Abstract
The central dogma of DNA-RNA-protein was established more than 40 years ago. However, important biological processes have been identified since the central dogma was developed. For example, methylation is important in the regulation of transcription. In contrast, proteins, are more complex due to modifications such as phosphorylation, glycosylation, ubiquitination, or cleavage. RNA is the mediator between DNA and protein, but it can also be modulated at several levels. Among the most profound discoveries of RNA regulation is RNA splicing. It has been estimated that 80% of pre-mRNA undergo alternative splicing, which exponentially increases biological information flow in cellular processes. However, an increased number of regulated steps inevitably accompanies an increased number of errors. Abnormal splicing is often found in cells, resulting in protein dysfunction that causes disease. Splicing of the survival motor neuron (SMN) gene has been extensively studied during the last two decades. Accumulating knowledge on SMN splicing has led to speculation and search for spinal muscular atrophy (SMA) treatment by stimulating the inclusion of exon 7 into SMN mRNA. This mini-review summaries the latest progress on SMN splicing research as a potential treatment for SMA disease.
Keywords: anti-sense oligonucleotide, SMA, SMN, splicing, trans-splicing
INTRODUCTION
Spinal muscular atrophy (SMA) is an autosomal recessive, early childhood disease that affects 20,000-30,000 patients in the US. Although SMA is an orphan disease, it is the most common genetic cause of infant mortality in the US and western Europe (Pearn, 1980). The carrier frequency of SMA is estimated to be as high as 1 in 40, leading to the disease in 1 in 6000 newborns (Pearn, 1980). Patients with SMA are intellectually normal but have problems with movement, muscle tone, and muscle-related functions such as breathing and swallowing. These symptoms result from abnormally low levels of the survival motor neuron (SMN) protein (Pearn, 1980), leading to degeneration of alpha-motor neurons in the anterior horn of the spinal cord. More recent studies have indicated that other organs such as the heart and liver are also damaged in patients with SMA (Bevan et al., 2010; Hua et al., 2011). Respiratory failure due to diaphragm weakness is the main cause of death in patients with SMA. SMA is categorized into four types based on severity. The mildest form of SMA is type IV, which is an adult SMA, with symptoms beginning in the middle 30s. Patients with the milder type III SMA start to experience muscle weakness from as early at 2 years of age to their teen years. They are frequently wheelchair bound when they reach their 20s. Patients with type III and type IV SMA have a normal lifespan. Patients with type II SMA usually experience muscle weakness at around 2 years of age, are wheelchair bound at early ages, and die before they reach 20 years of age. The most severe type of SMA is type I with onset at a few months old and patients usually die before 2 years of age.
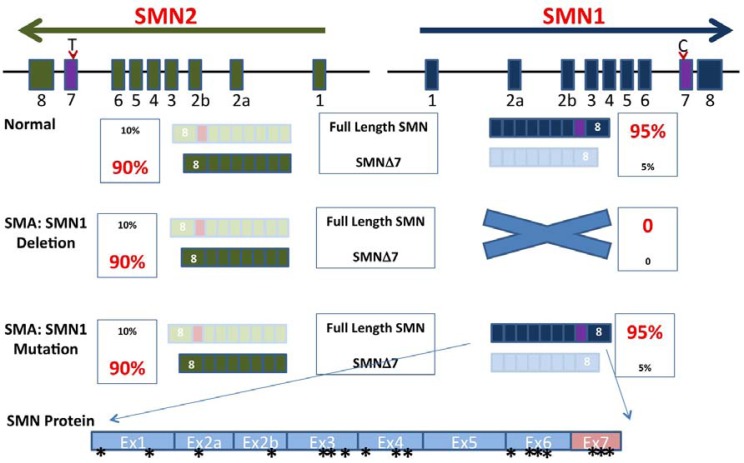
Based on linkage analysis and mapping, the survival motor neuron (SMN1) gene is responsible for SMA. Humans have two highly homologous copies of the SMN gene, SMN1 and SMN2, which are located on chromosome 5q13 as inverted repeats (Lefebvre et al., 1995) (Fig. 1). In > 95% of patients with SMA, the telomeric SMN1 copy is deleted or mutated (DiDonato et al., 1997; Lefebvre et al., 1995; Parsons et al., 1996; Wirth, 2000), whereas the SMN2 centromeric copy remains in most patients with SMA but predominantly (> 85%) produces exon7-skipped transcripts, generating truncated, unstable proteins that fail to compensate for the loss of SMN1 (Coovert et al., 1997; Lefebvre et al., 1995). The skipping of exon 7 in SMN2 is caused by alternative splicing of exon 7 (Lorson et al., 1999) (Fig. 1).
Fig. 1.
Survival motor neuron (SMN) genes and spinal muscular atrophy (SMA) disease. Two inverted SMN genes, SMN1 and SMN2 are located on chromosome 5. SMN1 and SMN2 have nine exons. Exon 7 is alternatively spliced. Under normal conditions (Normal), > 95% of SMN1 mRNA includes exon 7, whereas only 10% of SMN2 mRNA has exon 7. Most patients with SMA have a deletion of the SMN1 gene (SMA: SMN1 deletion). In these patients, 10% of full-length mRNA produced from the SMN2 gene is unable to compensate for the loss of the SMN1 gene. SMA can also be caused by point mutations (SMA: SMN1 mutation). Missense mutations have been identified across the gene (indicated by *).
SMN encodes a 294 amino acid protein with a molecular weight of 34 kD and is conserved across species. In mammalian cells, SMN forms a characteristic dot-like gem structure in the nucleus (Liu and Dreyfuss, 1996). SMN interacts with many cellular proteins (reviewed by Nicole et al., 2002) and is involved in the assembly of large macromolecular ribonucleoprotein particle (RNP) complexes (Buhler et al., 1999; Carvalho et al., 1999; Fischer et al., 1997; Massenet et al., 2002; Meister, 2001; Pellizzoni et al., 2002), playing an important role in RNA metabolism. We found that SMN is co-localized with the apoptosis-related proteins TIA-1/R and G3BP in stress granules (SGs) (Forch and Valcarcel, 2001; Hua and Zhou, 2004a; 2004b; Le Guiner et al., 2003), a cellular structure that stores translationally silenced snRNPs during the stress response. Additional studies have demonstrated that SMN initiates the stress response by facilitating the formation of SGs under stressful conditions (Hua and Zhou, 2004b). Therefore, SMN may function as an anti-apoptotic chaperone protein in SMN/SGs by stabilizing RNA/protein species against stress-induced apoptosis. Consistent with our results, SMN has been implicated in transporting specific mRNAs to neuron neurites (Pagliardini et al., 2000; Zhang et al., 2003), possibly in neuronal RNA granules (Bassell et al., 1998; Hu et al., 2003; Tiruchinapalli et al., 2003), a structure with a function reminiscent of SGs in other cell types (Wickens and Goldstrohm, 2003). SMN overexpression in cultured forebrain neurons induce granules that are actively transported to developing neurites and growth cones in a manner similar to that of RNA granules (Zhang et al., 2003). Based on these findings and our findings of SMN in SGs (Hua and Zhou, 2004b), we hypothesize that SMN plays a key role protecting and storing mRNAs/proteins in both neuronal cell bodies and processes (axons and dendrites). Low levels of SMN may fail to protect specific mRNA/protein species in long axons, particularly under stressful conditions, resulting in failure of synaptic plasticity.
SMA animal models
Unlike humans, mice contain one copy of the SMN gene, equivalent to the SMN1 gene in humans. Homozygous knockout of the mouse SMN gene is embryo lethal at a very early developmental stage (Schrank et al., 1997). Animals with an SMN−/− hSMN2 genotype were generated by crossing mice transgenic for human SMN2 into a SMN+/− heterozygous mouse (Monani et al., 2000). Because hSMN2 expresses a low level of the full length SMN protein, these mice do not die in utero, suggesting that human SMN2 can rescue the lethality of homozygous knockout. A single copy of human SMN2 was sufficient to restore mouse viability from SMN−/− lethality, but severe weakness developed, and the mice died within a few days of birth. In contrast, mice with eight copies of hSMN2 appeared normal, consistent with studies in human patients with SMA, in which SMN2 functions as a disease-modifying gene. and multiple copies of SMN2 gene can produce full-length SMN at a high enough level to compensate for the loss of SMN1; thus, explaining the correlation between disease severity and SMN2 copy number (Monani et al., 2000).
SMN homologues have also been identified from Caenorhabditis elegans (Miguel-Aliaga et al., 1999), Danio rerio (zebrafish) (Bertrandy et al., 1999; McWhorter et al., 2003), Schizosaccharomyces pombe (Hannus et al., 2000; Owen et al., 2000; Paushkin et al., 2000), and Drosophila (Miguel-Aliaga et al., 2000). The SMN proteins from these species retain a number of biochemical properties identified in the human SMN, including RNA binding activity and self-association. SMN models of C. elegans (Miguel-Aliaga et al., 1999), D. rerio (Bertrandy et al., 1999; McWhorter et al., 2003), and Drosophila (Miguel-Aliaga et al., 2000) show numerous SMA disease phenotypes. Defects of SMN proteins in C. elegans, D. rerio, and S. pombe affect cell viability and growth (Bertrandy et al., 1999; Hannus et al., 2000; McWhorter et al., 2003; Miguel-Aliaga et al., 1999; Owen et al., 2000; Paushkin et al., 2000). Drosophila SMN mutants (dSMN) result in abnormal neuromuscular junctions phenotypes (Chan et al., 2003). Ectopic overexpression of human SMN protein domains, which may function as dominant negatives to disrupt the endogenous dSMN protein, result in abnormally positioned wings and legs or pupal lethality (Miguel-Aliaga et al., 2000). Additionally, suppressing SMN expression in zebrafish induces motor axon-specific pathfinding defects, indicating that SMN functions in motor axon development, and that these early developmental defects may lead to subsequent motor neuron loss (McWhorter et al., 2003).
Mechanisms of RNA splicing and alternative splicing
Pre-mRNA splicing is a process in which introns are removed and exons are ligated together. Pre-mRNA splicing is divided into two steps. In the first step, the phosphate group at the 5′ splice site attacks the hydroxyl group of the adenine at the branch point to form a 2′-5′ phosphodiester bond. In the second step, the phosphate group at the 3′ splice site attacks the hydroxyl group at the 5′ splice site to form a 3′-5′ phosphodiester bond. The cleaved exons are ligated to form a mature RNA. Sequences required for pre-mRNA splicing include the 5′ splice site, the 3′ splice site, the branch point, and the polypyrimidine tract. These sequences are also called splicing signals. Pre-mRNA splicing of some introns requires a pre-mRNA enhancer region. Pre-mRNA splicing of some introns is regulated by pre-mRNA inhibitors.
Pre-mRNA splicing occurs in a large RNA protein complex called a spliceosome. A spliceosome contains two component parts: the first part is U1, U2, U4, U5, and U6 small nuclear RNA protein complexes (snRNPs); the second part includes proteins such as U2AF65 and SR. The formation of a spliceosome is a stepwise process. The first complex formed is an early spliceosome (complex E). In complex E, U1 snRNP is recruited into the pre-mRNA, and U1 snRNA in the U2 snRNP forms a base-pair with the 5′ splice site. The next complex formed is a pre-spliceosome. In the prespliceosome, U2 snRNP is recruited into the pre-mRNA, and the U2 snRNA in U2 snRNP forms a base-pair with the branch-point on pre-mRNA. U4/U5/U6 snRNPs are recruited into the mature spliceosome to perform catalytic activities.
In alternative splicing, different exons are included in different mRNAs. In humans, at least 75% of genes proceed by alternative splicing. Alternative splicing regulates almost every biological process in metazoans including signal transduction and energy transfer. A variety of diseases are caused by defects in pre-mRNA splicing; SMA is one example.
Is SMA caused by splicing?
Unlike many other diseases, SMA is a unique genetic disorder, because there are two copies of SMN genes in humans (Fig. 1). Yet, only when SMN1 is deleted or mutated, do patients suffer from the disease. SMN2 remains but is not able to produce sufficient full-length SMN protein to compensate for the loss of the SMN1 gene due to alternative splicing of exon 7. Strictly speaking, SMA is not a disease that is caused by splicing because it is the SMN1 gene deletions or mutations that cause the disease. However, the difference in splicing between SMN1 and SMN2 plays a critical role in how SMA disease develops, progresses, and is ultimately defined. For this reason, exon 7 splicing in SMN genes has been extensively studied during the last 15 years.
The primary sequences of SMN1 and SMN2 are rather similar. By examining mini-genes constructed from exon 6, intron 6, exon 7, intron 7, and exon 8, one of the major variations, a change in a C in SMN1 to a T in SMN2 at position 6 in exon 7 (Fig. 1), weakens the effect of SF2/ASF for enhancing inclusion of exon 7. This C6T change also creates a silencer element for hnRNP A1 to exclude exon 7. As a result, the C6T nucleotide largely determines the splicing patterns of the SMN1 and SMN2 genes, leading to 80–90% of SMN2 mRNA without exon 7 (see review by Singh, 2007). However, many studies have revealed that in addition to the C6T nucleotide in exon 7, several other elements regulate exon 7 alternative splicing in the SMN2 gene. On the positive site, 5′ and 3′ splicing sites, SF2/ASF binding sites in exon 7 of the SMN1 gene, and the “conserved tract”, which is located in the middle of exon 7, apparently stimulate the inclusion of exon 7, resulting in normal splicing of the exon in the SMN1 gene. In contrast, multiple negative elements are present in both the SMN1 and SMN2 genes, which overrun positive elements in the SMN2 gene. For example, a diversified intronic hnRNA A1 motif between SMN1 and SMN2 plays a role in exon 7 inclusion/skipping. Other negative elements including extended inhibitory context (Exinct), a 3′ cluster, the negative elements towards 5′ and 3′ ends of exon 7, two RNA secondary structures (terminal stem loop: TSL1 and TSL2) and more significantly intronic splicing silencer sequences which was identified by in vivo anti-sense oligo walking seem to have a great impact on regulation of exon 7 skipping (reviewed by Singh, 2007). Slight changes in any of these elements could tip the balance towards exclusion of exon 7 from SMN mRNA. How these elements regulate exon 7 has been comprehensively reviewed (Singh, 2007). In addition to cis-elements, multiple trans splicing factors such as hTra2β1 (Hofmann et al., 2000), SF2/ASF (Singh et al., 2004), hnRNP A1 (Harahap et al., 2012), and SRp30C (Young et al., 2002) have been identified and implicated to affect exon 7 splicing by directly or indirectly modulating silencers or enhancers.
Alternative splicing in SMN2 is targeted for SMA treatment
Unlike many other neurodegenerative diseases, SMA is a single gene disorder. More than 95% of SMA cases are directly caused by deletion or mutation defects in the SMN1 gene. Although there are only 20,000-30,000 patients in the US, this simple and unique genetic defect has lead to an extensive search for treating SMA disease. SMA is autosomal recessive; thus, exogenous expression of the SMN protein could be a viable approach for therapeutic treatment of patients with SMA. Not surprisingly, extensive efforts have been undertaken to examine constructs and vectors that can safely boost SMN expression in motor neurons of SMA models (Bevan et al., 2010; Dominguez et al., 2011; Foust et al., 2010; Glascock et al., 2011; Passini et al., 2010). This has been one of the main-stream objectives in the search for drugs to treat SMA. Significant progress has been made to deliver SMN products into SMA animal models during the last few years. Several groups have reported that SMN1 gene transfer into SMA models using self-complementary adeno-associated virus type 9 (scAAV9) or adeno-associated virus type 9 (AAV) can extend the life of mice with SMA from 15 to > 200 days without apparent disease symptoms.
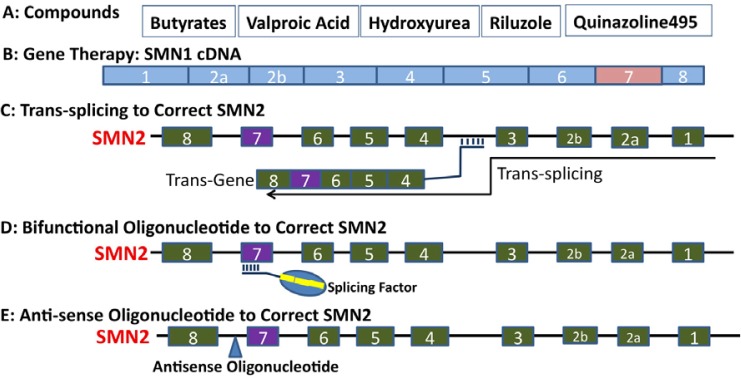
While gene therapy for SMA is promising, targeting exon 7 splicing of the SMN2 gene has been gaining momentum during the last few years. Initial efforts to identify drugs aimed at exon 7 in SMN2 were focused on small molecules. We developed a cell-based screen to identify small molecules capable of promoting inclusion of exon 7 in the mature SMN2 message using a mini-gene with minimal exon 7 modifications and a luciferase reporter fused to exon 8. We and others have used this cell-based assay and identified several compounds including sodium vanadate, aclarubicin, and indoprofen that increases inclusion of exon 7 (Andreassi et al., 2001; Lunn et al., 2004; Zhang et al., 2001). Identification of other compounds such as hydroxyurea (Grzeschik et al., 2005), valproate (Weihl et al., 2006), and phenylbutyrate (Andreassi et al., 2004) have been also described. In fact, several of these compounds have been examined for clinical trials in patients with SMA. However, selectivity and side effects of these compounds is still a concern. To increase specificities, multiple technologies to target aberrant splicing (Gallo et al., 2007) have been examined for SMN genes. One strategy that is employed to stimulate inclusion or exclusion of exon 7 is to use bifunctional oligoribonucleotides, which are molecules made of an anti-sense sequence that bind to an exonic element and a sequence tethering a trans factor, or link to a peptide domain mimicking a trans splicing factor (Cartegni and Krainer, 2003; Skordis et al., 2003; Villemaire et al., 2003). Bifunctional oligoribonucleotides effectively increase insertion of exon 7 into SMN2 mRNA. Relative to bifunctional oligoribonucleotides (Baughan et al., 2006; 2009; Dickson et al., 2008; Horne and Young 2009; Osman et al., 2012; Skordis et al., 2003; Voigt et al., 2010), the strategy of trans-splicing is new but has offered great promise to correct abnormal splicing of several other genes (Coady et al., 2007; Puttaraju et al., 1999; Rodriguez-Martin et al., 2005). Trans-splicing is based on studies showing that two pre-mRNAs can undergo trans-splicing if sufficient homologies between the two pre-mRNAs exist, leading to the first part of the mRNA from exons of premRNA1 and the second part from exons of pre-mRNA2. This strategy has been comprehensively explored by Lorson’s group during the last few years to correct SMN2 splicing. They optimized trans-splicing strategies and demonstrated that trans-splicing increases exon 7 insertion into SMN2 mRNA and extend the life of animals with SMA (Coady and Lorson, 2010; Coady et al., 2007; 2008; Shababi and Lorson, 2011; Shababi et al., 2011).
While all of these strategies that target splicing hold promise, the most intriguing results come from anti-sense oligonucleotides which target negative elements of exon 7 splicing in the SMN gene (Hua et al., 2007; 2008; 2010; 2011; Passini et al., 2011; Porensky et al., 2011; Singh et al., 2009). More interestingly, it has been shown that systemic administration of an antisense oligonucleotide (ASO-10-27) to neonates with SMA robustly rescues mice with severe SMA, much more effectively than intracerebroventricular administration, and subcutaneous injections extended median lifespan of animals with SMA 25-fold. These authors also demonstrated that not only the central nervous system but also peripheral tissues, in particular the liver, are involved in the pathogenesis of SMA. Restoration of liver function and other tissues may be essential for SMA treatment. Nonetheless, studies of SMN with documented methods to target exon 7 splicing provide multiple drug candidates for SMA therapy.
CONCLUSIONS
Can SMA be a model for disease treatment by targeting splicing?
Similar to transcription, splicing is a ubiquitous cellular process. Targeted splicing machinery may induce global effects on splicing events in cells. Therefore developing drugs that target splicing is difficult, if not impossible. However, compounds that were identified to regulate SMN splicing have shown relative selectivity to the SMN genes, leading scientists to believe that it may be possible to find safer drugs that can reverse abnormal splicing in disease (Andreassi et al., 2001). This promising perspective seems to be greatly boosted by recent developments in SMN/SMA studies in which technologies such as trans-splicing, bifunctional oligonucleotides, and anti-sense oligonucleotides very specifically increase the inclusion of exon 7 into SMN2 mRNA (Fig. 2) (Baughan et al., 2006; 2009; Coady and Lorson, 2010; Coady et al., 2007; 2008; Dickson et al., 2008; Hua et al., 2010; 2011; Lorson et al., 2010; Osman et al., 2012; Shababi and Lorson, 2011; Shababi et al., 2011; Skordis et al., 2003). In particular, the effectiveness of anti-sense oligos in animals with SMA has excited the scientific community so that SMA may be treatable in the near future. While clinical trials in humans are being developed, obstacles such as effectiveness and safety of these technologies in humans need to be overcome. In conclusion, SMA studies have set an example that splicing can be targeted for treatment of many diseases involved with abnormal splicing.
Fig. 2.
Potential therapeutic approaches for spinal muscular atrophy (SMA). (A) Compounds that have been or will be tested in clinical studies. (B) Gene therapy can be achieved by delivering full length SMN1 cDNA into patients with SMA; (C) Trans-splicing can be used to correct exon 7 skipping in the SMN2 gene; (D) A bifunctional oligonucleotide can recruit splicing factors to exon 7, leading to more exon 7 inclusion in SMN2 mRNA; (E) Anti-sense oligonucleotides would block negative elements in SMN splicing, stimulating exon 7 inclusion into SMN2 mRNA.
Acknowledgments
This work was supported by the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD) (JZ), P. R. China; Mid-career Researcher Program through a National Research Foundation (NRF) grant (2011-0000188 and 2011-0016757) funded by the Ministry of Education, Science, and Technology (MEST), Korea; the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, and Family Affairs (A100733-1102-0000100); and a Systems Biology Infrastructure Establishment grant provided by Gwangju Institute of Science and Technology (GIST) in 2011.
REFERENCES
- Andreassi C., Jarecki J., Zhou J., Coovert D.D., Monani U.R., Chen X., Whitney M., Pollok B., Zhang M., Androphy E., et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum. Mol. Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- Andreassi C., Angelozzi C., Tiziano F.D., Vitali T., De Vincenzi E., Boninsegna A., Villanova M., Bertini E., Pini A., Neri G., et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Zhang H., Byrd A.L., Femino A.M., Singer R.H., Taneja K.L., Lifshitz L.M., Herman I.M., Kosik K.S. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughan T., Shababi M., Coady T.H., Dickson A.M., Tullis G.E., Lorson C.L. Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol. Ther. 2006;14:54–62. doi: 10.1016/j.ymthe.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Baughan T.D., Dickson A., Osman E.Y., Lorson C.L. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum. Mol. Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrandy S., Burlet P., Clermont O., Huber C., Fondrat C., Thierry-Mieg D., Munnich A., Lefebvre S. The RNA-binding properties of SMN deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum. Mol. Genet. 1999;8:775–782. doi: 10.1093/hmg/8.5.775. [DOI] [PubMed] [Google Scholar]
- Bevan A.K., Hutchinson K.R., Foust K.D., Braun L., McGovern V. L., Schmelzer L., Ward J.G., Petruska J.C., Lucchesi P.A., Burghes A.H., et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler D., Raker V., Luhrmann R., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M.T., Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trulzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- Coady T.H., Lorson C.L. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J. Neurosci. 2010;30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady T.H., Shababi M., Tullis G.E., Lorson C.L. Restoration of SMN function: delivery of a trans-splicing RNA redirects SMN2 pre-mRNA splicing. Mol. Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- Coady T.H., Baughan T.D., Shababi M., Passini M.A., Lorson C.L. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS One. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T. W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Dickson A., Osman E., Lorson C.L. A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum. Gene. Ther. 2008;19:1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato C.J., Ingraham S.E., Mendell J.R., Prior T.W., Lenard S., Moxley R.T., 3rd, Florence J., Burghes A.H. Deletion and conversion in spinal muscular atrophy patients: is there a relationship to severity? Ann. Neurol. 1997;41:230–237. doi: 10.1002/ana.410410214. [DOI] [PubMed] [Google Scholar]
- Dominguez E., Marais T., Chatauret N., Benkhelifa-Ziyyat S., Duque S., Ravassard P., Carcenac R., Astord S., Pereira de Moura A., Voit T., et al. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu Q., Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Forch P., Valcarcelm J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis. 2001;6:463–468. doi: 10.1023/a:1012441824719. [DOI] [PubMed] [Google Scholar]
- Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gallo J.M., Noble W., Martin T.R. RNA and protein-dependent mechanisms in tauopathies consequences for therapeutic strategies. Cell Mol. Life Sci. 2007;64:1701–1714. doi: 10.1007/s00018-007-6513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glascock J., Osman E.Y., Wetz M.J., Krogman M.M., Shababi M., Lorson C. Decreasing disease severity in symptomatic Spinal Muscular Atrophy mice following scAAV9-SMN delivery. Hum. Gene Ther. 2012 doi: 10.1089/hum.2011.166. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik S.M., Ganta M., Prior T.W., Heavlin W.D., Wang C.H. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann. Neurol. 2005;58:194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- Hannus S., Buhler D., Romano M., Seraphin B., Fischer U. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 2000;9:663–674. doi: 10.1093/hmg/9.5.663. [DOI] [PubMed] [Google Scholar]
- Harahap I.S., Saito T., San L.P., Sasaki N., Gunadi, Nurputra D.K., Yusoff S., Yamamoto T., Morikawa S., Nishimura N., et al. Valproic acid increases SMN2 expression and modulates SF2/ASF and hnRNPA1 expression in SMA fibroblast cell lines. Brain Dev. 2012;34:213–222. doi: 10.1016/j.braindev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl. Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne C., Young P.J. Is RNA manipulation a viable therapy for spinal muscular atrophy? J. Neurol. Sci. 2009;287:27–31. doi: 10.1016/j.jns.2009.08.055. [DOI] [PubMed] [Google Scholar]
- Hu J.Y., Meng X., Schacher S. Redistribution of syntaxin mRNA in neuronal cell bodies regulates protein expression and transport during synapse formation and long-term synaptic plasticity. J. Neurosci. 2003;23:1804–1815. doi: 10.1523/JNEUROSCI.23-05-01804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Zhou J. Rpp20 interacts with SMN and is redistributed into SMN granules in response to stress. Biochem. Biophys. Res. Commun. 2004a;314:268–276. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- Hua Y., Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004b;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A. R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C. F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guiner C., Gesnel M.C., Breathnach R. TIA-1 or TIAR is required for DT40 cell viability. J. Biol. Chem. 2003;278:10465–10476. doi: 10.1074/jbc.M212378200. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO. J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson C.L., Rindt H., Shababi M. Spinal muscular atrophy mechanisms and therapeutic strategies. Hum. Mol. Genet. 2010;19:R111–R118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn M.R., Root D.E., Martino A.M., Flaherty S.P., Kelley B.P., Coovert D.D., Burghes A.H., Man N.T., Morris G.E., Zhou J., et al. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem. Biol. 2004;11:1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S., Pellizzoni L., Paushkin S., Mattaj I.W., Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 2002;22:6533–6541. doi: 10.1128/MCB.22.18.6533-6541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–932. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G.B.D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Culetto E., Walker D.S., Baylis H.A., Sattelle D. B., Davies K.E. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum. Mol. Genet. 1999;8:2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Chan Y.B., Davies K.E., van den Heuvel M. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett. 2000;486:99–102. doi: 10.1016/s0014-5793(00)02243-2. [DOI] [PubMed] [Google Scholar]
- Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossol W., Prior T. W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- Nicole S., Diaz C.C., Frugier T., Melki J. Spinal muscular atrophy: recent advances and future prospects. Muscle Nerve. 2002;26:4–13. doi: 10.1002/mus.10110. [DOI] [PubMed] [Google Scholar]
- Osman E.Y., Yen P.F., Lorson C.L. Bifunctional RNAs targeting the intronic splicing silencer N1 increase SMN levels and reduce disease severity in an animal model of spinal muscular atrophy. Mol. Ther. 2012;20:119–126. doi: 10.1038/mt.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen N., Doe C.L., Mellor J., Davies K.E. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum. Mol. Genet. 2000;9:675–684. doi: 10.1093/hmg/9.5.675. [DOI] [PubMed] [Google Scholar]
- Pagliardini S., Giavazzi A., Setola V., Lizier C., Di Luca M., De-Biasi S., Battaglia G. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum. Mol. Genet. 2000;9:47–56. doi: 10.1093/hmg/9.1.47. [DOI] [PubMed] [Google Scholar]
- Parsons D.W., McAndrew P.E., Monani U.R., Mendell J.R., Burghes A.H., Prior T.W. An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: further evidence for SMN as the primary SMA-determining gene. Hum. Mol. Genet. 1996;5:1727–1732. doi: 10.1093/hmg/5.11.1727. [DOI] [PubMed] [Google Scholar]
- Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O’Riordan C.R., Klinger K.W., Shihabuddin L.S., Cheng S. H. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Charroux B., Abel L., Perkinson R.A., Pellizzoni L., Dreyfuss G. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;1:919–922. doi: 10.1016/s0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K. D., Kaspar B.K., Wilton S.D., Burghes A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in the mouse. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr600. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M., Jamison S.F., Mansfield S.G., Garcia-Blanco M.A., Mitchell L.G. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin T., Garcia-Blanco M.A., Mansfield S.G., Grover A.C., Hutton M., Yu Q., Zhou J., Anderton B.H., Gallo J. M. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc. Natl. Acad. Sci. USA. 2005;102:15659–15664. doi: 10.1073/pnas.0503150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M., Lorson C.L. Optimization of SMN Trans-Splicing Through the Analysis of SMN Introns. J. Mol. Neurosci. 2012;46:459–469. doi: 10.1007/s12031-011-9614-3. [DOI] [PubMed] [Google Scholar]
- Shababi M., Glascock J., Lorson C.L. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum. Gene. Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- Singh R.N. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- Singh N.N., Androphy E.J., Singh R.N. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryot. Gene. Expr. 2004;14:271–285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli D.M., Oleynikov Y., Kelic S., Shenoy S.M., Hartley A., Stanton P.K., Singer R.H., Bassell G.J. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemaire J., Dion I., Elela S.A., Chabot B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J. Biol. Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- Voigt T., Meyer K., Baum O., Schumperli D. Ultra-structural changes in diaphragm neuromuscular junctions in a severe mouse model for Spinal Muscular Atrophy and their prevention by bifunctional U7 snRNA correcting SMN2 splicing. Neuromuscul. Disord. 2010;20:744–752. doi: 10.1016/j.nmd.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Weihl C.C., Connolly A.M., Pestronk A. Valproate may improve strength and function in patients with type III/IV spinal muscle atrophy. Neurology. 2006;67:500–511. doi: 10.1212/01.wnl.0000231139.26253.d0. [DOI] [PubMed] [Google Scholar]
- Wickens M., Goldstrohm A. Molecular biology. A place to die, a place to sleep. Science. 2003;300:753–765. doi: 10.1126/science.1084512. [DOI] [PubMed] [Google Scholar]
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- Zhang M.L., Lorson C.L., Androphy E.J., Zhou J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA potential therapy of SMA. Gene. Ther. 2001;8:1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]
- Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]