed with 150 μL 2× Binding Buffer and resuspended in 75 μL of 2× Binding buffer. Total RNA was added to the Dynabeads Oligo (dT)25 in Binding Buffer, mixed thoroughly, and annealed for 5 min at room temperature. The tube was kept on the magnet for at least 30 s and the supernatant was discarded. The beads in the tube were washed twice using 200 μL of washing buffer. Bead-bound RNA was reverse transcribed to bead-bound cDNA with SuperScript II reverse transcriptase according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). After 1 h, the beads were captured, the supernatant discarded, and the beads heated at 95°C in the presence of 2 mM EDTA. Random Prime Labeling (Invitrogen) was used to generate a radio-labeled target complementary to bead-bound cDNA with 2 μL Cy5/Cy3 dCTP (Amersham Biosciences, Sunnyvale, CA) for glass microarray slides.
Hybridization and post-hybridization stringency washes
Two-color microarray profiling was then used to characterize differential gene expression in crypt subsections. The 18,432 human cDNA clones in the gene set were grown overnight in 384-well microtiter plates. The inserts were PCR amplified using vector-specific primers M13F and M13R. PCR products were precipitated by ethanol, resuspended in 50% (v/v) DMSO, and printed on Poly-L-Lysine-treated glass slides as two separate arrays, one on each of two glass slides, and are referred to herein as data sets 1 and 2. The slides were then prepared for hybridization by blocking with succinic anhydride. Column purified Cy-3/Cy-5-labeled targets were dried and resuspended in 30 μL hybridization solution [20 μg denatured Cot1 DNA, 20 μg poly-d(A), 4× SSC, and 0.2% (w/v) SDS]. The hybridization solution was added to the microarray slide, covered with a glass coverslip, and hybridized at 65°C for 16–18 h in a humid environment to prevent drying. The coverslip was gently removed in solution (40 mL 2× SSC, 0.1% SDS) and the slide was then rinsed sequentially at room temperature with 40 mL 2× SSC, 0.1% SDS, 1× SSC, and finally 0.1× SSC, for 4 min each rinse. Slides were centrifuged at 500 rpm for 5 min to dry them.
Data acquisition and analysis
Microarray slides were scanned and the fluorescence intensity of each spot recorded using a ScanArray 5000 (Perkin Elmer, Boston MA), dual-color confocal laser scanner. Expression data were determined from paired (Cy5 and Cy3) 16-bit TIFF images using Quantarray. Specifically, the Cy5 and Cy3 images were used to quantify spot intensity following the adaptive method in the Quantarray 3.0 software (Perkin Elmer) to generate raw data files containing measurements of signal and background intensity. Background values based on signal intensities around each spot were subtracted and results were then normalized for differences in dye signal intensity using the Loess principle. Based on visual inspection of the scatterplots (Supplementary Fig. S2), all data below an intensity of 800 counts were considered insufficiently greater than background and were discarded (about 75% of genes). Each array contained duplicate spots for each gene. Therefore, all four spots were analyzed to assess replication. Data where all four spots did not possess an intensity within three standard deviations of each other, or did not show an up- or downregulation of 1.5-fold or greater, were discarded. This left a maximum of 2.13% of the data (415 genes) for consideration. For retained data, the mean intensity for each gene was quantified using all four spots. All further analysis was performed using the final gene list with these mean intensity values. Determination of the genes that showed differential expression was based on increased or decreased signal intensity of 1.5-fold or more between the two channels (Cy5 vs. Cy3). Graphic and informatic analyses of the data were performed using GeneSpring Software Version 6.1 (Silicon Genetics, Redwood City, CA). Gene ontology, annotation, and pathway analysis was performed using OntoGene (http://vortex.cs.wayne.edu:8080/ontoexpress/servlet/UserInfo) and EASE (www.david.niaid.nih.gov/david/ease1.htm).
Immunofluorescence staining of paraffin-embedded sections
Immunofluorescence studies were carried out (after IRB approval) on 5 μm paraffin sections of human colonic tissues from archival tissue blocks from Christiana Care Health System. The slides were deparaffinized in citra solv solution for 20 min to remove the embedding media, and washed twice in absolute ethanol for 5 min each. The slides were then gradually rehydrated gently in a series of 95% alcohol washes for 5 min each and were placed in distilled water for few seconds. Antigen retrieval was performed for tissue sections in 1× retrieval solution (Dako) for 25 s, at full power in a microwave and then for 12 min at low power. Slides were allowed to cool to room temperature and were rinsed in 1×-TBS buffer thrice for 5 min each. The slides were then incubated overnight in 1×-TBS buffer containing 10% normal goat serum and 3% bovine serum albumin (BSA) to block nonspecific antibody binding. This was followed by incubation of the slides with primary antibody (4°C, overnight), diluted 1:100 except for anti-STAMP, which was at 1:200. The primary antibodies used were anti-HOXA4 (Abcam 26097) and HOXD10 (Abcam 76897). Anti-ALDH (BD 611195), anti-CD166 (Abcam 49496), and anti-STAMP1 (Novus 68100) antibodies were the primary antibodies used in co-staining experiments. Next day, slides were washed thrice in 1X-TBS buffer for 5 min each. Tissues were then stained with anti-rabbit Alexa-fluor antibody (Invitrogen) (diluted at 1:1,000) and Hoechst 33342 (1:10,000) for 60 min. After final washes, slides were mounted with a coverslip using slow fade gold antifade reagent (Invitrogen S36936) and were sealed with clear nail polish.
Whole isolated colonic crypts were stained using anti-HOXA4 (Abcam 26097) and HOXD10 (Abcam 76897). Isolated colonic crypts were incubated in 0.2% Triton X-100 in 1× PBS for 5 min, followed by washing with 1× PBS. Blocking was performed using 3% BSA and 10% goat serum for 1 h at room temperature. After blocking, crypts were washed with 1× PBS. Primary antibody incubation was done for 1 h at room temperature. After washing crypts with 1× PBS, crypts were stained with anti-rabbit Alexa-fluor antibody (Invitrogen) (diluted at 1:1,000) and Hoechst 33342 (1:10,000) for 60 min. Stained crypts were then observed under a flouroscence microscope.
Real-time quantitative RT-PCR analysis
Total RNA from normal colonic crypts and colon tumor tissues were isolated using the TRIzol method. Equal amounts of RNA from both samples were reverse transcribed and amplified in a one-step reaction using Superscript III reverse transcriptase (Invitrogen 18080-44). Real-time PCR was performed using SYBR green PCR master mix (4309155 Applied Biosystem). Human HOXA4 and HOXD10 were amplified with gene-specific primers (HOXA4 sense primer: 5′-TCCCCATCTGGACCATAATAGG-3′; antisense primer: 5′-GCAACCAGCACAGACTCTTAACC-3′ and HOXD10 sense 5′-ATAAGCGCAACAAACTCATTTCG-3′ and anti- sense 5′-ATATCGAGGGACGGGAACCT-3′. β-actin (ACTB) was amplified as an internal control by using gene-specific primers, sense primer: 5′-TTGCCGACAGGATGCAGAA-3′; antisense primer: 5′-GCCGATCCACACGGAGTACT-3′. Cycling parameters used were denaturation at 95°C for 10 min, 40 cycles of 15 s at 94°C, 30 s at 60°C, 1 min at 72°C followed by a continuous melting curve. For real-time PCR validation, analysis was done using documented SC markers including ALDH1A1, LGR5, CD166 (ALCAM), DCLK1, and CD133 (PROM1). Human ALDH1a primers used were forward 5′-GTTGTCAAACCAGCAGAGCA-3′ and human ALDH1A reverse 5′-CTGTAGGCCCATAACCAGGA-3′. Lgr5 human primers used were forward 5′-CTTCCAACCTCAGCGTCTTC-3′ and reverse 5′-TTTCCCGCAAGACGTAACTC-3′. Human CD166 (ALCAM) primers were forward 5′-TAGCAGGAATGCAACTGTGG-3′ and reverse 5′-CGCAGACATAGTTTCCAGCA-3′. Human DCLK1 primers used were forward 5′-TGAACAAGAAGACGGCTCACTCC-3′ and reverse 5′-GCTGGTGGGTGATGGACTTGG-3′. Human CD133 (PROM1) primers used were 5′-GGACCCATTGGCATTCTC-3′ and reverse 5′-CAGGACACAGCATAGAATAATC-3′.
In this case, β-actin (ACTB) was the internal control, using sense primer 5′-CCCAGCACAATGAAGATCAA-3′ and 5′-ACATCTGCTGGAAGGTGGAC-3′ as antisense primer. Cycling parameters used for ALDH1A1, CD166 (ALCAM), and LGR5 were denaturation at 95°C for 10 min, 40 cycles of 15 s at 95°C, 60 s at 60°C. Cycling parameters used for CD133 (PROM1) were 50°C for 2 min, 95°C for 10 min, 40 cycles of 15 s at 95°C, 1 min at 60°C. Cycling parameters for DCLK1 were denaturation at 95°C for 10 min, followed by 35 cycles of 15 s at 95°C, 30 s at 65°C, 30 s at 72°C then 72°C for 10 min. The Ct value was then normalized to the internal housekeeping gene β-actin (ACTB). Fold change was determined for tumor samples compared to normal samples. This was calculated by the formula fold change=2^Avg.(Delta(Ct))Tumor/2^Avg.(Delta(Ct))Normal
Statistical analysis
Student's t-test was performed on real-time PCR data and P<0.05 was considered significant.
Results
Microarray analysis
RNA samples from crypt subsections (Supplementary Fig. S1) showed hybridization to ∼4,000 of the ∼18,500 genes analyzed (ie, to ∼22% of the clones on the array). An average correlation from a self-hybridization consistently yielded a value of ∼0.96 (Supplementary Fig. S2A). Each array was normalized using the Loess method [4,5]. This method centered the data around onefold expression ratios, and background cutoffs were calculated using transformed data (to log base 2) (Supplementary Fig. S2B). The experiment was set up as a loop design; therefore, normalization was performed between arrays using the samples. Raw intensity values ranged from 800 counts to 45,000 counts, showing that the experiment utilized the full dynamic range of the scanner. As expected, ∼95% of all genes showed no difference in expression between crypt subsections and no significant outliers or dye bias was detected. Using the log plot (Supplementary Fig. S2C) to determine the background cutoff, any value under 800 counts was not considered.
Bioinformatic analysis
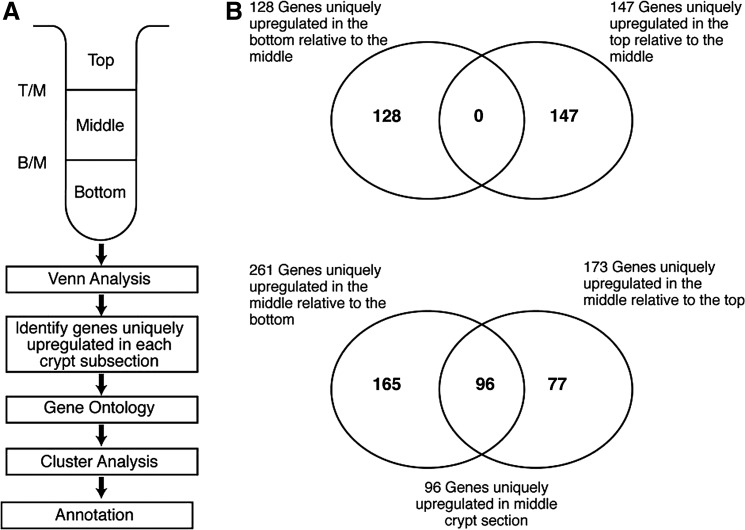
After normalization, data from any probe where all four values were not within 2 standard deviations of each other were not further considered. Data for probes that showed less than a 1.5-fold change were also not used. This left 2.13% of the original gene set to consider. Analysis of overlap was then used to determine which of these genes were up- or downregulated in each of the crypt subsections relative to the other two subsections. This is shown in Venn diagrams (Fig. 1A, B). In this analysis, we recognized that many genes are known to be expressed in a gradient fashion along the crypt axis. To determine which genes are preferentially expressed in the different crypt subsections, rather than in a gradient fashion, we made several comparisons. For example, there were 128 genes upregulated in the bottom relative to the middle (Fig. 1B). Forty of the 128 were also upregulated (but usually less so) in the middle relative to the top. This indicates that these 40 genes are expressed in a gradient fashion along the crypt axis, and that 88 genes are preferentially expressed in the bottom. Similarly, of the 96 genes upregulated in the middle relative to the top or the bottom (Fig. 1B), there were 28 that were upregulated in the bottom relative to the top or vice versa. Thus, we classified 68 genes (96−28=68) as being preferentially expressed in the middle. Using this approach, we found that 287 mRNA species were preferentially expressed in one of the three different crypt subsections: 88 in the crypt bottom; 68 in the crypt middle; 131 in the crypt top. Some (39%) of these transcripts are known genes (Supplementary Table S1), but many (61%) are expressed sequence tags and are as yet unidentified.
FIG. 1.
Identification of unique upregulated gene expression signatures for each crypt section by Venn analysis. (A) Comparisons were made between gene expression data for each crypt subsection. (B) Results from Venn analysis. The top Venn diagram shows that there is no crossover; none of the same genes were upregulated in both top and bottom. The bottom Venn diagram shows that a considerable number of genes are upregulated in the middle compared with both bottom and top.
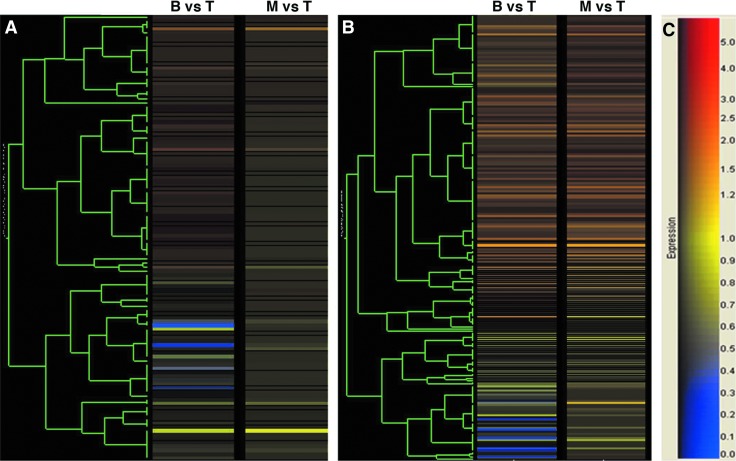
We then performed cluster analysis (Supplementary Fig. S3) as an exploratory visual approach. We analyzed the gene set from array 1 and called that dataset 1 (Supplementary Figs. S3A, S3C), and from array 2 and called that dataset 2 (Supplementary Figs. S3B, S3D). Both were randomly chosen gene sets from 10 different human tissues. K-means cluster analysis was performed on each of these data sets. No unique cellular pathway was apparent from this analysis. But, we did find that each dataset could be partitioned into the same four clusters (Supplementary Fig. S3C, S3D). Hierarchical clustering was performed on the three gene lists from the three crypt subsections, subgrouped, as noted above, into datasets 1 (Fig. 2A) and 2 (Fig. 2B). The data clearly showed that there is a gene expression signature in the crypt bottom that is unique relative to the middle and top. In dataset 1, many genes that are up in the bottom versus the top show relatively little change in the middle versus the top. In dataset 2, there are also many genes up in the bottom versus the top that are also up in the middle versus the top. These latter genes appear to be expressed as a gradient: highest in the bottom and decreasing toward the top of the crypt. In contrast, genes that are up in the bottom versus the top, but not up in the middle versus the top, appear to be selectively expressed in the bottom.
FIG. 2.
Hierarchical cluster analysis of our microarray results for genes expressed in the crypt bottom (bottom vs. top). (A) Data set 1. (B) Data set 2. (C) Color bar for expression ratios. This analysis shows that there are two sets of genes with increased expression (reddish brown color) in the crypt bottom: genes selectively expressed in the bottom (A); genes that are expressed in a gradient fashion (B) from highest in the crypt bottom to lowest in the crypt top. Color images available online at www.liebertpub.com/scd
Gene ontology was performed on all three gene lists (bottom, middle, and top) collectively to identify common themes, patterns, and/or molecular functions. Of these genes, 30% had a known function. The majority of the genes were involved in binding. Forty percent were genes whose functions were involved in protein binding. Several genes that are known to play a role in SC dynamics and/or development were identified (Supplementary Fig. S4), which is consistent with the fact that the crypt bottom is enriched in SCs. Indeed, among the genes upregulated in the crypt bottom, ∼30% were classified as growth and/or developmental genes. This includes two genes, HOXA4 and HOXD10, which are known to be involved in development—they were upregulated (more than twofold) in the crypt bottom. Also, there were several interesting cancer-related genes. Six of them mapped to the PI3 kinase pathway (Supplementary Fig. S5). Several also appeared to be part of the MAP kinase pathway. The gene showing the greatest upregulation (4.1-fold) in expression in the bottom relative to the top was STAMP1 (also known as PCANAP1 or STEAP2), which is a marker for prostate cancer.
Validation using real-time PCR and immunostaining
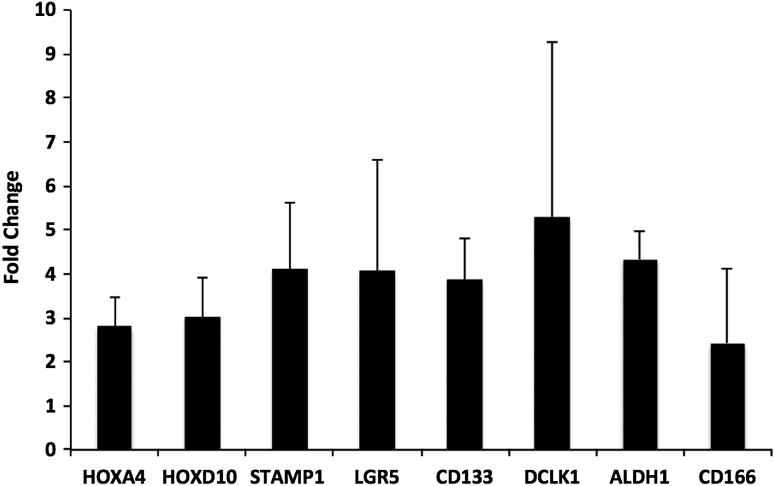
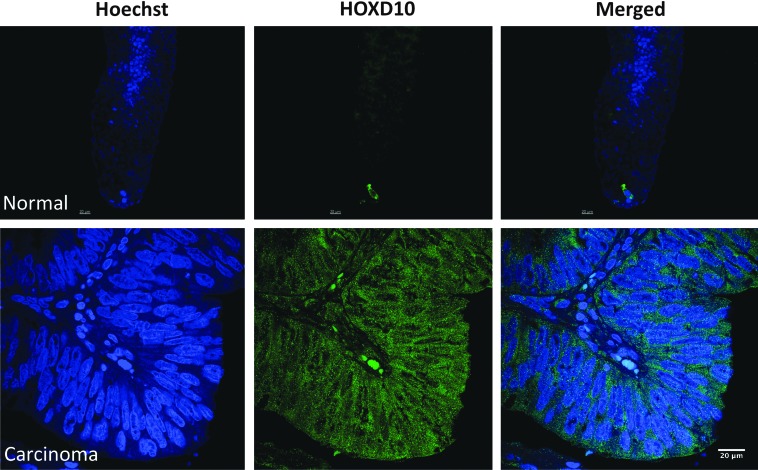
We performed real-time PCR analysis to validate our microarray analysis. We chose HOXA4, HOXD10, and STAMP1 as our target genes and β-actin as our endogenous control. Expression of five known SC markers ALDH1, CD166, CD133, LGR5, and DCLK1 was used as a positive control. Values were obtained for ΔΔCt, comparing the crypt subsections: bottom versus middle (Fig. 3). The real-time PCR data were consistent with the microarray results: HOXA4, HOXD10, and STAMP1 were each upregulated, as were the positive controls (ALDH1, CD166, CD133, LGR5, and DCLK1), in the bottom relative to the middle. As an independent method of validation, we did immunostaining, which confirmed that HOXD10 and HOXA4 are selectively expressed in the bottom of the colonic crypt (Supplementary Fig. S6 and Figs. 4 and 5). Co-staining studies using the SC markers CD166 and ALDH1 were done to determine whether HOXD10 and HOXA4 are expressed in SCs. Results showed that in normal colonic crypts, cells that stain for HOXA4 and HOXD10 co-stain for CD166 and ALDH1 (Fig. 6). Co-staining studies were also done using HOXD10 and STAMP1. Results showed that cells co-stain for HOXD10 and STAMP1 in the bottom of the colonic crypt. Results also showed an increased number of cells that co-stain for HOXD10 and STAMP1 in colon carcinomas (CRCs) (Supplementary Fig. S7).
FIG. 3.
Real-time PCR validation for selective expression of HOXA4, HOXD10, and STAMP1 in the crypt bottom. Positive controls (known SC markers) were LGR5, CD133, DCLK1, ALDH1A1, and CD166, which are known to be preferentially expressed in the crypt bottom. Each bar represents a ratio of gene expression in crypt bottom subsections divided by that in crypt middle subsections. Error bars represent SEM values. SC, stem cell.
FIG. 4.
HOXD10 expression in normal colon and carcinomas. Blue indicates nuclear staining with Hoechst dye; green indicates HOXD10 expression. Upper panels: normal colon. Lower panels: malignant colon. Scale bar=20 μm. Color images available online at www.liebertpub.com/scd
FIG. 5.
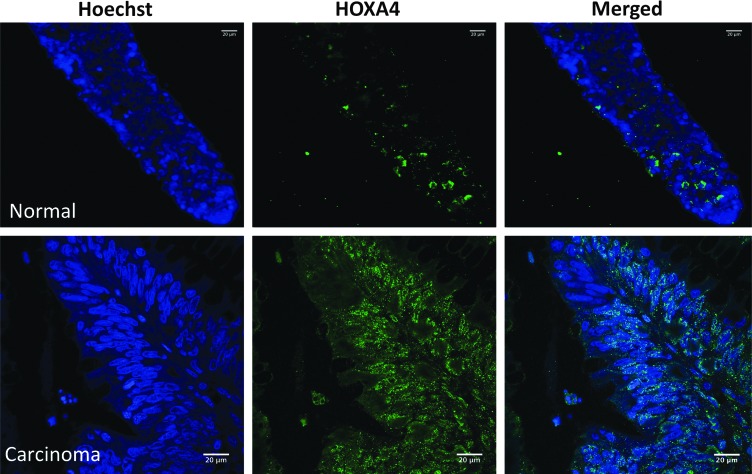
HOXA4 expression in normal colon and carcinomas. Blue indicates nuclear staining with Hoechst dye; green indicates HOXA4 expression. Upper panels: normal colon. Lower panels: malignant colon. Scale bar=20 μm. Color images available online at www.liebertpub.com/scd
FIG. 6.
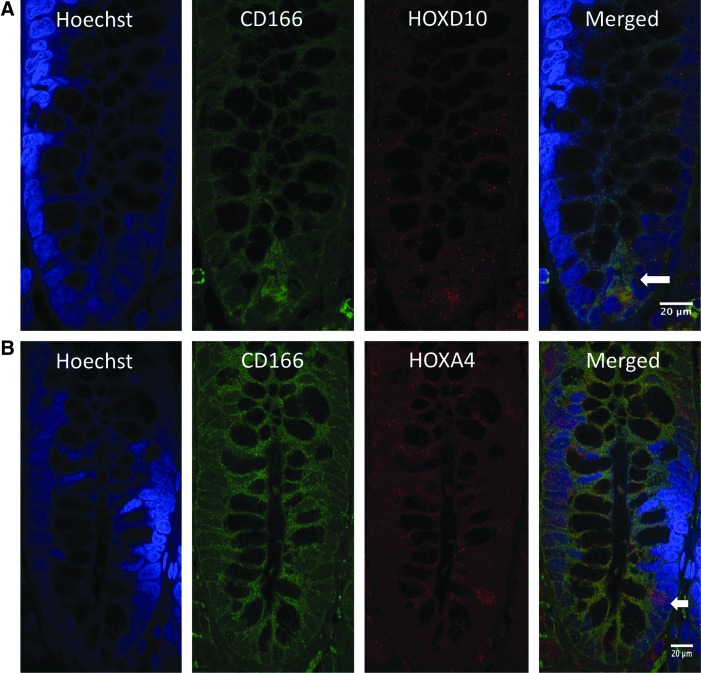
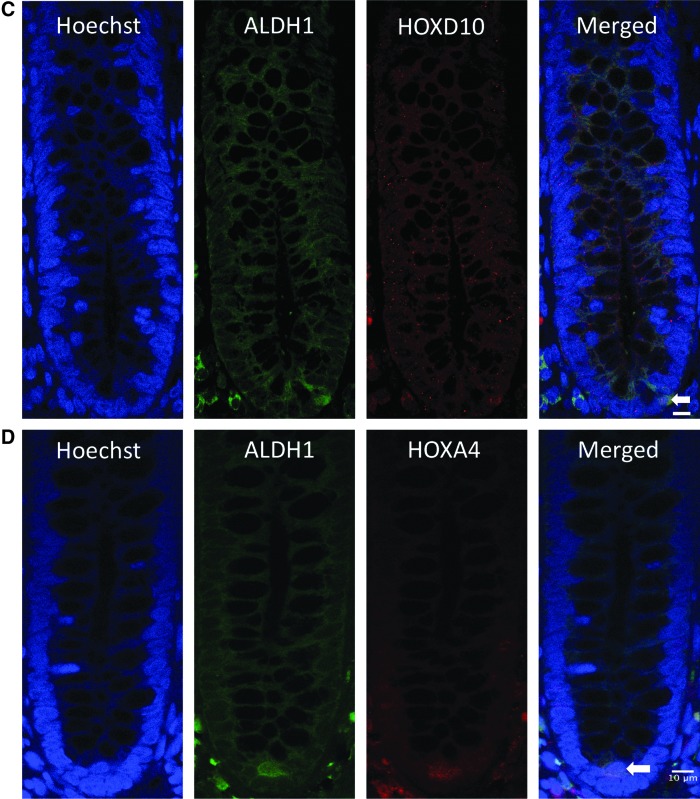
Co-staining images for HOXD10 and HOXA4 with SC markers (CD166 and ALDH1) in normal colonic crypts. (A, B) Co-staining for HOXD10, HOXA4, and CD166. Blue indicates nuclear staining with Hoechst dye; green indicates CD166 expression; red indicates HOXD10 and HOXA4 expression. CD166-positive cells co-stained with HOXD10 and HOXA4 at the bottom of the normal colonic crypt. Scale bar=20 μm. (C, D) Co-staining for HOXD10, HOXA4, and ALDH1 in normal colonic crypts. Blue indicates nuclear staining with Hoechst dye; green indicates ALDH1 expression; red indicates HOXD10 and HOXA4 expression. ALDH1-positive cells co-stained with HOXD10 and HOXA4 at the bottom of the normal colonic crypt. Scale bar=10 μm. Color images available online at www.liebertpub.com/scd
Analysis of gene expression in FAP crypts
Given that cancer SCs have been widely implicated in tumorigenesis in blood and several solid organs, we did a preliminary, hypothesis-generating study of gene expression changes in APC-mutant colon. In this pilot study, we analyzed gene expression in isolated whole FAP crypts and compared results to whole crypts isolated from healthy controls using a chip that contained 18,474 genes. These FAP patients had germline APC mutations that predispose them to develop CRC. For FAP, 6,159 genes were upregulated more than twofold, and 1,291 genes were downregulated more than twofold. We also isolated and analyzed crypts from an FAP patient treated with a nonsteroidal anti-inflammatory drug (NSAID), Sulindac, which is known to inhibit and even reverse adenoma development. In this patient, of the 18,474 genes, 271 were upregulated more than twofold; 421 were downregulated more than twofold compared with crypts from an untreated FAP patient (Supplementary Fig. S8). Thus, fewer than 4% of genes analyzed changed in expression with Sulindac treatment. We also did a survey of a subset of the genes (n=47) from the gene set that we had identified to be selectively expressed in the bottom of normal crypts. Among this subset of genes, 18 were overexpressed (more than twofold) in whole crypts from the untreated FAP patient compared with whole crypts from healthy controls. For all 18 genes, the level of expression was not lowered by Sulindac in the treated FAP case. Taken together, these data suggest that many genes selectively expressed in the crypt bottom of healthy controls are overexpressed in the SC niche of FAP crypts.
Expression of HOX genes in CRCs
Because colonic SC overpopulation occurs during colon tumorigenesis [1], we analyzed the expression of HOXD10 and HOXA4 in carcinomas. Immunostaining revealed that HOXD10 and HOXA4 expression in malignant colonic epithelium is substantially increased (both the number of cells staining and the intensity of the staining) in expression compared with normal colonic epithelium (Figs. 4 and 5). Quantitative PCR also showed that HOXA4 and HOXD10 are highly overexpressed in most CRCs (Fig. 7A). The SC markers CD166 and ALDH1 co-stained with HOXA4 or HOXD10 and there was more co-staining (both the number of cells co-staining and the intensity of the co-staining) in CRCs (Fig. 7B) compared with normal colonic epithelium (Fig. 6).
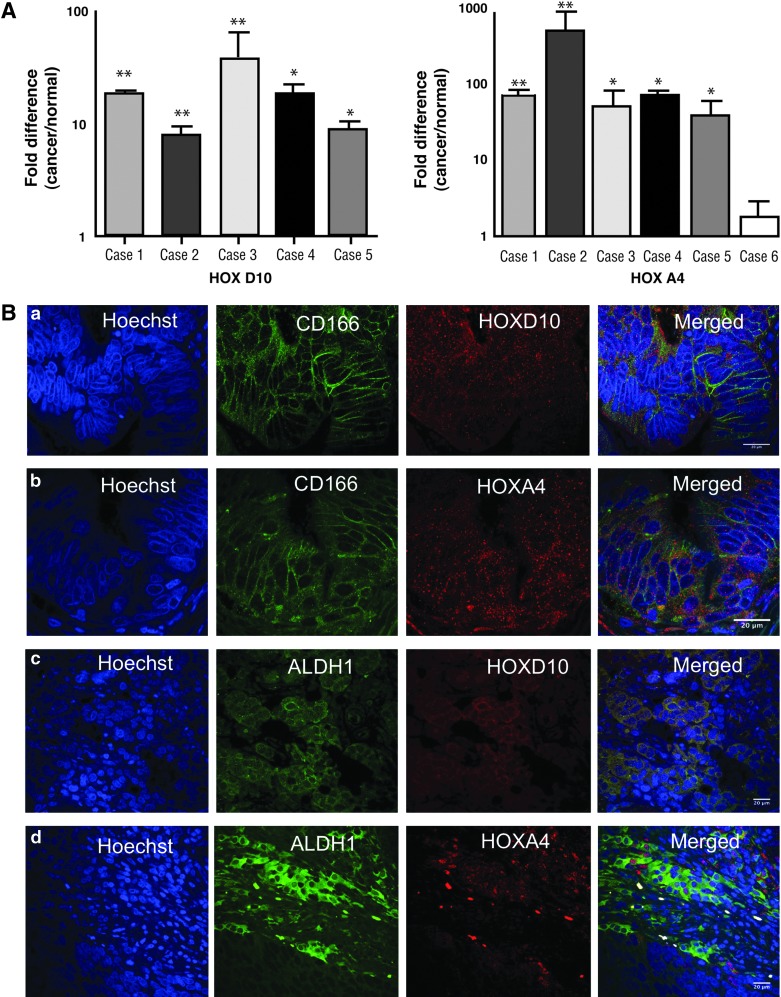
FIG. 7.
Expression of HOXD10 and HOXA4 in CRCs. (A) Overexpression of HOXD10 and HOXA4 in CRCs. Real-time PCR was done using expression of the indicated genes in colon samples (carcinomas and purified, normal colonic epithelium); n=6 for HOXA4; n=5 for HOXD10). Bars show the mean±SEM. (B) Co-staining for HOXA4 and HOXD10 with SC markers in CRCs. (a, b) Co-staining for HOXD10, HOXA4, and CD166 in CRCs. Blue represents nuclear staining with Hoechst dye; green represents CD166 expression; red represents HOXD10 and HOXA4 expression. Increased co-staining is seen for HOXD10, HOXA4, and the SC-marker CD166 in tumor tissues. Scale bar=20 μm. (c, d) Images of co-staining of HOXD10, HOXA4, and ALDH1 in CRC. Blue indicates nuclear staining with Hoechst dye; green indicates ALDH1 expression; red indicates HOXD10 and HOXA4 expression. Increased co-staining is seen for HOXD10, HOXA4, and ALDH1 in tumor tissues. *P<0.05; **P<0.005. Scale bar=20 μm. CRC, colon carcinoma. Color images available online at www.liebertpub.com/scd
Discussion
Main findings
Because colonic SC have unique functional characteristics—including lifetime proliferative capacity, capacity for self-renewal, anchorage, and multipotency—these cells should have a unique gene expression profile as should the region in which they reside, the SC niche at the crypt base. Our findings using microarray analysis demonstrate that there is indeed a unique gene expression pattern for the bottom 1/3 of human colonic crypts that distinguishes this region from the crypt middle and top.
Validity of our microarray methods
The use of an array representing a wide spectrum of genes is the most appropriate approach for this type of experiment because it ensures that novel genes are not missed by focusing our gene list too narrowly. Since we had little a priori knowledge regarding what genes might be uniquely over- or underexpressed in the crypt bottom, we used a robust normalization approach to adjust for outliers. However, since the availability of tissue was limited, and our sample (n=3) and replicate number (n=4) were thus relatively small, we used a very aggressive method for eliminating false positives. Indeed, we eliminated 95% of our data during outlier detection. Because we were comparing crypt subsections from the same patient, any variability from interpatient differences would not be encountered. Therefore, it is highly unlikely that there were many false positives left over in the gene list. While admittedly it is very likely that we eliminated a number of false negatives from further consideration, our goal was to identify SC-specific profiles, and it was therefore more important for us to identify true potential expression markers for each of the crypt subregions.
Rather than using an unsupervised visual exploratory approach, such as clustering, to determine interesting groups of genes, we simply looked for data that were consistent across all four replicates per sample and used the resulting list of upregulated genes to generate a list of potential markers for each crypt region.
Once we generated gene lists for each subregion, we used K-means clustering and hierarchical clustering to validate the lists. While we did not identify a functional pathway through K-means clustering, the similarity of the results from the two data sets (1 and 2) suggests that if we had not eliminated so much data, we might have found a potential pathway through this method. Likewise, the hierarchical clustering results show that there is clearly a unique gene expression signature in the bottom of the crypt compared with the top and middle. Indeed, we found that several of the upregulated genes in the crypt bottom map to the phosphatidylinositol-3-kinase (PI3K) pathway (discussed below).
Following cluster analysis, we performed gene ontology and annotation analysis to determine, through an evaluation of their known functions, whether our list of genes corresponded to SC-specific functions reported in the literature. Gene ontology on these lists showed that the upregulated genes from each of the gene lists play a role in growth and development.
To validate our findings that the expression results were truly representative of genes expressed in the SC niche, we analyzed, using real-time PCR, two groups of genes. Group 1 included HOXD10, HOXA4, and STAMP1. Group 2 included five known SC markers as positive controls (LGR5, CD166, CD133, DCLK1, and ALDH1). The analysis was done on bottom crypt subsections compared to middle crypt subsections. Results indicated that our microarray analysis method accurately identified genes whose expression was increased (Fig. 3) in the bottom relative to the middle crypt subsections.
Nature of the genes that showed differential expression: HOX, STAMP1, and PI3 kinase
Two HOX genes were identified in our microarray analysis as being upregulated in the crypt bottom. One was HOXD10 and the other HOXA4. Both these genes were validated by real-time analysis, and thus these HOX genes are putative markers for SC at the crypt bottom. Immunostaining confirmed the presence of HOXA4 and HOXD10 in the SC-enriched region at the bottom of the crypt. This is thus the first report showing that there is a unique gene expression pattern, which includes HOXA4 and HOXD10, in the SC niche at the crypt bottom. Our immunostaining data show that the expression of these genes is substantially increased in CRCs.
Both HOX genes have been reported to have an important role in embryonic development and in regulation of SCs. Disruption of HOXD10 expression causes severe hindlimb locomotor defects [6,7]. HOXA4 plays a role in SC regulation [8] and is important in proper development of the enteric gut [9]. Expression of the HOXA gene family is upregulated in the proliferative zone at the base of the crypt [10]. Aberrant expression of a few HOX genes has been previously reported in CRCs and CRC cell lines. For example, the HOX genes, HOX A9, HOX D11, and HOX B7 have all been found to be aberrantly expressed in primary CRCs [11]. Other studies showed upregulation of HOXB6, HOXB8, HOXC8, and HOXC9 in CRCs [12]. Given that HOX genes are important in embryonic development and organogenesis, a balance between the relative expression of different HOX genes has been considered to be essential to the maintenance of tissue-specific SCs [13].
There is little known about STAMP1, which we found to be the most upregulated gene at the crypt bottom. It has been implicated as a marker for the progression of prostate cancer and co-localizes with EEA1 in the Golgi apparatus [14]. STAMP1 expression is exclusively expressed in epithelial cells of the prostate and is increased in prostate cancer compared with normal prostate [15]. Given that STAMP1 is a cell surface antigen (with a six transmembrane domain), it may be useful for isolating colonic SC or maybe even serve as a therapeutic target.
Among the set of genes that we found were upregulated in the crypt bottom relative to the other two subsections were genes that mapped to the PI3K pathway. The PI3K pathway has been implicated in the cell's decision between survival, proliferation, and differentiation [16]. PI3K is a key signaling molecule in this pathway. PI3K is frequently mutated in human cancers, resulting in unregulated activation of PI3K signaling [17]. Interestingly, one third of CRCs showed somatic mutations in PI3K [18].
Pilot analysis of FAP crypts
Our pilot study was designed to compare expression of putative crypt SC genes in whole normal versus whole APC-mutant (FAP) crypts. This involved a survey of a subset of genes (n=47) from our main study, which established a signature for the crypt bottom of healthy controls. The data showing that 18 of these genes are overexpressed in colonic crypts from FAP patients suggest that germline APC mutations can cause an increase in expression of those genes that are selectively expressed in the normal SC niche. This finding is consonant with our previous studies that showed that SC overpopulation occurs in FAP colon: (1) our mathematical modeling studies [19,20] of the known proliferative change [21–23] in FAP crypts implicating SC overproduction in CRC initiation, (2) our immunohistochemical study [24] on crypts from FAP patients showing that APC mutations cause expansion of the crypt base cell population, (3) our proteomic (2D PAGE) analyses [25,26] showing that germline APC mutations alter protein expression in colonic crypts and fibroblasts from FAP patients. Moreover, we did a study [27] that used SC markers (ALDH1, CD44, and CD133) and quantitative immunohistochemical mapping to track SC population size in colonic crypts during colon tumorigenesis. This latter study showed that the number of SCs increased (by 2.5-fold) in normal-appearing FAP crypts, and increased even further (by 5.2-fold) in adenomatous crypts from FAP patients.
Expression of HOX genes in CRCs
Because we saw increased expression of HOXA4 and HOXD10 genes in the SC-enriched population at the crypt bottom, we wanted to determine whether these genes are actually expressed in colonic SC. CD166 and ALDH1 are known to be specific markers commonly used to identify colon SC [27–30]. Consequently, we determined whether HOXA4 and HOXD10 are co-expressed in cells that stain for the SC markers CD166 and ALDH1. Indeed, our results showed that HOXA4 and HOXD10 genes are co-expressed with CD166 and ALDH1 in cells within the SC niche of the normal colonic crypt. Co-staining was considerably increased in CRCs. Thus, our co-staining results also validate our data on gene expression from microarray analysis. This leads us to hypothesize that HOXA4 and HOXD10 genes play a role in maintenance of SC populations in normal colon, and that upregulation of these genes contributes to the crypt SC overpopulation [1,19,20,24,27] that drives CRC development.
HOX genes in development and SC self-renewal
Our microarray analysis showed 18 genes were uniquely expressed at the base of the crypt (a SC-enriched region) and these genes were classified as being involved in development and cell growth or maintenance. We found that HOXA4 and HOXD10 genes are expressed in colon SCs. This made us hypothesize as follows: (1) HOX genes are involved in colon SC self-renewal pathways and (2) their aberrant expression contributes to the SC overpopulation that drives CRC development.
Conclusions
Using an innovative “comparative” gene expression profiling approach to identify genes selectively expressed in the stem cell-enriched crypt region (bottom) compared to those genes expressed in crypt regions containing mostly proliferating cells (middle) or terminally differentiated cells (top), we discovered a unique signature for the crypt bottom that contains many genes that have functions consistent with known biologic roles involving SCs. Indeed, two of the genes are HOXA4 and HOXD10, which are known to be important in development. Their expression in the crypt bottom suggests that HOX genes play a role in maintenance of SC populations in the SC niche of the normal crypt. Overexpression of HOXA4 and HOXD10 in colon tumors suggests that HOX genes contribute to crypt SC overpopulation [1,19,20,24,27], which drives development of CRC in humans.
Taken together, our findings indicate that “comparative” expression profiling of different cell subpopulations within any given human tissue is a valuable tool for identifying genes that are expressed in normal SCs and determining which will become overexpressed during tumorigenesis. In this way, genes might be identified that contribute to SC overpopulation and drive development of cancers in humans. For example, this approach might help determine the mechanisms that lead to increased symmetric division of SCs, which is the mechanism thought to underlie development of cancer SC overpopulation [31]. Identifying such mechanisms may also suggest more effective molecular targets for new approaches to cancer treatment and chemoprevention.
Supplementary Material
Acknowledgments
We thank Dr. Kirk Czymmek, Delaware Bioimaging Center, University of Delaware for help in imaging, Dr. Zhengqiang Gao for technical assistance with isolating crypt subsections, and Dr. Nicholas Petrelli for his support. This study was supported in part by an NIH grant (R21DK062146) to B.M.B. It was also funded in part by a generous gift from Gregg & Stacey Bacchieri, and from Cancer B Ware organization.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Boman BM. and Huang E. (2008). Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J Clin Oncol 26:2828–2838 [DOI] [PubMed] [Google Scholar]
- 2.Potten CS,, Wilson JW. and Booth C. (1997). Regulation of and significance of apoptosis in the stem cells of the gastrointestinal epithelium. Stem Cells 15:82–93 [DOI] [PubMed] [Google Scholar]
- 3.Wildrick DM,, Lointier P, Nicholas DH, Roll R, Quintanilla B. and Boman BM. (1997). Isolation of normal human colonic mucosa: comparison of methods. In Vitro Cell Dev Biol Anim 33:18–27 [DOI] [PubMed] [Google Scholar]
- 4.Yang YH,, Dudoit S, Luu P. and Speed TP. (2001). Normalization for cDNA microarray data. In Microarrays: Dorsel A. N, and Dougherty E. R. (eds), Proceedings of SPIE, vol. 4266, pp. 141–152 [Google Scholar]
- 5.Yang YH,, Dudoit S, Luu P, Lin DM,, Peng V, Nagai J. and Terence SP. (2002). Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Wang G, Scott SA. and Capecchi MR. (2008). Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development 135:171–182 [DOI] [PubMed] [Google Scholar]
- 7.Carpenter EM,, Goddard JM, Davis AP, Nguyen TP. and Capecchi MR. (1997). Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development 124:4505–4514 [DOI] [PubMed] [Google Scholar]
- 8.Packer AI,, Mailutha KG, Ambrozewicz LA. and Wolgemuth DJ. (2000). Regulation of Hoxa4 and Hoxa5 genes in embryonic mouse lung by retinoic acid and TGFbeta1: implications for lung development and patterning. Dev Dyn 217:62–74 [DOI] [PubMed] [Google Scholar]
- 9.Pitera JE,, Smith VV, Thorogood P. and Milla PJ. (1999). Coordinated expression of 3' Hox genes during murine embryonal gut development: an enteric Hox code. Gastroenterology 117:1339–1351 [DOI] [PubMed] [Google Scholar]
- 10.Freschi G, Taddei A, Bechi P, Faiella A, Gulisano M, Cillo C, Bucciarelli G. and Boncinelli. E. (2005). Expression of HOX homeobox genes in the adult human colonic mucosa (and colorectal cancer?.). Int J Mol Med 16:581–587 [PubMed] [Google Scholar]
- 11.De Vita G, Barba P, Odartchenko N, Givel J, Freschi G, Bucciarelli G, Magli MC, Boncinelli E. and Cillo C. (1993). Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur J Cancer 29A:887–893 [DOI] [PubMed] [Google Scholar]
- 12.Vider BZ,, Zimber A, Chastre E, Gespach C, Halperin M, Mashiah P, Yaniv A. and Gazit A. (2000). Deregulated expression of homeobox-containing genes, HOXB6, B8, C8, C9 and Cdx-1, in human colon cancer cell lines. Biochem Biophys Res Commun 272:513–518 [DOI] [PubMed] [Google Scholar]
- 13.Shah N. and Sukumar S. (2010). The Hox genes and their roles in oncogenesis. Nat Rev Cancer 10:361–371 [DOI] [PubMed] [Google Scholar]
- 14.Korkmaz KS,, Elbi C, Korkmaz CG, Loda M, Hager GL. and Saatcioglu F. (2002). Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J Biol Chem 277:36689–36696 [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Jin Y, Arnoldussen YJ, Jonson I, Qu S, Maelandsmo GM, Kristian A, Risberg B, Waehre H, Danielsen HE. and Saatcioglu F. (2010). STAMP1 is both a Proliferative and an Antiapoptotic factor in Prostate Cancer. Cancer Res 70:5818–5828 [DOI] [PubMed] [Google Scholar]
- 16.Dancey JE. (2004). Molecular targeting: PI3 kinase pathway. Ann Oncol Suppl 4:iv233–239 [DOI] [PubMed] [Google Scholar]
- 17.Chalhoub N. and Baker SJ. (2009). PTEN and the PI3-kinase pathways in cancer. Annu Rev Pathol Mech Dis 4:127–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz SD. and Bertagnolli MM. (2009). Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med 361:2449–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boman BM,, Fields JZ, Bonham-Carter O. and Runquist OA. (2001). Computer modeling implicates stem cell overproduction in colon cancer initiation. Cancer Res 61:8408–8411 [PubMed] [Google Scholar]
- 20.Boman BM,, Fields JZ, Cavanaugh KL, Guetter A. and Runquist OA. (2008). How dysregulated colonic crypt dynamics cause stem cell overpopulation and initiate colon cancer. Cancer Res 6:3304–3313 [DOI] [PubMed] [Google Scholar]
- 21.Deschner E, Lewis CM. and Lipkin M. (1963). In vitro study of human rectal epithelial cells.I Atypical zone of H3 thymidine incorporation in mucosa of multiple polyposis. J Clin Invest 42:1922–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lightdale C, Lipkin M. and Deschner E. (1982). In vivo measurements in familial polyposis: kinetics and location of proliferating cells in colonic adenomas. Cancer Res 142:4280–4283 [PubMed] [Google Scholar]
- 23.Potten CS,, Kellett M, Rew DA. and Roberts SA. (1992). Proliferation in human gastrointestinal epithelium using bromodeoxyuridine in vivo: data for different sites, proximity to a tumour and polyposis coli. Gut 33:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boman BM,, Walters R, Fields JZ, Kovatich AJ, Zhang T, Isenberg GA, Goldstein SD. and Palazzo JP. (2004). Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population. Am J Pathol 165:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung AT,, Patel BB, Li XM, Seeholzer SH, Coudry RA, Cooper HS, Bellacosa A, Boman BM, Zhang T, et al. (2008). One-hit effects in cancer: altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. Cancer Res 68:7579–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel BB,, Li XM, Dixon MP, Blagoi EL, Nicolas E, Seeholzer SH, Cheng D, He YA, Coudry RA, et al. (2011). APC+/- alters colonic fibroblast proteome in FAP. Oncotarget 3:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang EH,, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fiels JZ, Wicha MS. and Boman BM. (2009). Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69:3382–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin TG,, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR. and Wong MH. (2010). Characterization of the intestinal cancer stem cell marker, CD166/ALCAM, in the human and mouse gastrointestinal tract. Gastroenterology 139:2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien CA,, Pollett A, Gallinger S. and Dick JE. (2007). A human colon cancer cell capable of initiating tumor growth in immunodeficient mice. Nature 445:106–110 [DOI] [PubMed] [Google Scholar]
- 30.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C. and Maria RD. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445:111–115 [DOI] [PubMed] [Google Scholar]
- 31.Boman BM,, Wicha MS, Fields JZ. and Runquist OA. (2007). Symmetric division of cancer stem cells—a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther 81:893–898 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.