Abstract
Abscisic acid stress ripening (ASR1) protein is a small hydrophilic, low molecular weight, and stress-specific plant protein. The gene coding region of ASR1 protein, which is induced under high salinity in rice (Oryza sativa Ilmi), was cloned into a yeast expression vector pVTU260 and transformed into yeast cells. Heterologous expression of ASR1 protein in transgenic yeast cells improved tolerance to abiotic stresses including hydrogen peroxide (H2O2), high salinity (NaCl), heat shock, menadione, copper sulfate, sulfuric acid, lactic acid, salicylic acid, and also high concentration of ethanol. In particular, the expression of metabolic enzymes (Fba1p, Pgk1p, Eno2p, Tpi1p, and Adh1p), antioxidant enzyme (Ahp1p), molecular chaperone (Ssb1p), and pyrimidine biosynthesis-related enzyme (Ura1p) was up-regulated in the transgenic yeast cells under oxidative stress when compared with wild-type cells. All of these enzymes contribute to an alleviated redox state to H2O2-induced oxidative stress. In the in vitro assay, the purified ASR1 protein was able to scavenge ROS by converting H2O2 to H2O. Taken together, these results suggest that the ASR1 protein could function as an effective ROS scavenger and its expression could enhance acquired tolerance of ROS-induced oxidative stress through induction of various cell rescue proteins in yeast cells.
Keywords: abiotic stress, ASR1 gene, redox homeostasis, stress tolerance, yeast
INTRODUCTION
Common environmental stress factors such as dehydration, high salinity, and low temperature influence plant growth and development as well as place major limits on plant productivity in cultivated areas. To overcome these limitations and improve crop yield under stress conditions, it is important to improve the stress tolerance of crops. The responses of plant cells to sudden and adverse environmental changes by certain mechanisms are important since these not only initiate the repair of macromolecular damage but also establish a tolerant state for the prevention of further damage (Jain et al., 2009). Especially, plants use a number of molecular mechanisms to cope with the effects of stress on protein structure, since stress has multiple effects on cellular components and especially biopolymers like proteins. These defensive mechanisms include increasing the conformational stability of proteins, preventing protein aggregation, and degradation of irreversibly damaged proteins. Proteins stability and aggregation prevention can be achieved by accumulating molecular chaperones and other stabilizing proteins as well as low-molecular weight proteins known as hydrophilins including LEA (late embryogenesis abundant) proteins (Konrad and Bar-Zvi, 2008).
Abscisic acid-, stress-, and ripening-induced (ASR) proteins are small and heat-stable proteins that have a high hydrophilicity and act as downstream components of a common transduction pathway for sugar (Yang et al., 2005). Among ASR proteins, ASR1 is a plant-specific, highly charged low molecular weight and heat-stable protein whose expression in plants is induced by high salinity, dehydration, and treatment with the plant hormone ABA (Goldgur et al., 2007). The molecular mechanism behind its biological activity cannot be simply deduced based on sequence homology with other known proteins. ASR1 belongs to a small gene family, with homologs cloned from vegetative (leaf, shoot, and stem) and reproductive (fruit and pollen) tissues in monocot and dicot plant species. Low accumulation of ASR1 mRNA and its protein product has been detected in the roots and shoots of irrigated tomato plants. However, following NaCl, polyethylene glycol (PEG) or ABA treatment, a rapid increase in ASR1 levels occurs (Goldgur et al., 2007). In addition, up-regulation of an ASR1 homolog was also observed in the roots of salt-stressed maize plants. In two rice varieties with different salt tolerance, the ASR1 homolog was highly expressed upon exposure to excess salt for 3 and 6 h. ASR1 expression was also shown to be developmentally regulated since expression levels of ASR1 mRNA increase during fruit ripening (Rom et al., 2006). In addition, ASR proteins share properties (protective properties against freezing and dehydration) with LEA proteins as they are considered to be group 7 LEA proteins (Battaglia et al., 2008).
Budding yeast S. cerevisiae is considered as an excellent model for studying the mechanisms underlying tolerance, particularly oxidative stress, due to the high degree of evolutionary conservation of stress responses between higher and lower eukaryotes (Hur et al., 2011). Furthermore, S. cerevisiae has been used to investigate the effects of mangrin stress tolerance, based on its utility as a model organism for the analysis of eukaryotic genes (Jain et al., 2009). Here, to investigate the in vitro function of rice ASR1, we used a yeast model system to determine whether or not stress tolerance is mediated by ASR1. We were able to obtain evidence of the protective role of the rice ASR1 protein against oxidative stress in yeast cells. Constitutive expression of ASR1 under the control of the ADH promoter resulted in increased tolerance, following an alleviated redox state towards H2O2 during vegetative cell differentiation. In addition, ASR1-expressing yeast cells exhibited markedly enhanced resistance to ROS-induced oxidative stress caused by pro-oxidants, heavy metals, and SDS. This study provides insights into the novel mechanism underlying the protective effects of ASR1 expression and will help to better understand the genetic mechanisms that control plant growth, development and the response to environmental stress.
MATERIALS AND METHODS
Plant materials
Rice seeds (Oryza sativa L. ilmi) were germinated in half-strength Murashige-Skoog (MS) solid medium in a dark growth chamber for 2 days at 32°C. The seeds were transplanted into paddy soil pots and grown in a glasshouse (16 h light/8 h dark cycle) at 28–32°C. Three-week-old rice seedlings were placed in 100 mM NaCl solution for 8 days. The phenotypes of the plants that continued to grow were photographed.
Construction of the ASR1-expressing recombinant yeast strain
Total RNA was isolated from rice leaves. The cDNA probe was prepared by reverse transcriptase-polymerase chain reaction (RT-PCR). The ASR1-coding region was amplified from cDNA by PCR using Taq and Pfu polymerase (Roche). The PCR reaction conditions were as follows: initial denaturation cycle was 94°C for 3 min, followed by 30 cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min, and a final extension step for 7 min at 72°C. The sense and antisense primers utilized for PCR cloning of the ASR1 gene were 5′-TGTCACTTCCATCTC ATTCTCC-3′ and 5′-TTTGGAGAAAGTCAAGGTTCG-3′, respectively. The PCR product of the ASR1 gene naturally contained an NcoI site in the front part of the start codon. The PCR product was inserted into a TOPO TA cloning vector (Invitrogen). The cloned plasmid was sequenced using the M13 primer set to confirm that no PCR-induced mutations were introduced. The cloned plasmid DNA was digested with NcoI and BamHI restriction endonuclease. The ASR1 DNA fragment, which was approximately 850 kb in size, was ligated into yeast expression vector pVTU260 (EUROSCARF) and E. coli expression vector pKM260 was digested with the appropriate restriction enzymes. The E. coli DH5α strain (endA, RedA, hsd, deoR, and LacZ M15), transformed with pVTU260::ASR1 and pKM260::ASR1 plasmids, was used to amplify the plasmid DNA, which was isolated using a plasmid isolation kit (Nucleogen). S. cerevisiae BY4741 cells (MATa, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0) were grown overnight in YPD medium for 4–5 h at 30°C until the cultures reached an OD600 of approximately 1.0. The cells were, then transformed with pVTU260::ASR1 via the PEG/LiCl method (Gietz and Woods, 2001). Transformants were selected as growing cells on yeast synthetic drop-out agar medium containing 2% glucose without uracil at 30°C for 2–3 days. Colonies were then restreaked and incubated under the same conditions. Positively transformed colonies with pVTU260::ASR1 were able to grow on medium lacking uracil. The pKM260::ASR1 plasmid was transformed into E. coli BL21. Transformants were selected as growing cells on LB agar plates, which were supplemented with 50 μg/ml of ampicillin for 24 h at 37°C.
Yeast growth conditions and stress tolerance assay
Yeast cells were cultured at 30°C with shaking (160 rpm). To monitor the growth rate under abiotic stress, cells (1 × 105 cells per ml) were grown overnight and inoculated into YPD medium supplemented with 4.0 mM H2O2. The optical density (OD) was measured at 600 nm at 2 h intervals for the indicated time. For the spotting assay, once mid-log phase (A600 = 1.5) was reached, cell aliquots were exposed to 20 mM H2O2, 0.1 mM menadione (MD), 50 mM sulfuric acid (H2SO4), 0.5 M lactic acid, 0.15 M salicylic acid, 10 mM copper sulfate (CuSO4), 15% ethanol, and 1.5 M NaCl for 1 h at 30°C (having been properly shaken), and serially diluted 10-fold with distilled water. Then, 5.0 μl of each diluted solutions was loaded onto the YPD agar medium. To induce heat shock, cells were treated for 30 min at 50°C. Each experiment was carried out three times and representative results are shown.
Semi-quantitative RT-PCR
Total RNA from mid-log phase yeast cells (OD600 = 1.0) and rice leaves was obtained using the Total Isolation Kit (Promega). First-strand cDNA was synthesized by incubating 1 μg of total RNA with RT-PreMix (Bioneer) and oligo(dT)18 at 42°C for 1 h in a 20 μl reaction volume. Subsequently, the ASR1 gene was amplified from synthesized cDNA using a gene-specific primer. The oligonucleotides used were ASR1-F and ASR1-R with sequences of 5′-ACCACCTGTTCCACCACAAG-3′ and 5′-CGCTCTTGTGGTCCTTCTTC-3′, respectively. The PCR reaction conditions were as follows: initial denaturation cycle at 94°C for 3 min, followed by 25 cycles at 94°C for 30 s, 54°C for 30 s, 72°C for 1 min, and a final extension step for 7 min at 72°C. The PCR amplicon was separated in a 1.2% agarose gel in 0.5X TBE buffer, stained with ethidium bromide and then, visualized and photographed. The PCR amplicons of PDA1 (Wenzel et al., 1995) and tubulin (TUB) were used as housekeeping controls for yeast and rice, respectively. The tubulin primer sets were as follows: TUB-F: 5′-TACCGTGCCCTTACTGTTCC-3′ and TUB-R: 5′-CGGTGGAATGTCACAGACAC-3′.
Protein extraction and 2-D electrophoresis analysis from rice and yeast
For rice, leaves were ground in liquid nitrogen and suspended in cold extraction buffer containing 0.7 M sucrose, 0.1 M KCl, 0.5 M Tris-HCl, pH 7.5, 50 mM EDTA, 2% β-mercaptoethanol and 1 mM PMSF as well as an equal of volume of phenol saturated with Tris-HCl at pH 7.5. The mixture was incubated for 30 min at 4°C and centrifuged at 5,000 rpm for 30 min. An equal volume of extraction buffer was then added to the upper phenolic phase, incubated for 30 min at 4°C and centrifuged at 5,000 rpm for 30 min at 4°C. This step was repeated twice. To precipitate the protein, five volumes of cold 0.1 M ammonium acetate in methanol was added to the collected phenol phase, stored at −20°C overnight, and centrifuged at 5,000 rpm at 4°C for 30 min. To wash the pellet, two volumes of ice-cold methanol was added to the cleared supernatant, mixed gently, and centrifuged for 10 min at 8,000 rpm at 4°C. This step was repeated three times (Isaacson et al., 2006). The pellet was then vaccum-dried, resuspended in sample buffer containing 9.5 M urea, 4% CHAPS, 40 mM Tris, 0.1 M DTT and 0.2% Bio-Lyte (3–10; Bio-Rad) at room temperature with shaking. The sample was then, centrifuged at 15,000 rpm for 30 min. The cleared supernatant was carefully collected, and protein concentration was measured using a modified Bradford assay (Ramagli and Rodriguez, 1985). 250 μg of proteins was loaded onto preparative gels and run with pH 4–7 IPG strips (7 cm, linear; Bio-Rad). The focusing conditions were as follows: 250 V for 1 h, 250 V to 4,000 V for 3 h, and 4,000 V for 7 h. Yeast cells were grown to mid-log phase and exposed to H2O2 for 1 h at 30°C with shaking. Cells were then harvested by centrifugation, washed twice with cold-PBS buffer, suspended in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 2% sodium dodecyl sulfate (SDS), 5% glycerol, 2% β-mercaptoethanol (ME), 2 mM EDTA and 1 mM PMSF, added to an equal volume of glass beads (452–600 μm; Sigma), and disrupted with a MicroMixer. To inactivate protease activity, cells were boiled for 5 min and cooled on ice for 5 min. Protein crude extracts were collected by high speed centrifugation (13,000 rpm, 4°C, 10 min), and incubated with a DNase/RNase/Mg mixture on ice for 15 min. Samples were then precipitated using 10% TCA on ice for 1 h and centrifuged at 13,000 rpm for 10 min at 4°C. The pellets were washed three times with cold-ethanol containing 0.1% ME, vaccum-dried, resuspended in sample buffer containing 9.5 M urea, 4% CHAPS, 40 mM Tris, 0.1 M DTT and 0.2% Bio-Lyte (3–10; Bio-Rad) at room temperature with shaking, and centrifuged at 15,000 rpm for 30 min. The cleared supernatants were carefully collected. Then, 750 mg of proteins were loaded onto preparative pH 4–7 IPG strips (17 cm, linear; Bio-Rad) and rehydrated for 16 h at 20°C under passive conditions. The focusing conditions were as follows: 250 V for 1 h, 250 V to 10,000 V for 5 h, and 10,000 V for 9 h. After isoelectric focusing (IEF), the gel strips were equilibrated. SDS-PAGE was carried out in 12% gels at 10 mA per gel for the first 1 h followed by 25 mA per gel, stained with Coomassie Brilliant Blue R-250 (CBB R-250), and then destained. Spots with significant changes were considered to be accumulated proteins. Interesting spots were excised from the gel. In-gel digestion and MALDI-TOF MS analysis was then carried out. Protein identification was conducted using the MASCOT program (www.Matrixscience.com).
Yeast protein preparation and immunoblotting blot analysis
Crude protein extracts were prepared using the glass bead method. Cells treated with or without 20 mM H2O2 for 1 h were washed three times with cold-PBS buffer containing 0.85% NaCl and resuspended in lysis buffer containing 50 mM HEPES, pH 7.5, 5% glycerol, 1 mM PMSF, 2 μM pepstatin A, and protease inhibitor cocktail with an equal volume of glass beads (425–600 μm; Sigma). After vigorously vortexing on ice four times for 1 min each at a 2-min interval, the cleared protein extracts were collected by centrifugation at 13,000 rpm for 20 min at 4°C. The protein concentration was determined using a Bradford assay with the Protein Dye Reagent (Bio-Rad). The protein extract (30 μg) was loaded onto 15% SDS-PAGE gel, and run at 50 V. The gel was electrophoretically transferred to a PVDF membrane after SDS-PAGE. The membranes were then blocked for 1 h at room temperature and incubated overnight at 4°C with anti-His tag primary antibody (Millipore). Anti-tubulin (SantaCruz) antibody was used as a housekeeping control. Blots were then washed and incubated with either anti-rabbit or anti-mouse secondary antibodies coupled to horseradish peroxidase for 1.5 h at room temperature. After washing, the signal was visualized using an ECL Western blotting detection reagent (GE Healthcare).
Redox state analysis
To measure cytosolic ROS, exponential cells were incubated for 20 min at 30°C with 50 μM dichlorodihydrofluorescein diacetate (DCFHDA; Invitrogen), exposed to 20 mM H2O2 for 1 h, washed twice with PBS buffer, and then resuspended in the same PBS buffer. Cells loaded with fluorescent probes were imaged by fluorescence microscopy (excitation, 488 nm; emission, 525 nm).
ASR1 protein purification and ROS scavenging and molecular chaperone activities
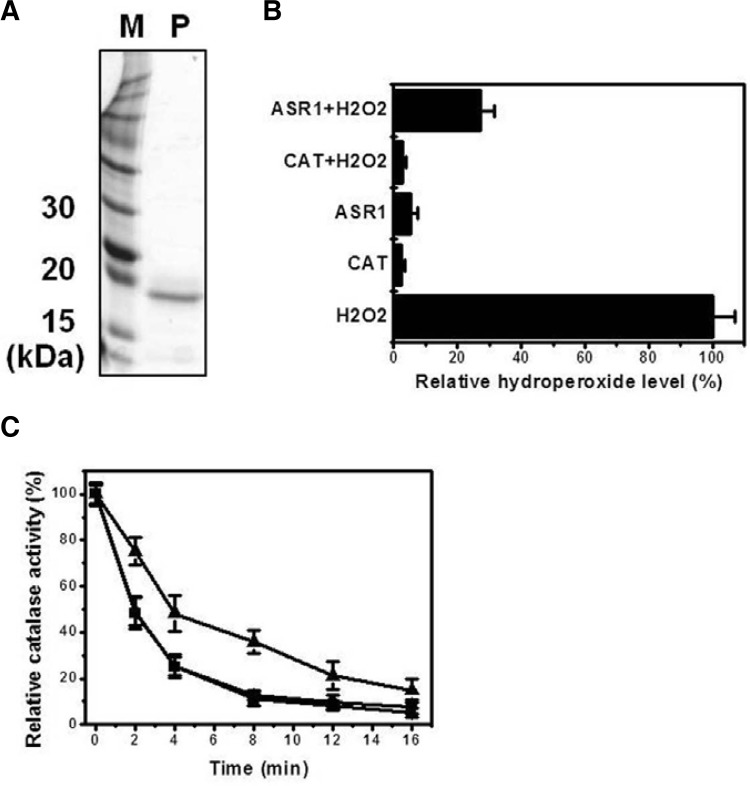
For expression analysis, E. coli BL21 (DE3) harboring the pKM260::ASR1 plasmid was grown in LB-medium supplemented with 100 μg/ml of ampicillin with vigorous shaking (200 rpm) at 37°C. When the culture reached mid-log phase (A660 = 0.4), isopropyl-β-D-thiogalactopyranoside (IPTG) was added at a final concentration of 0.4 mM, and the culture was grown for another 3 h at 37°C. Cells were harvested by centrifugation at 5,000 rpm for 10 min, and then washed twice with cold-PBS buffer. The resulting cell pellet was resuspended in lysis buffer (50 mM phosphate buffer, pH 7.5, 0.3 M NaCl, 10 mM imidazole, 1 mM DTT, and 1 mM PMSF) and sonicated to disrupt the cells. The homogenate was then centrifuged at 12,000 rpm for 20 min at 4°C to remove cell debris. The cleared supernatant containing soluble protein was purified using immobilized metal affinity chromatography (IMAC). The clear supernatant was loaded by gravity onto a column containing a pre-equilibrated Ni-NTA affinity resin (Quiagen). The column was washed twice with wash buffer (50 mM phosphate buffer, pH 7.5, 0.3 M NaCl, and 50 mM imidazole). Bounded ASR1 protein was eluted with elution buffer (50 mM phosphate buffer, pH 7.5, 0.3 M NaCl, and 0.25 M imidazole). The resulting purified ASR1 protein (2 μg) was loaded onto 15% SDS-PAGE, run at 50 V, stained with CBB R-250, and then destained. ROS scavenging activity was measured using the FOX reagent or catalase. The hydroperoxide level was determined by ferrous ion oxidation in the presence of a ferric ion indicator, xylenol orange (Deiana et al., 1999).
Five micrograms each of ASR1 and catalase (Sigma) were soaked in a 17 μM H2O2 solution and incubated at 25°C for 15 min. Then, 50 μl of the reaction mixtures was added to 950 μl of FOX reagent containing 100 μM xylenol orange, 250 μM ammonium ferrous sulfate, 100 mM sorbitol, and 25 mM sulfuric acid. The mixtures were incubated at room temperature for 30 min and then centrifuged to remove any flocculants before measuring the absorbance at 560 nm. The ROS scavenging activity was represented by setting the percentage of the 17 μM H2O2 solutions without enzyme at 100%. Chaperone-like activity was also measured to investigate the molecular chaperone capacity of the ASR1 protein. Catalase (0.9 μg) and ASR1 protein solutions (0.1 μg) were added to the reaction mixture containing 50 mM potassium phosphate buffer, pH 7.5, and heated at 50°C for 16 min. Glucose-6-phosphate dehydrogenase (G6PDH; 0.1 μg) solution containing catalase (0.9 μg) was used as a negative control. Each sample was taken at the indicated times. Catalase activity was measured at 240 nm for 1 min. Enzyme activity was set at 100% for the catalase solution.
Statistical analysis
Expression intensity as measured by semiRT-PCR and immunoblotting analysis was determined using the ImageJ software (http://rsbweb.nih.gov/ij/). All experiments were repeated at least three times independently. Results were expressed as mean ± standard deviation (SD). For the spotting assay and growth curve, data reported were representative values of at least two independent growth experiments carried out under identical conditions, as indicated in the figure legends.
RESULTS
Identification of rice ASR1 protein under high salinity condition
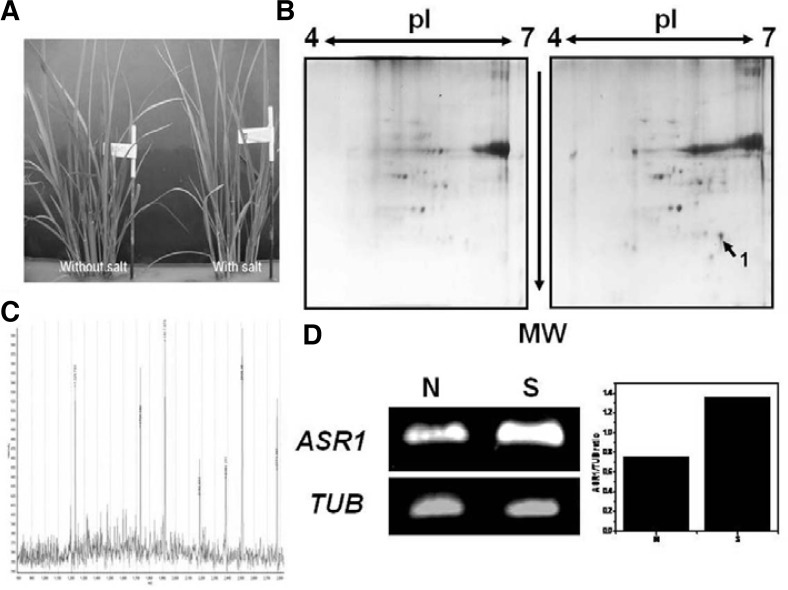
To defend against salt stress, plants can their change gene expression pattern and accumulate certain protein. The leaf is the main organ involved in the salt stress response of plants, and salt stress-responsive genes are more strongly induced in the leaf in response to environmental stimuli than any other organ. Rice is not only an important crop but also a model plant for the study of stress tolerance. For example, the International Rice Genome Sequencing Project using the Nipponbare cultivar is nearly complete. The resulting sequence information has facilitated proteomic studies, especially protein identification by mass spectrometry (MS) (Yan et al., 2005). Therefore, we investigated the salt stress-responsive proteins in the leaves of rice (Oryza sativa L. ilmi). Three-week-old rice seedlings were treated with 100 mM NaCl for 8 days. The seedlings displayed certain stress symptoms such as shriveled leaves, but they survived under salt stress challenge (Fig. 1A). Comparative proteomic analysis was used to investigate the protein profiles under salt stress. To distinguish stress responses from protein accumulation, both normal and salt-treated leaves were harvested. The total protein content of leaves was extracted and separated by 2-DE using pH 4–7 IPG strips IEF (Sarma et al., 2008). The 2-DE maps are shown in Fig. 1B. One protein spot showing significant changes in abundance (Fig. 1B) was analyzed by MALDI-TOF (Fig. 1C). Using MS analysis, this spot was determined to be an abscisic acid- and stress-inducible protein (ASR1). MS coverage, molecular weight and pI of the identified protein ASR1 were 43%, 15.45, and 6.2, respectively. We then performed semi-quantitative RT-PCR to confirm whether or not ASR1 transcription was induced under salt conditions. As shown in Fig. 1D, transcription of ASR1 increased by at least 2-fold in the salt-treated sample compared to the control sample. This result shows that expression of the ASR1 protein was strongly induced under salt conditions in rice, which also contains three motifs (motifs 1, 2 and 3) with high degrees of conservation within the region responsible for LEA group 7 (ASR1) active sites (Supplementary Fig. S2).
Fig. 1.
Induction of the ASR1 gene in the presence of salt stress. (A) Three-week-old rice seedlings were incubated in a 100 mM NaCl solution for 8 days. Up-regulation of ASR1 gene was confirmed by 2-DE (B) and MALDI-TOF analysis (C). To identify this gene, semiRT-PCR was carried out (D). Amplicon of the rice TUB gene was used as a housekeeping standard (left). Expression intensity was expressed as the ratio of ASR1 to TUB (right). N, without salt treatment; S, with salt treatment.
Construction and expression of Oryza sativa ASR1 recombinant yeast
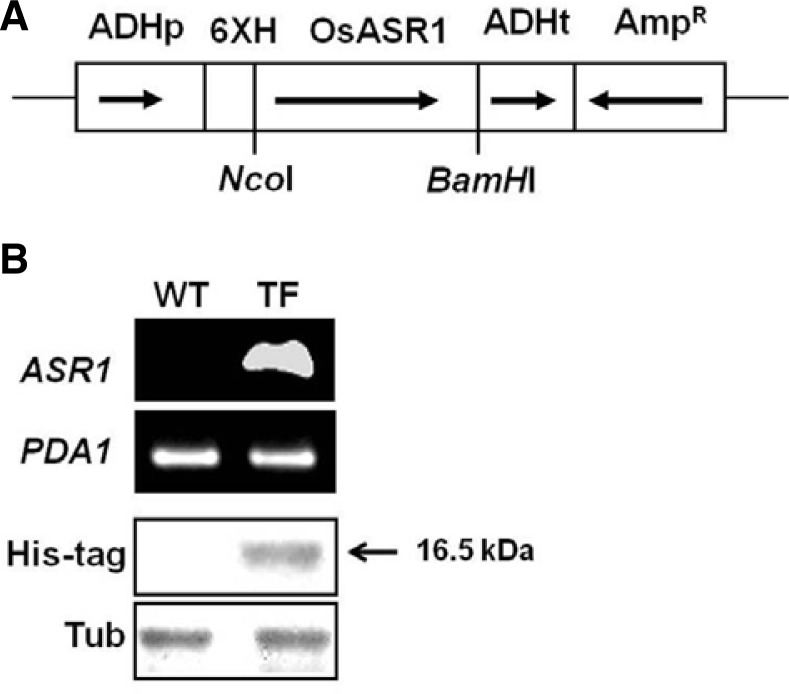
Previous studies have shown that the accumulations of antioxidants and antioxidant enzymes is a crucial mechanism for overcoming oxidative stress (Xue et al., 2009). Since cell metabolism involves a complex and diverse array of mechanisms, other unknown antioxidant systems may exist in plant cells. Based on these facts, we examined whether or not ASR1 overexpression had an effect on acquired tolerance against oxidative stress in yeast cells. cDNA containing the ASR1 open reading frame (ORF) region from Oryza sativa L. Ilmi (OsASR1) was cloned into the yeast expression vector pVTU260, which allows constitutive expression of genes of interest under the control of the yeast ADH1 promoter (Fig. 2A). To make sure that the ASR1 gene can be effectively expressed in yeast, we conducted semiRT-PCR analysis. One 351 bp DNA fragment corresponding to the region between the ASR1 ORF was amplified in ASR1-expressing transformed yeast cells (TF cells), whereas no amplification signal was detected in wild-type cells transformed with only the vector (WT cells) (Fig. 2B). To determine whether or not the ASR1 gene was properly expressed at the translational level, we also performed immunoblotting analysis. As shown in Fig. 2C, the expression of ASR1 in TF cells was confirmed since the anti-His tag antibody was reactive in TF cells but not in WT cells. These results indicate that the ASR1 protein was properly expressed in yeast cells.
Fig. 2.
Heterologous expression of the ASR1 gene in transgenic yeast cells. (A) Schematic diagram showing the over-expression of ASR1 in yeast. ADHp, alcohol dehydrogenase promoter; 6XH, six histidine-tagged residue; OsASR1, ASR1 gene of Oryza sativa (rice); ADHt, alcohol dehydrogenase terminator; AmpR, selectable marker. Arrows indicate the direction of each gene. (B) Confirmation of ASR1 gene expression by semi-quantitative RT-PCR. The PCR product of the PDA1 gene was used as a standard control. (C) Expression of the ASR1 protein was measured by Western-blot analysis using an anti-His tag antibody. An anti-Tubulin (Tub) antibody was used as a housekeeping control. WT, yeast cells with empty vector alone; TF, ASR1-expressing transgenic yeast cells.
Stress tolerance to hydrogen peroxide and redox state in ASR1-expressing transformed yeast cells
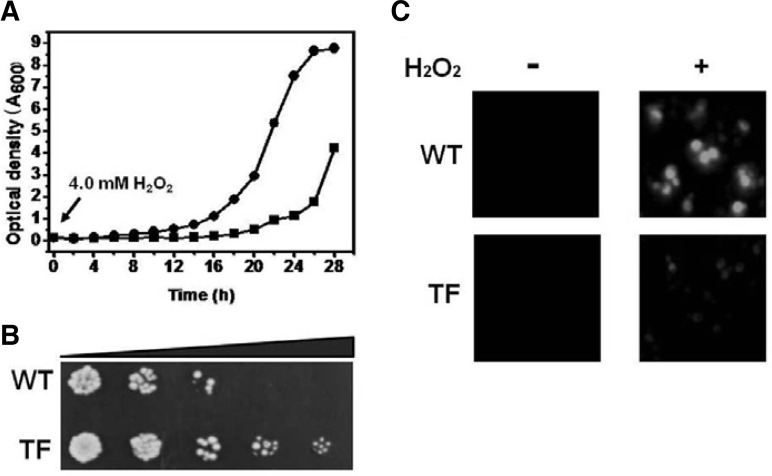
The role of ASR1 protects in oxidative stress in plants was explored based on previous results, which demonstrated that the ASR1 protein was involved in the response against ROS-induced oxidative stress caused by NaCl, freezing, and dehydration (Battaglia et al., 2008). The effects of oxidative challenge on the growth rate and survival were evaluated in yeast cells transformed with the pVTU260 plasmid containing either the ASR1 gene or vector alone. The pVTU260 plasmid is a vector used for constitutive expression of a gene of interest under the ADH promoter. The growth kinetics of TF yeast cells was higher than those of WT cells when the cells were cultured in liquid medium containing 4.0 mM H2O2. During the first 8 h, growth of TF yeast cells occurred at almost the same rate as that of WT yeast cells. This was followed by a lag, after which growth resumed more rapidly. TF yeast cells entered the stationary phase at 24 h. WT yeast cells had a longer lag phase period (16 h) compared to the rapid restoration of growth observed in the TF yeast cells. Complete entrance into the log growth phase was achieved 24 h after initial inoculation (Fig. 3A). There was no difference in the growth rates between TF and WT yeast cells under normal conditions (data not shown). Stress tolerance was also measured using the spotting assay with serial dilutions. Cell recovery of mid-log TF yeast cells exposed to 20 mM H2O2 for 1 h was higher than that of WT yeast cells (Fig. 3B). Therefore, heterologous ASR1 expression in yeast cells improved tolerance to H2O2-induced oxidative stress. To dertermine whether or not the stress tolerance in TF yeast cells conferred by ASR1 expression result in improved redox homeostasis, the cellular hydroperoxide level was measured in the absence and presence of H2O2. The levels of intra cellular hydroperoxides were evaluated by fluorescence microscopy using a cytosolic oxidant-sensitive probe, DCFHDA. An increase in DCF fluorescence was observed in both WT and TF yeast cells upon exposure to 20 mM H2O2 for 1 h. However, the fluorescence intensity was more pronounced in the WT yeast cells than the TF yeast cells. In addition, we observed a moderate release of the probe from the WT yeast cells (Fig. 3C). The alleviated redox state was supported by the cell viability, recovery and the ROS level in TF yeast cells under oxidative stress.
Fig. 3.
Stress tolerance and redox state analyses. (A) The growth rate was spectrophotometrically monitored at 600 nm for 28 h in the presence of 4.0 mM H2O2. Results are representative of at least two independent growth experiments. Square, WT cells; circle, TF cells. (B) For the spotting assays, mid-log phase cells were exposed to 20 mM H2O2 for 1 h. Ten-fold series dilution was carried out, and 5 μl of each dilution was spotted onto an YPD agar plate. (C) Redox state analysis was carried out using the oxidant-sensitive probe DCFHDA in the absence (−) and presence (+) of 20 mM H2O2 for 1 h.
Activation of cell rescue proteins in ASR1-expressing yeast cells against oxidative stress
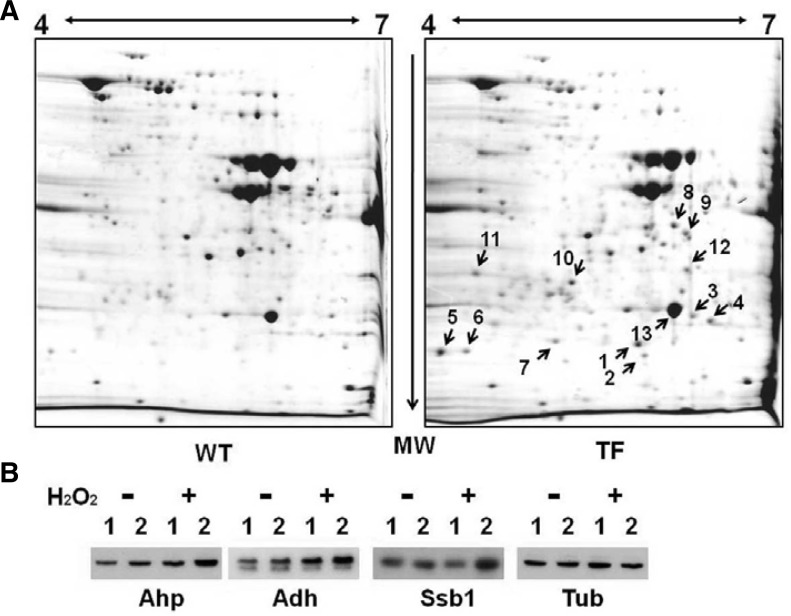
Oxidant-induced protective responses are often due to the coordinated activation of proteins involved in ROS neutralization, since cellular antioxidants act in a concerted manner. Therefore, we investigated the changes in the activities of the major cell rescue proteins induced by heterologous ASR1 expression. The experiment was conducted using exponential growth phase yeast cells in the presence of H2O2 and analyzed by 2-DE and immunoblotting. As shown in Fig. 4A and Supplementary Table S1, the expression of proteins involved in energy-generating systems via glycolysis including fructose-1,6-biphosphate aldolase (Fba1p), triosephospahte isomerase (Tpi1p, No.13), 3-phosphogly-cerate kinase (Pgk1p, No.4), enolase (Eno2p, No.8), alcohol dehydrogenase (Adh1p, No.12), antioxidative enzyme (thioredoxin peroxidase; Ahp1p, No.5,6), molecular chaperone (heat shock protein 70 family; Ssb2p, No.7), and pyrimidine biosynthesis (dihyroorotate dehydrogenase; Ura1p, No.9) was up-regulated in TF yeast cells in response to oxidative stress when compared to WT yeast cells. Western blot analysis clearly supported the MALDI-TOF MS data, following 2-DE. Protein expression of Ahp1, Adh1, and Ssb in the TF cells was 2.0-fold higher than that in WT cells under H2O2-induced oxidative stress (Fig. 4B). Over-expression of these proteins in TF yeast cells under oxidative stress can lead to increased levels of energy-generating proteins (NADPH and ATP) and protein homeostasis following increased expression of chaperones with protein-folding abilities. In contrast, the difficulty of maintaining redox homeostasis in WT yeast cells could increase the vulnerability to oxidative stress caused by H2O2. Therefore, our results show that expression of heterologous ASR1 protein activated redox homeostasis and proteostasis by up-regulating a variety of antioxidative proteins in the presence of H2O2.
Fig. 4.
Comparative analysis of whole cellular proteins between WT and ASR1-expressing strains of S. cerevisiae under H2O2 stress by 2-D gel electrophoresis and immunoblot analysis. (A) Total soluble protein samples were obtained from exponential growth phase cells, which were treated with 20 mM of H2O2 for 1 h. The protein loading and running conditions are described in the “Materials and Methods”. After SDS-PAGE and CBB staining, spots of interest were cut out. After digestion with trypsin, MALDI-TOF MS analysis was carried out. (B) To verify the MS results, Western blot was performed using anti-thioredoxin peroxidase (Ahp), anti-alcohol dehydrogenase, and anti-heat shock protein 70 (Ssb1) antibodies in the absence (−) and presence of 20 mM H2O2 (+). Anti-Tubulin (Tub) antibody was used as a housekeeping control. 1, WT cells with empty vector alone; 2, TF cells with pVTU260::ASR1.
Function of ASR1 as a ROS scavenger and molecular chaperone in vitro
To further explore the biochemical properties of ASR1 in rice, a recombinant His-ASR1 fusion protein was constructed and expressed in E. coli BL21 (DE3). The ASR1 protein was purified via the IMAC method as shown in (Fig. 5A), and used in vitro as a ROS scavenger. To determine the in vitro efficiency of ASR1 as a ROS scavenger as well as its ability to convert H2O2 to H2O as compared with that of catalase (Magalhaes et al., 2008), H2O2 levels were evaluated using the FOX reagent. As shown in Fig. 5B, absorbance of the ASR1 solution (30 μg) containing 17 μM H2O2 was 4-fold lower compared to the solution containing only 17 μM H2O2. Absorbance of the catalase solution (30 μg) containing 17 μM H2O2 was similar to the blank value, which indicates that the catalase enzyme completely converted H2O2 to H2O. We also determined whether or not the ASR1 protein displayed molecular chaperone activity to prevent protein aggregation. The ASR1 protein solution containing catalase displayed 2-fold higher catalase activity against heat shock than the catalase solution without the ASR1 protein. In contrast, the G6PDH solution plus catalase, which was used as a negative control, displayed similar catalase activity as that of the catalase solution without any protein (Fig. 5C). Although the scavenging ability of ASR1 was lower than that of catalase, this result demonstrates that ASR1 functions as an antioxidant and molecular chaperone. Therefore, ASR1-expression in yeast cells improved the alleviated redox state (Fig. 3C) and proteostasis (Fig. 4) in the presence of H2O2 when compared to WT cells. Proteins stability and prevention against protein aggregation can be achieved by the accumulation of molecular chaperones, other stabilizing proteins and low-molecular weight organic molecules known as hydrophilins such as LEA proteins (late embryogenesis abundant) and ASR protein (Konrad and Bar-Zvi, 2008). Therefore, it could be concluded that ASR1 acts as an important endogenous cell rescue protein that responds to ROS stress in a direct manner.
Fig. 5.
Purification and ROS-scavenging ability assay of ASR1 protein. (A) SDS-PAGE (15%) analysis after ASR1 protein purification. 2 μg of the purified protein was loaded. Line M, protein marker; line P, purified ASR1 protein. (B) ROS-scavenging ability of ASR1 protein was measured spectrophotometically. Cata-lase was used as a positive control. Absorbance of the sample solution with only H2O2 was set at 100%. (C) To observe chaperone-like activity, each sample solutions without and with the ASR1 protein was collected within the specified time at 50°C, after which catalase activity was measured. G6PDH solution plus catalase was used as a negative control. Enzyme activity was calculated to be 100% of the catalase solution. Triangle, catalase solution with ASR1 protein; circle, catalase solution with G6PDH protein; square, catalase solution without ASR1 protein.
Effect of ASR1 over-expression on stress tolerance under diverse conditions
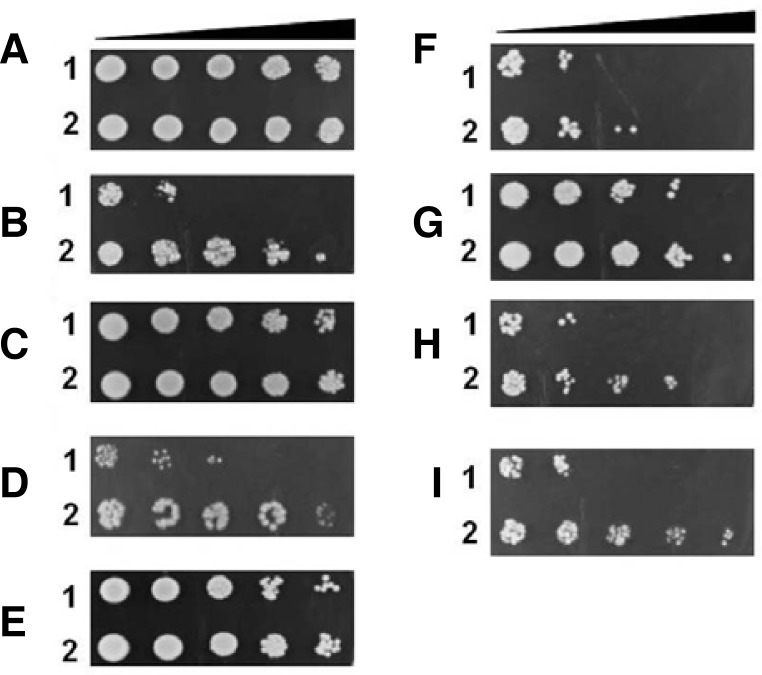
Since resistance to H2O2 was achieved in TF yeast cells, we decided to assess the involvement of ASR1 expression in acquired tolerance to different kinds of stressors, such as prooxidants (menadione, heat shock, and ethanol), heavy metals (iron, zinc, cobalt, and copper), high salinity (NaCl), and acid stress (lactic acid, sulfuric acid, benzoic acid, and salicylic acid) in yeast cells. A comparative analysis of stress tolerance was carried out using the spotting assay. TF yeast cells constitutively expressed the ASR1 protein under regulation of the ADH promoter, which was observed by serial dilutions of yeast cells exposed to 0.1 mM menadione (MD), 15% ethanol, 50 mM cobalt chloride, 0.1 M zinc chloride, 20 mM copper sulfate, 50 mM sulfuric acid, 0.5 M lactic acid, 0.15 M salicylic acid, 25 mM benzoic acid, and 1.5 M NaCl for 1 h. For heat shock, cells were incubated at 50°C for 30 min. The recovery ability of TF yeast cells was more rapid than that of WT yeast cells in the presence of heat shock, MD, ethanol, copper, sulfuric acid, lactic acid, and salicylic acid (Fig. 6), whereas cell viability of TF yeast cells decreased in the presence of cobalt, zinc, iron, and benzoic acid when compared to that of WT yeast cells (data not shown). This valuable information on the ASR1 protein has not only been reported in yeast cells but also in some plants under favorable conditions. Thus, these results shows that ASR1-expression in transgenic yeast enhanced the acquisition of tolerance to pro-oxidants, heat shock, heavy metal, acid stressors, salicylic acid, and NaCl, although the level of resistance was somewhat dependent on stress types.
Fig. 6.
Stress tolerance assay of heterologous ASR1-expressing yeast cells in the presence of stress stimuli. Yeast cells that reached mid-log phase were exposed to no stress (A), heat shock (B), 0.1 mM menadione (C), 10 mM CuSO4 (D), 1.5 M NaCl (E), 15% ethanol (F), 50 mM H2SO4 (G), 0.5 M lactic acid (H), and 0.15 M salicylic acid (I) for 1 h at 30°C with shaking, and then properly diluted from 100 to 10−4 with distilled deionized water. A diluted sample (5 μl) was spotted onto an YPD agar plate. 1, WT cells with empty vector alone; 2, TG cells expressing ASR1.
DISCUSSION
Oxidative stress results from an imbalance between ROS production and ROS scavenging ability, and it causes damage to various cellular components such as carbohydrates, lipids, proteins, and nucleic acids (Lushchak, 2011). ROS-induced oxidative stress limits agricultural production yields worldwide (Ashraf, 2009) and causes various diseases such as aging, cancer, arteriosclerosis, and diabetes in humans (Lushchak, 2011). To cope with and adapt to different exogenous stimuli, aerobic organisms have developed a variety of defense mechanisms. These include activation of cascades or network events, which begin with sensing stress and end with the expression of several effector genes (Bae et al., 2011). Antioxidant defense systems, including non-enzymatic and enzymatic antioxidants, have been reported to play important roles in the response to ROS-induced oxidative stress (Gill and Tuteja, 2010). In plants, previous studies have shown that the regulation of antioxidant accumulation and antioxidant enzyme activities is crucial for overcoming oxidative stress (Xue et al., 2009). However, due to the complexity and diversity of cellular metabolism, other unknown antioxidant systems may exist in plant cells. Based on these facts, a less complex eukaryotic organism, S. cerevisiae, was used for the functional analysis of stress-responsive and/or stress-inducible genes.
ASR1 (Accession No. AF039573 or Os11g0167800) was identified in NaCl-treated rice leaves by two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionization-time-of flight mass spectrometry (MALDI-TOF MS) (Fig. 1). The molecular weight (MW) and isoelectric point (pI) of the ASR1 protein were determined to be 15.45 kDa and 6.20, respectively. The ASR proteins in rice (ASR1) are small, heatstable, intrinsically unstructured and contain three highly conserved regions (Supplementary Fig. S2). In addition, these proteins are similar to other LEA proteins in terms of physiochemical properties (Battaglia et al., 2008). ASR gene expression has been observed in different plant species, as well as different organs and growth stages (Battaglia et al., 2008). As shown in Fig. 1, the ASR gene responded to environmental stresses, such as water deficit, salinity, low temperature, and limited light (Battaglia et al., 2008). Although the functions of ASR1 are still unknown, there is no evidence to suggest that this protein participates in the adaptive response to stress. To analyze the physiological and functional roles of ASR1 in the oxidative stress response, we incorporated ASR1 (836 bp) into yeast expression vector pVTU260, which was under the regulation of the ADH1 promoter (Fig. 2). Heterologous overexpression of ASR1 conferred TF yeast cells with enhanced tolerance to ROS-induced oxidative stresses (Fig. 6), especially H2O2 (Figs. 3A and 3B). The H2O2 level in TF yeast cells was only half that in WT yeast cells (Fig. 3C). We also observed that ASR1 expression in TF yeast cells increased the expression of other proteins in the presence of H2O2. We identified eight of these proteins using 2-DE followed by MALDI-TOF MS analysis. These proteins were found to be involved in the metabolic pathway (Fba1p, Eno2p, Adh1p, Pgk1p, and Tpi1p), antioxidant system (Ahp1p), and protein folding (Ssb2p) (Supplementary Table S1). Ahp1p, a thiol specific peroxiredoxin, highly accumulated in TF cells in the presence of H2O2 when compared to WT cells, reduces the sensitivity to hydroperoxides, which improves the redox homeostasis of yeast (Iwai et al., 2010). Under H2O2-induced oxidative conditions, metabolic enzymes such as enolase (Eno2), alcohol dehydrogenase (isoforms 1 and 2), phosphoglycerate mutase (Gpm or Pgk), triosephosphate isomerase (Tpi1p) and fructose-1,6-biphosphate aldolase (Fba1p), as well as Hsp70 molecular chaperones, were also upregulated in the in TF yeast cells relative to the WT cells. Yeast cells lacking Adh1 activity grew poorly on glucose, while over-expression of ADH1 was able to enhance the formaldehyde resistance of yeast cells (Grey et al., 1996). Tpi1p is an abundant glycolytic enzyme that makes up approximately 2% of the soluble cellular protein. In humans, deficiency of TPI caused neurological disorder (Olah et al., 2002). Fba1p, which encodes fructose-1,6-biphosphate aldolase was required for glycolysis and gluconeogenesis and located in mitochondrial outer membrane surface upon oxidative stress (Rinnerthaler et al., 2006). Pgk1p was abundantly expressed in cells grown in glucose, and transcription was increased by heat shock (Piper et al., 1986) and regulated by the transcription factors Rab1p, Abf1p, and Reb1p (Packham et al., 1996). In particular, enolase and Ssb protein are major targets of protein damage in WT yeast cells exposed to oxidative stress (Cabiscol et al., 2000; Reverter-Branchat et al., 2004). Down-regulation of glycolytic enzymes by irreversible protein oxidation impairs glucose utilization and this effect is correlated with enhanced gluconeogenic and energy storage pathways, and causes an imbalance in proteostasis, which plays a role in protein translocation, folding, and assembly (Reverter-Branchat et al., 2004). Therefore, a high accumulation of metabolic enzymes in the TF yeast cells is important for the production of ATP and NADPH because NADPH serves as a reducing agent in thioredoxin systems containing Ahp1p, whereas ATP is used as a substrate of molecular chaperones such as Ssb2 protein to prevent protein aggregation or refolding of damaged proteins during H2O2 stress.
As seen in yeast, the over-expression of the ASR protein increases stress tolerance in several plants (Goldgur et al., 2007). For example, over-expression of tomato ASR1 and lily ASR increases salt tolerance in tobacco (Kalifa, 2004) and Arabidopsis (Yang et al., 2005), respectively. ASR1-overexpressing transgenic (TG) tobacco plants showed increased expression of different genes encoding NAD+-dependent malate dehydrogenase, proline-rich protein, secretory peroxidase, lipid transfer protein, fructose biphosphate aldolase (Fbap), raffinose synthase, chloroplast ATPase, 4-coumarate-coenzyme A ligase, and NADP+-dependent ferredoxin reductase under salt stress conditions. As a result, TG plants over-expressing these proteins showed improved tolerance towards salt stress (Kalifa, 2004). Thus, acquired stress tolerance in TF yeast cells altered metabolic and chaperone activities and this affected the redox state and proteostasis, which was not observed in WT yeast cells.
To further elucidate the functions of ASR1 protein, we conducted in vitro assays similar to those used to determine the roles of other protective molecules such as ROS-scavengers. Purified ASR1 protein from Escherichia coli BL21 exhibited antioxidative ability in vitro in the absence of antioxidants. It also showed chaperone-like activity by reducing protein aggregation under heat stress (Fig. 4). Cytosolic tomato ASR1 (SlASR1) has chaperone-like activity, and is able to confer stability to a number of proteins in response to heat and freeze-thaw cycle (Konrad and Bar-Zvi, 2008). Previous studies have also demonstrated the cryoprotective function of LEA group 7 (ASR1) proteins (Battaglia et al., 2008). Although ASR proteins play a role in the modulation of gene expression (Kalifa, 2004), no study has examined the antioxidant activity of the ASR1 protein. Therefore, based on the results of this study, we can conclude that rice the ASR1 protein plays a role in scavenging toxic ROS and preventing protein denaturation caused by abiotic stress (Reyes et al., 2005).
Yeast and plant growth is severely affected by abiotic stress (Gill and Tuteja, 2010). Therefore, augmenting plant resistance to various environmental stresses for improving growth and productivity has been the aim of genetic engineers. In summary, the ASR1 protein in TF yeast cells might play a critical role as a ROS-scavenger and chaperone-like protein in response to stress until other protective proteins or mechanisms are activated, which increased improved stress tolerance by altering the expression of the stress-responsive gene ASR1 in TF yeast cells. Therefore, our results provide an important clue to improving stress-tolerant yeast or plants in response to exogenous stimuli.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008115), Rural Development Administration, Republic of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ashraf M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009;27:84–93. doi: 10.1016/j.biotechadv.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Bae Y.S., Oh H., Rhee S.G., Yoo Y.D. Regulation of reactive oxygen species generation in cell signalling. Mol. Cells. 2011;32:491–509. doi: 10.1007/s10059-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E., Piulats E., Echave P., Herrero E., Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- Deiana L., Carru C., Pes G., Tadolini B. Spectrophotometric measurement of hydroperoxides at increased sensitivity by oxidation of Fe2+ in the presence of xylenol orange. Free Radic. Res. 1999;31:237–244. doi: 10.1080/10715769900300801. [DOI] [PubMed] [Google Scholar]
- Gill S.S., Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Goldgur Y., Rom S., Ghirlando R., Shkolnik D., Shadrin N., Konrad Z., Bar-Zvi D. Desiccation and zinc binding induce transition of tomato abscisic acid stress ripening 1, a water stress- and salt stress-regulated plant-specific protein from unfolded to folded state. Plant Physiol. 2007;143:617–628. doi: 10.1104/pp.106.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey M., Schmidt M., Brendel M. Overexpression of ADH1 confers hyper-resistance to formaldehyde in Saccharomyces cerevisiae. Curr. Genet. 1996;29:437–440. [PubMed] [Google Scholar]
- Hur J.Y., Park M.C., Suh K.Y., Park S.Y. Synchronization of cell cycle of Saccharomyces cerevisiae by using a cell chip platform. Mol. Cells. 2011;32:483–488. doi: 10.1007/s10059-011-0174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Damasceno C.M., Saravanan R.S., He Y., Catala C., Saladie M., Rose J.K. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006;1:769–774. doi: 10.1038/nprot.2006.102. [DOI] [PubMed] [Google Scholar]
- Iwai K., Naganuma A., Kuge S. Peroxiredoxin Ahp1 acts as a receptor for alkylhydroperoxides to induce disulfide bond formation in the Cad1 transcription factor. J. Biol. Chem. 2010;285:10597–10604. doi: 10.1074/jbc.M109.090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., Roy N., Chattopadhyay D. CaZF, a plant transcription factor functions through and parallel to HOG and calcineurin pathways in Saccharomyces cerevisiae to provide osmotolerance. PLoS One. 2009;4:e5154. doi: 10.1371/journal.pone.0005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y., Perlson E., Gilad A., Konard Z., Scolnik P.A., Bar-Zvi D. Overexpression of the water and salt-stress regulated Asr1 gene confers an increased salt tolerance. Plant Cell Environ. 2004;27:1459–1468. [Google Scholar]
- Konrad Z., Bar-Zvi D. Synergism between the chaper-one-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta. 2008;227:1213–1219. doi: 10.1007/s00425-008-0693-5. [DOI] [PubMed] [Google Scholar]
- Lushchak V.I. Adaptive response to oxidative stress: Bacteria, fungi plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011;153:175–190. doi: 10.1016/j.cbpc.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Magalhaes L.M., Segundo M.A., Reis S., Lima J.L. Methodological aspects about in vitro evaluation of antioxidant properties. Anal. Chim. Acta. 2008;613:1–19. doi: 10.1016/j.aca.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Oláh J., Orosz F., Keserü G.M., Kovári Z., Kovács J., Hollán S., Ovádi J. Triosephosphate isomerase deficiency a neurodegenerative misfolding disease. Biochem. Soc. Trans. 2002;30:30–38. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Packham E.A., Graham I.R., Chambers A. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol. Gen. Genet. 1996;250:348–356. doi: 10.1007/BF02174393. [DOI] [PubMed] [Google Scholar]
- Piper P.W., Curran B., Davies M.W., Lockheart A., Reid G. Transcription of the phosphoglycerate kinase gene of Saccharomyces cerevisiae increases when fermentative cultures are stressed by heat-shock. Eur. J. Biochem. 1986;161:525–531. doi: 10.1111/j.1432-1033.1986.tb10474.x. [DOI] [PubMed] [Google Scholar]
- Ramagli L.S., Rodriguez L. Quantitation of microgram amounts of proteins in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis. 1985;6:559–563. [Google Scholar]
- Reverter-Branchat G., Cabiscol E., Tamarit J., Ros J. Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae common targets and prevention by calorie restriction. J. Biol. Chem. 2004;279:31983–31989. doi: 10.1074/jbc.M404849200. [DOI] [PubMed] [Google Scholar]
- Reyes J.L., Rodrigo M.J., Colmenero-Flores J.M., Gil J.V., Garay-Arroyo A., Campos F., Salamini F., Bartels D., Covarrubias A.A. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 2005;28:709–718. [Google Scholar]
- Rinnerthaler M., Jarolim S., Heeren G., Palle E., Perju S., Klinger H., Bogengruber E., Madeo F., Braun R.J., Breitenbach-Koller L., et al. MMI1 (YKL056c, TMA19), the yeast orthologue of the translationally controlled tumor protein (TCTP) has apoptotic functions and interacts with both microtubules and mitochondria. Biochim. Biophys. Acta. 2006;1757:631–638. doi: 10.1016/j.bbabio.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Rom S., Gilad A., Kalifa Y., Konrad Z., Karpasas M.M., Goldgur Y., Bar-Zvi D. Mapping the DNA- and zinc-binding domains of ASR1 (abscisic acid stress ripening), an abiotic-stress regulated plant specific protein. Biochimie. 2006;88:621–628. doi: 10.1016/j.biochi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Sarma A.D., Oehrle N.W., Emerich D.W. Plant protein isolation and stabilization for enhanced resolution of two-dimensional polyacrylamide gel electrophoresis. Anal. Biochem. 2008;379:192–195. doi: 10.1016/j.ab.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Xue T., Li X., Zhu W., Wu C., Yang G., Zheng C. Cotton metallothionein GhMT3a, a reactive oxygen species scavenger increased tolerance against abiotic stress in transgenic tobacco and yeast. J. Exp. Bot. 2009;60:339–349. doi: 10.1093/jxb/ern291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Tang Z., Su W., Sun W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005;5:235–244. doi: 10.1002/pmic.200400853. [DOI] [PubMed] [Google Scholar]
- Yang C.Y., Chen Y.C., Jauh G.Y., Wang C.S. A Lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiol. 2005;139:836–846. doi: 10.1104/pp.105.065458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.