Abstract
Autophagy, or self-consuming of cytoplasmic constituents in a lytic compartment, plays a crucial role in nutrient recycling, development, cell homeostasis, and defense against pathogens and toxic products. Autophagy in plant cells uses a conserved machinery of core Autophagy-related (Atg) proteins. Recently, research on plant autophagy has been expanding and other components interacting with the core Atg proteins are being revealed. In addition, growing evidence suggests that autophagy communicates with other cellular pathways such as the ubiquitin-proteasome system, protein secretory pathway, and endocytic pathway. An increase in our understanding of plant autophagy will undoubtedly help test the hypothesized functions of plant autophagy in programmed cell death, vacuole biogenesis, and responses to biotic, abiotic, and nutritional stresses. In this review, we summarize recent progress on these topics and suggest topics for future research, after inspecting common phenotypes of current Arabidopsis atg mutants.
Keywords: autophagosome, NBR1, p62, selective autophagy
INTRODUCTION
Autophagy is an intracellular pathway for the bulk degradation of cytoplasmic materials, delivering autophagic cargoes to a lytic compartment, for example, the lysosome in animal cells and the vacuole in yeast (Saccharomyces cerevisiae) and plant cells. Autophagy determines the basal turnover of cytoplasm, renovates cells during cell differentiation, recycles old molecules for reuse, and protects cells from their own dangerous products and even from unwanted visitors. It is not surprising, therefore, to find defective autophagy associated with various disease symptoms in animals (reviewed by Levine and Kroemer, 2008; Mizushima and Komatsu, 2011).
Of the 3 major types of autophagy, namely, macroautophagy, microautophagy, and chaperone-mediated autophagy, macroautophagy is better characterized. Macroautophagy involves the formation of cytoplasmic autophagosomes sequestering a portion of cytoplasm (Fig. 1). An autophagosome is a double-membrane organelle developed from the phagophore, an early autophagic structure whose membrane origin is unknown. After the autophagosome is formed, the outer membrane of the autophagosome fuses with the membrane of the vacuole (or lysosome), and autophagic cargoes are rapidly degraded by the resident hydrolytic enzymes. Since vacuoles (particularly plant vacuoles) are bigger than autophagosomes, many autophagic bodies, or autophagosome-derived vesicles released in the vacuole, can be observed if stabilized by inhibitors of vacuolar degradation (e.g. E-64) or acidification (e.g. concanamycin A). In microautophagy, the lysosomal/vacuolar membrane is invaginated to sequester cytoplasmic constituents, whereas chaperone-mediated autophagy involves selective translocation of substrates into the vacuole.
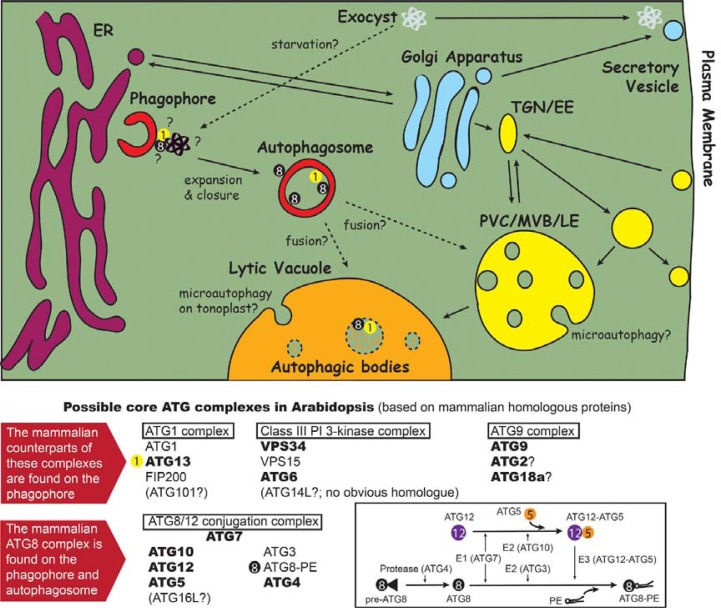
Fig. 1.
Plant autophagic pathway, its interacting organelles, and genes encoding core autophagic complexes. Colors represent intracellular compartments: ER (purple), Golgi and secretory vesicle (cyan), endosomal compartments (yellow), autophagic vesicles (red), and lytic vacuole (orange). Autophagic bodies in the lytic vacuole are outlined with dashed circles. Solid arrows indicate known transport or maturation, and dashed arrows symbolize hypothetical fusion or translocation. Two billiard balls, numbered 1 and 8, show the locations of GFP-ATG8 and YFP-ATG1a puncta, respectively, in Arabidopsis cells. Hypothetical components of 4 core ATG complexes are shown below. The components are based on the conserved members of metazoan homologues. Genes encoding the proteins in bold were characterized by corresponding knockout or knock-down plants (see Table 1 and text). Inset, ATG8 and ATG12 conjugation pathway in Arabidopsis. PE, phosphatidylethanolamine PVC/MVB/LE, prevacuolar compartment/multivesicular body/late endosome. TGN/EE, trans-Golgi network/early endosome.
Elucidation of the functions of macroautophagy (hereafter referred to as autophagy) was facilitated by the isolation of the Autophagy-related (Atg) genes from yeast mutants (Matsuura et al., 1997; Tsukada et al., 1993). Subsequent biochemical and cell biological characterization of Atg genes revealed a model of autophagic processes that are controlled by several classes of Atg gene (Fig. 1). There are 4 main classes of core Atg proteins that are highly conserved in various eukaryotes (Yang and Klionsky, 2010) including plants such as Arabidopsis (references in Table 1), rice and maize (Chung et al., 2009; Shin et al., 2009); (i) components of the Atg1 kinase complex, (ii) components of an autophagy-speci-fic phosphatidylinositol (PI) 3-kinase complex, (iii) components of a complex containing transmembrane Atg9 protein, and (iv) proteins involved in Atg8 and Atg12 conjugation. The Atg1 kinase complex containing Atg1, Atg13, FIP200, and Atg101 regulates the induction of autophagy in response to nutrient limitation. The Atg1 kinase complex is located on a specific ER subdomain and regulates an autophagy-specific PI 3-kinase complex containing Vps34 kinase, Vps15, Atg6, and Atg14. Atg14 is an autophagy-specific component (Itakura et al., 2008; Kametaka et al., 1998), but a plant Atg14 homologue has not been reported. The PI 3-kinase complex is enriched in phagophores and generates PI3P, which is recognized by various PI3P effectors such as Atg18 proteins interacting with Atg2. The function of Atg9 complex is not yet clear, although it moves from the trans-Golgi network to the phagophore during starvation (Young et al., 2006). Atg8 is a ubiquitin-fold protein that is conjugated to phosphatidylethanolamine (PE) by E1 (ubiquitin-activating enzyme)-like Atg7, E2 (ubiquitin-conjugating enzyme)-like Atg3, and the E3-like Atg12-Atg5-Atg16 complex (Fig. 1, inset). The Atg4 protease removes extra C-terminal residues of an Atg8 precursor and thereby exposes a specific glycine residue for conjugation. Atg12 is another ubiquitin-fold protein conjugated to Atg5 by Atg7 (E1) and Atg10 (E2). By conjugation to PE, Atg8 is targeted to the phagophore and required for the expansion and closure of the autophagic vesicle to form an autophagosome. Atg8 is used as a standard marker for autophagy, since it is located in most autophagic membranes - the phagophore, autophagosome, and autophagic bodies (Mizushima et al., 2010).
Table 1.
Phenotypes of common atg mutants and transgenic plants in Arabidopsis
| Allele name | Autophagic vesicles | ATG8-PE formation | Other phenotypes | References |
|---|---|---|---|---|
| atg mutants of the ATG8 and ATG12 conjugation pathway | ||||
| atg4a-1 atg4b-1 double mutant | V-EM (−) V-8 (−) C-8 (−/+) |
GFP-ATG8-PE (−) | ES | Yoshimoto et al. (2004) |
| atg4a-2 atg4b-2 double mutant | ATG8-PE (−) | ES, HC, HN | Chung et al. (2010) | |
| atg5-1 (protein null) | V-8 (−) V-NR (−) C-8 (−/+) |
ATG8-PE (−) GFP-ATG8-PE (−) |
ES, HC, HN, DT, ECD |
Chung et al. (2010) Inoue et al. (2006) Kwon et al. (2010) Lenz et al. (2011) Phillips et al. (2008) Thompson et al. (2005) Yoshimoto et al. (2009) |
| atg7-1 (protein null) | V-8 (−) C-LT (↓) |
ES, HC, HN, DCD |
Doelling et al. (2002) Hofius et al. (2009) Thompson et al. (2005) |
|
| atg7-2 (protein null) | V-8 (−) C-8 (−/+) |
ATG8-PE (−) GFP-ATG8-PE (−) |
ES, HC, HN |
Chung et al. (2010) Hofius et al. (2009) Suttankakul et al. (2011) |
| atg10-1 | V-8 (−) | ATG8-PE (−) | ES, HC, HN, ECD |
Chung et al. (2010) Lenz et al. (2011) Phillips et al. (2008) Wang et al. (2011) |
| atg12a-1 atg12b-1 double mutant | V-8 (−) | ATG8-PE (−) | ES, HC, HN | Chung et al. (2010) |
| Other atg mutants | ||||
| atg2-1 | V-NR (↓) | ES, ECD |
Inoue et al. (2006) Wang et al. (2011) Yoshimoto et al. (2009) |
|
| atg2-2 | V-8 (−) C-8 (−) |
ES, ECD | Wang et al. (2011) | |
| atg6-1 | homozygotes not recovered due to male sterility |
Fujiki et al. (2007) Qin et al. (2007) |
||
| atg6 antisense | C-MDC (−) C-LT (↓) |
ES, HC, HN, Dev, ECD | Patel and Dinesh-Kumar (2008) | |
| atg9-1 | ES, HC, HN, DCD |
Hanaoka et al. (2002) Hofius et al. (2009) |
||
| atg9-2 | V-NR (+) | Inoue et al. (2006) | ||
| atg13a-1 atg13b-1 double mutant | V-8 (−) C-8 (−) |
ATG8-PE (+) | ES, HC, HN | Suttankakul et al. (2011) |
| atg18a RNAi | C-MDC (−) C-8 (−) |
ES, HC, HN, HDS, HSS, ECD |
Lenz et al. (2011) Liu et al. (2009) Xiong et al. (2005) |
|
Abbreviations: C-8, presence (+) or absence (−) of cytoplasmic GFP-ATG8 bodies (i.e. autophagosomes), with (−/+) representing ambiguous data (see text); ↓ (C-LT), less accumulation of cytoplasmic bodies stained with LysoTracker than a wild-type control; C-MDC, presence (+) or absence (−) of cytoplasmic bodies stained with monodansylcadaverine; V-8, presence (+) or absence (−) of vacuolar GFP-ATG8 bodies with the treatment of concanamycin A (i.e. autophagic bodies); V-EM, presence (+) or absence (−) of vacuolar autophagic bodies assessed by an electron microscopy; V-NR, absence (−), less accumulation than a wild-type control (↓), or a wild-type level (+) of vacuolar bodies stained with neutral red; ATG8-PE, presence(+)/absence(−) of ATG8-PE by the ATG8 immunoblot analysis; GFP-ATG8-PE, presence(+)/absence(−) of GFP-ATG8-PE by the GFP immunoblot analysis; DCD, delayed cell death by infection (Hofius et al., 2009); Dev, multiple developmental phenotypes including stunted growth; ECD, enhanced cell death by infection (Lenz et al., 2011; Patel and Dinesh-Kumar, 2008; Wang et al., 2011); ES, early senescence; HC, hypersensitivity to carbon limitation; HN, hypersensitivity to nitrogen limitation; HDS, hypersensitivity to drought stress; HSS, hypersensitivity to salt stress; DT, delayed differentiation of tracheary elements.
Recent reviews have summarized the genetic components of autophagy in Arabidopsis and other plants (Avin-Wittenberg et al., 2012; Thompson and Vierstra, 2005), as well as tools available for plant autophagy research (Chung, 2011; Mitou et al., 2009). Here, we describe recent progress in our knowledge on core Atg genes in plants, and draw conclusions from the comparison of various Arabidopsis atg mutants. We introduce the plant proteins that interact with Atg8 and discuss the possibility of communication between autophagy and other cellular processes, such as proteasome-mediated proteolysis and endocytosis. Finally, functions of plant core Atg genes are proposed based on available genetic data. Readers may also refer to specific reviews that we will cite in the respective sections below.
Genetics of core ATG genes in Arabidopsis
Initial molecular genetics studies on plant autophagy relied on sequence similarity to Atg homologues in yeast, although electron microscopic studies identified ultrastructures indicating microautophagy and macroautophagy in a number of plant species, prior to the discovery of the Atg genes (reviewed by Bassham et al., 2006). Investigation of mutations in the core Atg genes in Arabidopsis (Fig. 1 and Table 1) revealed the physiological roles of the Atg genes in plant development and under nutrient limitation and various stresses. So far, all canonical atg mutants have shown phenotypes of accelerated leaf senescence and hypersensitivity to carbon and nitrogen limitation (Table 1). However, no big difference in plant architecture was identified from the mutants, except for atg6 null mutants and antisense plants (see Table 1). This seemingly contrasts with Atg7 knockout mice, which showed multiple developmental defects (Komatsu et al., 2005; reviewed by Mizushima and Levine, 2010). However, this does not necessarily indicate the unimportance of plant Atg genes in plant development (see below). For instance, recent research has shown that tracheary element differentiation is delayed in atg5-1 mutants (Kwon et al., 2010).
A phenotypic difference between atg6 and other atg mutants is also seen in metazoans. While atg7 and other mutants defective in the Atg8 and Atg12 conjugation survive the embryonic stage, Becline-1/Atg6 knockout mice have more severe phenotypes and are embryo-lethal (Qu et al., 2003; Yue et al., 2003). The difference supports the view that Atg6 and Ulk1/Atg1 act upstream of the Atg8 conjugation system and are responsible for an unconventional form of autophagy that is independent of the Atg8 conjugation system (Nishida et al., 2009). Alternatively, it may indicate that plant and metazoan Atg6 genes play a role in cellular processes other than autophagy. Various forms of mammalian class III PI 3-kinase complexes, containing Atg6 as a common component, are involved in multiple steps of autophagy and potentially in the endocytic pathway (reviewed by Funderburk et al., 2010). Notably, Arabidopsis atg6 knockout mutants (Fujiki et al., 2007; Qin et al., 2007) are phenotypically similar to vps34 knockout mutants (Lee et al., 2008).
The latter view was supported by a recent report that autophagy-defective mutants lacking 2 Arabidopsis ATG13 homologues are viable and share common phenotypes with ATG8 conjugation mutants (Suttangkakul et al., 2011) (see Table 1). However, this cannot exclude the possibility that ATG1/ATG13-independent autophagy may still exist in plant cells. Notably, the double mutants are capable of conjugating ATG8 to PE, indicating that ATG8 lipidation is required but not sufficient for autophagy. Another interesting finding is that ATG13 and ATG1 are rapidly targeted into the vacuole for degradation when wildtype Arabidopsis plants are starved (Suttangkakul et al., 2011). This raises the possibility that the Arabidopsis ATG1/ATG13 complex is both a regulator and a target of autophagy.
Plant autophagy markers
Recent reviews have extensively discussed tools for autophagy research (Mitou et al., 2009; Mizushima et al., 2010). Here we focus on fluorescent markers of autophagy in plant cells and highlight precautionary notes to be considered by researchers.
First, autophagosomes can accumulate not only when autophagy is induced but also when a later step of autophagy is inhibited. For instance, if the fusion of autophagosomes with lysosomes is inhibited or delayed, autophagosomes will accumulate (reviewed by Mizhushima et al., 2010). Concanamycin A can be used in combination to determine whether an observed accumulation of autophagic structures is due to the induction of autophagy.
Second, neutral red, Lysotracker, and monodansylcadaverine (MDC) are markers for acidic compartments but not specific for autophagosomes (Mizushima, 2004). Thus, it is not recommended to use these dyes as the only tool for measuring autophagic activities.
Finally, cytoplasmic GFP-ATG8 markers may not always indicate autophagosomes, at least in mutants defective in ATG8 and ATG12 conjugation. For example, GFP-ATG8 foci have been detected in the cytoplasm of atg4a-1 atg4b-1 double mutants (Yoshimoto et al., 2004), atg7-2 (Suttangkakul et al., 2011) and atg5-1 (unpublished data by Chung T and Vierstra RD) (see “C-8 (−/+) in Table 1). The nature of the cytoplasmic GFP-ATG8 foci in the mutants is not yet clear, but they may represent immature, nonfunctional autophagosomes. Alternatively, the foci may be the aggregates of GFP-ATG8 fusion proteins, since GFP-ATG8 in atg4a-1 atg4b-1 double mutants was solubilized more effectively by ionic detergent than by non-ionic detergent, salt, or urea (Yoshimoto et al., 2004). Notably all published plant GFP-ATG8 markers are ectopically expressed by the cauliflower mosaic virus 35S promoter. In any case, it is noteworthy that the mutants consistently lack autophagic bodies in the vacuole, which are visualized by GFP-ATG8 signals in the presence of concanamycin A (see “V-8 (−)” in Table 1; Chung et al., 2010; Phillips et al., 2008; Thompson et al., 2005; Yoshimoto et al., 2004). In conclusion, vacuolar GFP-ATG8 puncta are a better indicator for ATG7-dependent autophagy than cytoplasmic GFP-ATG8 dots, at least under the conditions in which GFP-ATG8 forms protein aggregates (Chung, 2011).
GFP-ATG8 is known to be an aggregation-prone protein. How can we avoid potential artifacts, such as GFP-ATG8 protein aggregates? A transgenic line that produces an endogenous or a moderate level of GFP-ATG8 fusion proteins may be useful as an autophagy marker (Moore and Murphy, 2009). Unfortunately, efforts to generate GFP-ATG8 transgenic lines using a native promoter to drive transgene expression have been unsuccessful, because of the low level of fluorescent proteins that may rapidly and continuously be degraded by autophagy (unpublished data by Chung T and Vierstra RD). Instead, a modest level of mCherry-ATG8 and GFP-ATG8 expression was obtained (Suttangkakul et al., 2011; unpublished data by Chung T and Kim S) using the ubiquitin gene promoter (Geldner et al., 2009).
Beyond the core
Metazoan genes related to autophagy were initially identified by amino acid sequence similarity among conserved Atg genes. Recently, metazoan-specific genes have been discovered by multiple approaches such as a protein-interactome study combined with siRNA technology (Behrends et al., 2010), a genome-wide siRNA screen (Lipinski et al., 2010; Orvedahl et al., 2011), and a forward genetic screen in nematodes (Tian et al., 2010). These studies confirmed a genetic network containing core Atg genes and revealed novel network components that are not found by the homology-based approach. A sub-network comprising Atg8 homologues is central to the network and interacts with the Atg8/Atg12 conjugation system and many new interacting proteins (Behrends et al., 2010).
Although no comparable network for plant autophagy has yet been described, a part of the metazoan autophagy network is expected to be conserved in plant genomes. So far, only a few plant proteins have been shown to interact with ATG8 (Table 2).
Table 2.
Plant proteins shown to interact with ATG8
| Interacting protein | Method of detection and confirmation | Domains and motifs | Mammalian homologue | References |
|---|---|---|---|---|
| Arabidopsis NBR1 | IVPD, PCLVV | PB1, ZZ, AIM, UBA | p62, NBR1 | Svenning et al. (2011) |
| Tobacco Joka2 | Y2H, IVPD, PCLVV | PB1, ZZ, AIM, UBA | p62, NBR1 | Zientara-Rytter et al. (2011) |
| Arabidopsis ATI1 | Y2H, BiFC, PCLVV (rare) | TM, AIM | not found | Honig et al. (2012) |
| Tomato Adi3 | Y2H | STK, PIF | ? | Devarenne (2011) |
Abbreviations: AIM, ATG8-interacting motif; BiFC, in vivo interaction by bimolecular fluorescence complementation; FW, 4 tryptophan domain; IVPD, in vitro pull-down assay; PB1, Phox-Bem1 domain; PCLVV, partial co-localization with GFP-ATG8 or CFP-ATG8 in vivo; PIF, Pdk1-interacting fragment; STK, serine/threonine kinase domain; TM, trans-membrane domain; UBA, ubiquitin-associated domain; Y2H, yeast two-hybrid interaction; ZZ, ZZ-type zinc finger domain.
Interactions of ATG8 with plant homologues of p62/NBR1 have recently been described (Svenning et al., 2011; Zientara-Rytter et al., 2011). Mammalian p62 and its paralogue NBR1 (neighbor of BRCA1 gene 1) are considered to be receptors for autophagic cargoes, interacting with LC3/Atg8 through its LC3-interacting region (LIR; also known as an ATG8-interacting motif, or AIM) (Kirkin et al., 2009; Komatsu et al., 2007; Pankiv et al., 2007). Mammalian p62 and NBR1 are thought to play a critical role in selective autophagy, in which autophagic cargoes are selectively recognized and sequestered by autophagic vesicles and targeted to the lysosome for degradation (see below). The Arabidopsis NBR1 homologue has biochemical and cell biological properties that are very similar to those of its mammalian counterpart (Svenning et al., 2011). The Arabidopsis NBR1 homologue has a functional AIM and 2 ubiquitin-associated (UBA) domains at the C terminal. The NBR1 homologue also contains a PB1 domain for homo-polymerization, and is targeted to the vacuole for degradation in an ATG7-dependent manner. A tobacco NBR1/p62 homologue has also been reported and contains a similar combination of conserved domains (Zientara-Rytter et al., 2011).
ATI1 and ATI2 are plant-specific genes that were identified from a yeast two-hybrid screen for ATG8f-interacting proteins (Honig et al., 2012). When overexpressed, GFP-ATI1 forms spherical structures called ATI1 bodies adjacent to the ER network, despite the lack of apparent N-terminal signal peptide in the deduced ATI1 and ATI2 amino acid sequences. The ATI1 bodies are highly dynamic, increase in number during carbon starvation, and accumulate in the central vacuole in the presence of concanamycin A, indicating that the ATI bodies are finally degraded in the vacuole. The ATI1 bodies did not colocalize with any organelle markers tested in non-stressed conditions. Although a BiFC experiment confirmed the in vivo interaction of ATG8f with the 2 ATI homologues, only a small fraction of the ATI1 bodies emitted an mRFP-ATG8f signal during carbon starvation. Thus, the ATI1 bodies may represent an unknown compartment that functionally relates to autophagy in plant cells. It is unlikely that ATI1 bodies simply represent protein aggregates resulting from the ectopic expression of the GFP-ATI1 transgene, because GFP-ATI1 fluorescence and immunoreactive signals are mostly found on the surface of the ATI1 bodies, not in their lumen. In addition, the ATI1 bodies were seen in transmission images of confocal microscope and were identical to spherical bodies also found in carbon-starved, non-transgenic plants (Honig et al., 2012). Future studies may focus on the functions of ATI1 and ATI2 in selective autophagy and reveal the nature of ATI1 bodies and their relationship with canonical autophagosomes, or ATG8 bodies. It is not known whether mRFP-ATG8f co-localizes with stabilized ATI1 bodies in vacuoles treated with concanamycin A. It will be interesting to see whether ATG genes are required for the transport of ATI1 bodies to the vacuole.
Adi3 in tomato is a serine/threonine AGC kinase that interacts with Pto and AvrPto, and suppresses programmed cell death. Adi3 was also shown to interact with tomato Atg8h by a yeast two-hybrid screen (Devarenne, 2011). It remains to be seen whether Adi3 is targeted by autophagy and degraded in the vacuole.
Arabidopsis TSPO (tryptophan-rich sensory protein) appears to be degraded by ATG5-dependent autophagy, since TSPO protein levels increase through the actions of PI 3-kinase inhibitors and by atg5 mutation (Vanhee et al., 2011). The Arabidopsis TSPO has an AIM-like consensus sequence, and the YFP-TSPO signal partially overlapped with that of GFP-ATG8e, although further work is needed to show a direct interaction between ATG8 and TSPO. Human TSPO homologues have not yet been shown to interact with LC3/Atg8.
Plant cell compartments relating to autophagy
An analysis of autophagy in mammalian cells recently revealed communication between the autophagic pathway and endomembrane transport systems such as the endocytic pathway (reviewed by Rusten and Stenmark, 2009). For example, inactivation of Endosomal Sorting Complex Required for Transport (ESCRT) complexes blocks endocytosis and also leads to the accumulation of autophagosomes, and it appears that ESCRT complexes are required for the maturation and fusion of autophagosomes with various endocytic organelles (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007) (see Fig. 1). Evidence supports the view that this is also the case in plant cells. Human AMSH (Associated Molecule with the SH3 domain of STAM) deubiquitinating enzyme hydrolyzes K63-linked ubiquitin chains, which have a role in endocytosis (McCullough et al., 2004). Recently Arabidopsis AMSH3 was shown to cleave K48- and K63-linked ubiquitin chains and to be required for endocytosis. amsh3 mutant seedlings show a phenotype of arrested growth, accumulate ubiquitin conjugates, and have small, fragmented vacuoles. Furthermore, electron microscopic analysis indicated that autophagosomes accumulate in amsh3 mutants (Isono et al., 2010; Rusten and Stenmark, 2009). Human AMSH and Arabidopsis AMSH3 differ somewhat in their domain composition, substrate specificity, and subcellular localization. Nevertheless, the phenotypes of amsh3 mutants suggest that endocytosis is related to autophagy in plant cells. Since the Arabidopsis genome has genes encoding ESCRT complexes (reviewed by Reyes et al., 2011; Winter and Hauser, 2006), investigation of plant mutants containing a defective ESCRT complex will provide a mechanistic explanation of the link between endocytosis and autophagy.
In yeast and slime mold (Dictyostelium discoideum), several core Atg genes are required for the unconventional secretion of Acyl-coA binding protein (ACBP) lacking an N-terminal signal peptide (Duran et al., 2010; Manjithaya et al., 2010). Autophagy induction in astrocytes also leads to the secretion of mammalian ACBP (Loomis et al., 2010). A model of ACBP “exophagy” postulates the fusion of an endosome with an autophagosome containing ACBP as a cargo (reviewed by Manjithaya and Subramani, 2011). Arabidopsis ACBP3, an ACBP homologue, is indeed secreted to apoplast. However, unlike yeast and mammalian ACBP, Arabidopsis ACBP3 has an N-terminal signal peptide, and a mutant ACBP3 lacking the signal peptide is not secreted (Manjithaya and Subramani, 2011; Xiao et al., 2010). Interestingly, when a gene encoding Arabidopsis ACBP3 is overexpressed, leaf senescence is accelerated and MDC-stained bodies are disrupted, suggesting that autophagy is inhibited in the overexpression line (Manjithaya and Subramani, 2011; Xiao et al., 2010). Notably, distribution of cytoplasmic GFP-ATG8e puncta does not appear to be affected by ACBP3 overexpression. It remains to be established whether the overexpression blocks the accumulation of GFP-ATG8-labeled autophagic bodies in the vacuole.
Exocyst is an octameric complex mediating post-Golgi vesicle trafficking and fusion with the plasma membrane. Exocyst was first described in yeast but is conserved in metazoans and plants. Using a fluorescent protein fused to an Arabidopsis Exocyst component called Exo70E2, Wang et al. (2010) defined a new compartment called EXPO (Exocyst-positive organelle). EXPO is a cytoplasmic, double-membrane-bound organelle not overlapped by known markers for the Golgi apparatus, endosomes, and autophagosomes. EXPO is not affected by inhibitors of secretory and endocytic pathways, but still fuses with the plasma membrane and has thus been proposed to mediate unconventional secretion in plant cells (Wang et al., 2010). EXPO is probably not the same compartment as the ATI bodies, since carbon starvation decreases EXPO (Wang et al., 2010) but increases ATI bodies (Honig et al., 2012). More recently, a role for Exocyst in autophagosome formation was identified in human cells (Bodemann et al., 2011). Exocyst members were found to interact with multiple components of the class III PI 3-kinase complex, Atg1/Atg13 complex, and Atg8 conjugation complex. Exocyst exists in a perinuclear region prior to autophagy induction, but upon starvation, it was detected at cytosolic puncta co-localized with phagophore markers and PI3P. These data suggest that Exocyst acts as a scaffold for autophagosome assembly (see Fig. 1). An immediate question is whether plant EXPO plays a role in autophagosome formation. One possibility is that EXPO represents the perinuclear Exocyst complex before autophagy induction. It will be meaningful to see whether fluorescent proteins fused with other Exocyst components are co-localized with phagophores and autophagosomes.
Autolysosomes in tobacco culture cells are yet another structure that participates in autophagic processes (Moriyasu and Ohsumi, 1996; Takatsuka et al., 2011). The autolysosomes increase in number when tobacco cells are cultured without sucrose and treated with a cysteine protease inhibitor. Like animal autolysosomes, tobacco autolysosomes contain acid hydrolases and partially degraded cytoplasmic materials within an acidic environment. The autolysosomes are generally bigger (1–6 μm in diameter; with a protease inhibitor) than cytoplasmic structures displaying GFP-ATG8 (typically less than 2 μm). It is not clear whether autolysosome-like structures exist in intact plant cells under physiological conditions. The same research group identified autolysosome-like structures in young Arabidopsis root cells treated with a cysteine protease inhibitor, although autolysosomes in mature root cells appear rare (Inoue et al., 2006). Working with starved Arabidopsis suspension-cultured cells, Marty and colleagues also observed a double-membrane-bound structure (0.6–2.5 μm in diameter; without a protease inhibitor) called an autophagic vacuole (Rose et al., 2006). Interestingly, at least some of the autolysosome membrane appears to come from the plasma membrane (Takatsuka et al., 2011). The relationship of autolysosomes and autophagic vacuoles with similar organelles such as endocytic multivesicular bodies (also called prevacuolar compartments), ATI bodies (Honig et al., 2012), and senescence-associated vacuoles (Otegui et al., 2005) may be a subject for future research.
UPS and selective autophagy
The ubiquitin-proteasome system (UPS) and autophagy are 2 important degradation systems conserved in eukaryotes. Substrates of UPS are tagged by a ubiquitin chain, the addition of which is tightly regulated by the specific interaction of the substrate with a particular E3 ligase complex. UPS can only break down soluble polypeptides, while autophagy destroys virtually any cytoplasmic materials including insoluble proteins. Autophagy was thought to capture a portion of cytoplasm randomly, but there is a selective type of autophagy; cargoes for selective autophagy include protein aggregates, mitochondria, ribosomes, peroxisomes, and so on. According to a current model of selective autophagy, ubiquitinated autophagic cargoes are recognized by a UBA domain of p62/NBR, which interacts with ATG8 on the phagophore, thereby targeting autophagic cargoes to the autophagosome (reviewed by Johansen and Lamark, 2011). Thus, both UPS and selective autophagy appear to use ubiquitin as a tag for destruction.
Although the model of p62-mediated selective autophagy is supported by biochemical studies, evidence suggests that p62 action may not be simple. Firstly, p62 is also involved in the selective autophagy of non-ubiquitinated substrates (Gal et al., 2009; Watanabe and Tanaka, 2011). Secondly, p62 is required for pexophagy (Kim et al., 2008) and xenophagy (Zheng et al., 2009) but is dispensable for mitophagy (Narendra et al., 2010). Finally, while Atg7 or Atg5 knockout mice contain an elevated level of ubiquitinated proteins, p62 single knockout mice do not accumulate a normal level of ubiquitinated proteins. If this results from genetic redundancy by NBR1, a ubiquitin immunoblot analysis of p62 nbr1 double mutants will be helpful. The fact that Arabidopsis has only one gene encoding p62/NBR may be advantageous, although the Arabidopsis gene may be essential (Svenning et al., 2011).
A crosstalk between UPS and autophagy has recently been proposed. For example, autophagy is enhanced when proteasome activity is blocked by inhibitors (Pandey et al., 2007; Takeda et al., 2010). The induction of autophagy could compensate for insufficient proteasome activity, since up-regulation of autophagy was shown to protect cells from proteasome inhibition (Pandey et al., 2007). By contrast, when autophagy is blocked genetically or pharmacologically, protein degradation by UPS is inhibited (Korolchuk et al., 2009), or not affected (Komatsu et al., 2006; 2007), depending on tissue type and assay method. A plausible explanation for the inhibition of UPS and the accumulation of ubiquitinated proteins in atg7 mutants is that the impairment of autophagy leads to the accumulation of p62, which in turn competes with other ubiquitin-binding proteins for the delivery of ubiquitinated substrates to the proteasome, thereby inhibiting UPS (reviewed by Korolchuk et al., 2010). Consistent with this hypothesis, p62 is an autophagic cargo, and overexpression of p62 is sufficient to inhibit the UPS (Korolchuk et al., 2009). This hypothesis is not mutually exclusive to the simple model of p62 for selective autophagy, but also explains why atg7 p62 double knockout mice showed a lower level of ubiquitinated proteins than atg7 single knockout mice (Komatsu et al., 2007).
It is not known whether a similar crosstalk between UPS and autophagy exists in plant cells. However, evidence suggests that autophagy compensates for UPS in plants. ATG18a RNAi plants are more sensitive to methyl viologen, an inducer of photo-oxidative stress, and accumulate a higher level of oxidized proteins than non-transgenic controls (Xiong et al., 2007). Furthermore, numerous MDC-labeled structures were detected when either hydrogen peroxide or methyl viologen was added to plant medium (Xiong et al., 2007). Since UPS is assumed to eliminate oxidized proteins and strong oxidative stresses inactivate UPS, this result is consistent with a possible compensation by autophagy during UPS inactivation. Confirmation by using other UPS inhibitors may be performed in future to support the proposed role of autophagy.
Roles of Atg genes in biotic and abiotic stresses
Plants that lack mobile immune cells solely rely on the innate immune system for defense by recognizing the pathogen-derived nonself molecules. The plant immune receptors can be categorized into 2 distinct groups; surface and intracellular immune receptors. A surface receptor, also called a pattern recognition receptor (PRR), recognizes a pathogen-associated molecular pattern (PAMP), which is a microbial structural or functional unit, and activates the PAMP-triggered immunity (PTI). The latter, also called Resistance (R) protein, either directly or indirectly interacts with an effector that is in general translocated into host cells by pathogens to disturb the host immune responses. Their interactions lead to the activation of effector-triggered immunity (ETI), which often results in the host plant cell death, referred to as hypersensitive response (HR) (Dodds and Rathjen, 2010; Jones and Dangl, 2006).
Since the well-known phenotype of autophagy mutant plants is early senescence accompanied by accelerated cell death (Hayward and Dinesh-Kumar, 2011; Talbot and Kershaw, 2009) and since HR cell death plays a key role in plant disease resistance, probably by limiting the pathogen spread, autophagy is thought to be somehow associated with plant defense responses, particularly with ETI. Indeed, atg5, atg10, and atg18a plants show retained immune responses of stomatal closure, MAPK phosphorylation, ethylene (ET) production, callose deposition, and defense gene expression by flg22, an active bacterial PAMP (Lenz et al., 2011). This suggests that autophagy is unrelated to PTI. The link between cell death control by autophagy and plant defense responses was first reported in Nicotiana benthamiana by Liu et al. (2005). The virus-induced gene silencing (VIGS) of Atg6/Beclin-1 resulted in uncontrolled cell death spread beyond infection sites of tobacco mosaic virus (TMV). The same cell death spread in Atg6/Beclin-1-deficient N. benthamiana by transient co-expression of an effector and its cognate R protein (TMV p50/tobacco N, Pseudomonas syringae AvrPto/tomato Pto, or Cladosporium fulvum Avr9/tomato Cf-9) indicates that this failed restriction of plant cell death is caused by ETI. The involvement of autophagy in ETI was further supported by a finding that autophagosome formation in Arabidopsis is induced by detectable effector (AvrRpm1 or AvrRps4)-expressing P. syringae but not by a virulent P. syringae DC3000 (Hofius et al., 2009).
The early senescence and pathogen-induced HR cell death spread observed in light-grown autophagy mutant plants requires salicylic acid (SA) signaling, because these phenotypes are suppressed by inhibiting SA biosynthesis/accumulation or by inhibiting SA signaling (Wang et al., 2011; Yoshimoto et al., 2009). Indeed, SA is highly accumulated in autophagy mutant plants (Lenz et al., 2011; Yoshimoto et al., 2009), suggesting that autophagy negatively regulates SA signaling. Pathogen infection and treatment with an SA analogue benzo-(1,2,3)-thiadiazole-7-carbothioic acid (BTH) also induce the expression of Atg genes and the formation of autophagosomes in plants (Hofius et al., 2009; Lai et al., 2011; Liu et al., 2005; Patel and Dinesh-Kumar, 2008; Wang et al., 2011; Yoshimoto et al., 2009). This suggests that autophagy and SA signaling form a negative feedback to control each other. However, some autophagy-dependent phenotypes such as dark-induced senescence and starvation-triggered growth inhibition in atg5 plants are not suppressed by depletion of SA (Yoshimoto et al., 2009). Similarly, increased cell death caused by a pathogenic powdery mildew fungus, Golovinomyces cichoracearum, in atg2 plants is not fully suppressed by inactivation of SA signaling (Wang et al., 2011). These suggest that autophagy functions both SA-dependently and -independently. The SA-independent activity of autophagy can be explained by elevated levels of reactive oxygen species (ROS) observed in autophagy mutant plants, which is partly dependent on SA (Lenz et al., 2011; Wang et al., 2011; Yoshimoto et al., 2009).
SA is a key plant defense hormone against biotrophic pathogens that colonize obligately in living plant cells (Koornneef and Pieterse, 2008; Spoel and Dong, 2008). In addition, SA is antagonistic to jasmonic acid (JA) and ethylene, which are required for resistance to necrotrophic pathogens that kill plant cells for colonization (Koornneef and Pieterse, 2008; Spoel and Dong, 2008). Since the SA level is upregulated in autophagy mutant plants, it is simply expected that they are more resistant to biotrophs but hyper-susceptible to necrotrophs. Indeed, it was reported that the growth of necrotrophic fungal pathogens such as Alternaria brassicicola and Botrytis cinerea is markedly enhanced in atg5, atg7, atg10, and atg18a plants (Lai et al., 2011; Lenz et al., 2011). The expression of PDF1.2 (a marker gene of JA signaling) is reduced and not prolonged, and the expression pattern of PR1 (a marker gene of SA signaling) is unchanged. Therefore, the compromised resistance to necrotrophs in autophagy mutant plants by B. cinerea infection (Lai et al., 2011) may due to the suppression of JA responses by elevated SA levels. However, the role of autophagy in defense against biotrophic pathogens remains unclear. While it was reported that the growth of biotrophic bacteria was increased, particularly at earlier time points (Hofius et al., 2009; Patel and Dinesh-Kumar, 2008), it was also reported that bacterial growth was either not significantly changed (Yoshimoto et al., 2009) or even reduced in autophagy mutant plants (Lenz et al., 2011).
Interestingly, SA-dependent HR cell death spread in autophagy mutant plants depends on plant age, because unlimited HR cell death can be observed only in older plants (7- to 8-week grown) not in young plants (4- to 5-week grown) (Yoshimoto et al., 2009). The fact that Hofius et al. (2009) examined 3- to 4-week-old plants may explain why Hofius et al. (2009) failed to see HR cell death spread beyond infection sites. These age-dependent phenotypic variations related to SA levels may also explain the contrasting observation of bacterial growth in autophagy mutant plants. Alternatively, other differences in experimental methods between the studies may be responsible for the apparent discrepancies with regard to the role of autophagy in plant cell death. For instance, a pro-death role of autophagy is evident when only cells at or near the infection sites are used for cell-death analysis (reviewed by Hofius et al., 2011). To summarize, for a particular host-pathogen interaction, a pro-death role of autophagy can, if it exists, be apparent only in a limited region during a specific period of time (possibly early and transiently), while pro-survival and cytoprotective roles of autophagy dominate in other cells for most of the time (especially when cells and tissues are not young).
Atg genes were also tested to determine whether they are implicated in the responses to abiotic stresses. Multiple atg mutants are known to be hypersensitive to salt and drought stresses (Liu et al., 2009; Shin et al., 2009) in addition to oxidative stress (described above). The mechanism for this hypersensitivity is not clear, but ROS may be an important link. ROS is a common mediator of plant responses to nutritional and environmental stresses (Mittler et al., 2004; Shin and Schachtman, 2004). ROS appears to induce autophagy in plant cells (Xiong et al., 2007), as well as in mammalian cells (Scherz-Shouval et al., 2007). Thus, it is possible that nutritional, biotic, and abiotic stresses induce ROS production, which in turn activates autophagy. Conversely, under a presumably non-stressed condition, atg2 and atg5 mutant leaves accumulate a higher level of hydrogen peroxide than wild-type leaves, suggesting that autophagy normally limits ROS (Yoshimoto et al., 2009). ROS accumulation in the mutant may indicate cellular damage due to the lack of the housekeeping role of autophagy (e.g. cleaning up protein aggregates and damaged organelles), and additional abiotic stresses can elevate ROS to a lethal level.
Developmental roles
Accumulation of autophagosomes is a morphological criterion for classifying programmed cell death (PCD) during plant development (van Doorn and Woltering, 2005; van Doorn et al., 2011). However, it is not yet established whether ATG7-dependent autophagy is required for developmental PCD in plants (van Doorn et al., 2011). In animals, Atg7 mutants showed several morphological defects and changes in specific PCD, including neonatal lethality in mice and delayed pupal development in the fruit fly (reviewed by Meléndez and Neufeld, 2008; Mizushima and Levine, 2010). As mentioned earlier, no change in plant architecture was apparent, not only in several Arabidopsis mutants defective in ATG8-PE conjugation but also in the atg13a atg13b double mutants (see Table 1). Leaf senescence is accelerated in all viable atg mutants (see Table 1), refuting an argument that autophagy exclusively executes PCD in senescing leaves. Delayed differentiation of atg5 tracheary elements (Kwon et al., 2010) is the only atg phenotype related to developmental PCD. This finding also suggests that careful examination of atg mutants may reveal subtle developmental roles played by Atg genes.
Autophagy has been proposed to play a role in vacuole biogenesis (Marty, 1999). At least 2 types of vacuole, lytic vacuoles and protein storage vacuoles (PSVs), are present in plant cells and perform lytic and storage functions, respectively. Available data indicate multiple ways to generate plant vacuoles, while the mechanism of vacuole biogenesis remains elusive (reviewed by Zouhar and Rojo, 2009). This is partly due to the lethality of mutants without a functional central vacuole (Isono et al., 2010; Rojo et al., 2001). Thus, the viability of various atg mutants is not consistent with the view that autophagy is essential for vacuole biogenesis. Protoplasts prepared from atg7 contain a central vacuole that is indistinguishable from the wild-type vacuole (Doelling et al., 2002). Nevertheless, more thorough analysis of the vacuole in autophagy-defective mutants may reveal a possible delay of vacuole biogenesis or transient abnormal structures such as small fragmented vacuoles. Devacuolated miniprotoplasts (i.e. protoplasts lacking a central vacuole) in tobacco BY-2 cells can rapidly regenerate, which involves an autophagy-like process that was insensitive to inhibitors of PI 3-kinases (Yano et al., 2007). It will be interesting for various Arabidopsis mutants to be tested for their ability to regenerate vacuoles from miniprotoplasts.
Autophagy is also implicated in the formation of PSVs in monocot seeds. Trafficking of protein bodies containing prolamine to wheat PSVs resembles an autophagy-like process (Levanony et al., 1992). Autophagic vesicles containing seed storage proteins were also detected in the cytoplasm and PSV of maize aleurone cells, although the vesicles do not contain Atg8 proteins (Reyes et al., 2011).
CONCLUSIONS
It is now clear that plants have highly conserved, core Atg proteins that are functionally similar to yeast and metazoan counterparts. Plant genomes also encode Atg8-interacting proteins that are either conserved (e.g. p62/NBR homologue) or plant-specific (e.g. ATI1). Although not mentioned here in detail, the fact that selective autophagy has also been demonstrated in plants (reviewed by Reumann et al., 2010) indicates unexplored functions of plant autophagy. Future research should employ multiple strategies in order to answer outstanding questions with regard to the regulation of autophagy, Atg8-independent autophagy, and the mechanism of selective autophagy.
Since null Arabidopsis mutants for various ATG genes (particularly those encoding the ATG8/ATG12 conjugation pathway) are available, it is often preferable to analyze multiple mutants from more than one ATG locus and exclude nonspecific phenotypes. Nevertheless, more autophagy-defective mutants beyond the ATG8 conjugation pathway are needed to clarify the role of autophagy in plant development and responses to nutritional, biotic, and abiotic stresses. Large-scale analysis of the autophagic network has been successful in finding novel members of the network, and we expect more to come from both metazoan and plant genomes and proteomes in next few years.
Acknowledgments
We apologize to colleagues whose work has not been mentioned because of space limitations. We thank Dr. Richard Vierstra and Kwang Deok Shin for kind advice. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2011-0010683) and by a grant from the Next-Generation BioGreen 21 Program (No. PJ009004), Rural Development Administration, Republic of Korea. This work was also supported by the Research Fund Program of Research Institute for Basic Sciences, Pusan National University, Korea, Project No. RIBS-PNU-2010-0613255, and by Pusan National University Research Grant No. 2010-0613324.
REFERENCES
- Avin-Wittenberg T., Honig A., Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma. 2012;249:285–299. doi: 10.1007/s00709-011-0296-z. [DOI] [PubMed] [Google Scholar]
- Bassham D.C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L.J., Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy. 2006;2:2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemann B.O., Orvedahl A., Cheng T., Ram R.R., Ou Y.H., Formstecher E., Maiti M., Hazelett C.C., Wauson E.M., Balakireva M., et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophago-some assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T. See how I eat my greens - autophagy in plant cells. J. Plant Biol. 2011;54:339–350. [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149:220–234. doi: 10.1104/pp.108.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- Devarenne T.P. The plant cell death suppressor Adi3 interacts with the autophagic protein Atg8h. Biochem. Biophys. Res. Commun. 2011;412:699–703. doi: 10.1016/j.bbrc.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Mal-erod L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshimoto K., Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk S.F., Wang Q.J., Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J., Strom A.L., Kwinter D.M., Kilty R., Zhang J., Shi P., Fu W., Wooten M.W., Zhu H. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J. Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. Rapid combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 2011;49:557–576. doi: 10.1146/annurev-phyto-072910-095333. [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Hofius D., Munch D., Bressendorff S., Mundy J., Petersen M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011;18:1257–1262. doi: 10.1038/cdd.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig A., Avin-Wittenberg T., Ufaz S., Galili G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell. 2012;24:288–303. doi: 10.1105/tpc.111.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. AtATG genes, homologs of yeast autophagy genes are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–1652. doi: 10.1093/pcp/pcl031. [DOI] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.D., Geldner N., Chory J., Schwechheimer C. The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22:1826–1837. doi: 10.1105/tpc.110.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M., Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjorkoy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M. Cross talk in defense signaling. Plant Physiol. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Mansilla A., Menzies F.M., Rubinsztein D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Menzies F.M., Rubinsztein D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Jung J.H., Yoshimoto K., Shirasu K., Park O.K. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010;64:151–164. doi: 10.1111/j.1365-313X.2010.04315.x. [DOI] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66:953–968. doi: 10.1111/j.1365-313X.2011.04553.x. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim E.S., Choi Y., Hwang I., Staiger C.J., Chung Y.Y., Lee Y. The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 2008;147:1886–1897. doi: 10.1104/pp.108.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H.D., Haller E., Melzer E., Kober K., Wurster K., Stahl M., Bassham D.C., Vierstra R.D., Parker J.E., Bautor J., et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011;66:818–830. doi: 10.1111/j.1365-313X.2011.04546.x. [DOI] [PubMed] [Google Scholar]
- Levanony H., Rubin R., Altschuler Y., Galili G. Evidence for a novel route of wheat storage proteins to vacuoles. J. Cell Biol. 1992;119:1117–1128. doi: 10.1083/jcb.119.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski M.M., Hoffman G., Ng A., Zhou W., Py B.F., Hsu E., Liu X., Eisenberg J., Liu J., Blenis J., et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates program-med cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- Loomis W.F., Behrens M.M., Williams M.E., Anjard C. Pregnenolone sulfate and cortisol induce secretion of acyl-CoAbinding protein and its conversion into endo-zepines from astrocytes. J. Biol. Chem. 2010;285:21359–21365. doi: 10.1074/jbc.M110.105858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Subramani S. Autophagy: a broad role in unconventional protein secretion? Trends Cell Biol. 2011;21:67–73. doi: 10.1016/j.tcb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions and autophagosome formation. J. Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F. Plant vacuoles. Plant Cell. 1999;11:587–600. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez A., Neufeld T.P. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitou G., Budak H., Gozuacik D. Techniques to study autophagy in plants. Int. J. Plant Genomics. 2009;2009:451357. doi: 10.1155/2009/451357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I., Murphy A. Validating the location of fluorescent protein fusions in the endomembrane system. Plant Cell. 2009;21:1632–1636. doi: 10.1105/tpc.109.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyasu Y., Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R., Jr., Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.S., Martínez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 2005;41:831–844. doi: 10.1111/j.1365-313X.2005.02346.x. [DOI] [PubMed] [Google Scholar]
- Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neuro-degeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S.P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.J., Gu H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007;17:249–263. doi: 10.1038/cr.2007.7. [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Voitsekhovskaja O., Lillo C. From signal transduction to autophagy of plant cell organelles: lessons from yeast and mammals and plant-specific features. Protoplasma. 2010;247:233–256. doi: 10.1007/s00709-010-0190-0. [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Chung T., Holding D., Jung R., Vierstra R., Otegui M.S. Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell. 2011;23:769–784. doi: 10.1105/tpc.110.082156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Gillmor C.S., Kovaleva V., Somerville C.R., Raikhel N.V. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell. 2001;1:303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- Rose T.L., Bonneau L., Der C., Marty-Mazars D., Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell. 2006;98:53–67. doi: 10.1042/BC20040516. [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Stenmark H. How do ESCRT proteins control autophagy? J. Cell Sci. 2009;122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R., Schachtman D.P. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.H., Yoshimoto K., Ohsumi Y., Jeon J.S., An G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells. 2009;27:67–74. doi: 10.1007/s10059-009-0006-2. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. Making sense of hormone cross-talk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. doi: 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka C., Inoue Y., Higuchi T., Hillmer S., Robinson D.G., Moriyasu Y. Autophagy in tobacco BY-2 cells cultured under sucrose starvation conditions isolation of the autolysosome and its characterization. Plant Cell Physiol. 2011;52:2074–2087. doi: 10.1093/pcp/pcr137. [DOI] [PubMed] [Google Scholar]
- Takeda K., Yoshida T., Kikuchi S., Nagao K., Kokubu A., Pluskal T., Villar-Briones A., Nakamura T., Yanagida M. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc. Natl. Acad. Sci. USA. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N.J., Kershaw M.J. The emerging role of autophagy in plant pathogen attack and host defence. Curr. Opin. Plant Biol. 2009;12:444–450. doi: 10.1016/j.pbi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Vierstra R.D. Autophagic recycling lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., Woltering E.J. Many ways to exit? Cell death categories in plants. Trends Plant Sci. 2005;10:117–122. doi: 10.1016/j.tplants.2005.01.006. [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., Beers E.P., Dangl J.L., Franklin-Tong V.E., Gallois P., Hara-Nishimura I., Jones A.M., Kawai-Yamada M., Lam E., Mundy J., et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011;18:1241–1246. doi: 10.1038/cdd.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi: 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ding Y., Wang J., Hillmer S., Miao Y., Lo S.W., Wang X., Robinson D.G., Jiang L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell. 2010;22:4009–4030. doi: 10.1105/tpc.110.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011;68:74–87. doi: 10.1111/j.1365-313X.2011.04669.x. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J. Cell Sci. 2011;124:2692–2701. doi: 10.1242/jcs.081232. [DOI] [PubMed] [Google Scholar]
- Winter V., Hauser M.T. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006;11:115–123. doi: 10.1016/j.tplants.2006.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q.F., Chan S.W., Zheng S.X., Ma J., Wang M., Welti R., Chye M.L. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22:1463–1482. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42:535–546. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Mammalian autophagy core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Hattori M., Moriyasu Y. A novel type of autophagy occurs together with vacuole genesis in miniprotoplasts prepared from tobacco culture cells. Autophagy. 2007;3:215–221. doi: 10.4161/auto.3739. [DOI] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Kochl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. Auto-phagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawr-zynska A., Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selec-tive autophagy cargo receptors. Autophagy. 2011;7:1145–1158. doi: 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J., Rojo E. Plant vacuoles: where did they come from and where are they heading? Curr. Opin. Plant Biol. 2009;12:677–684. doi: 10.1016/j.pbi.2009.08.004. [DOI] [PubMed] [Google Scholar]