Abstract
We previously reported that one of the brassinosteroid-insensitive mutants, bri1-9, showed increased cold tolerance compared with both wild type and BRI1-overexpressing transgenic plants, despite its severe growth retardation. This increased tolerance in bri1-9 resulted from the constitutively high expression of stress-inducible genes under normal conditions. In this report, we focused on the genes encoding class III plant peroxidases (AtPrxs) because we found that, compared with wild type, bri1-9 plants contain higher levels of reactive oxygen species (ROS) that are not involved with the activation of NADPH oxidase and show an increased level of expression of a subset of genes encoding class III plant peroxidases. Treatment with a peroxidase inhibitor, salicylhydroxamic acid (SHAM), led to the reduction of cold resistance in bri1-9. Among 73 genes that encode AtPrxs in Arabidopsis, we selected four (AtPrx1, AtPrx22, AtPrx39, and AtPrx69) for further functional analyses in response to cold temperatures. T-DNA insertional knockout mutants showed increased sensitivity to cold stress as measured by leaf damage and ion leakage. In contrast, the overexpression of AtPrx22, AtPrx39, and AtPrx69 increased cold tolerance in the BRI1-GFP plants. Taken together, these results indicate that the appropriate expression of a particular subset of AtPrx genes and the resulting higher levels of ROS production are required for the cold tolerance.
Keywords: brassinosteroid-insensitive 1, cold stress, plant-specific class III peroxidases, stress response
INTRODUCTION
As an alternative to physical escape, plants adjust to extreme environments via metabolic and developmental modifications. In these instances, plants may utilize various signaling events that link the perception of environmental stress to the modulation of stress-inducible genes. Environmental stresses are often complex; plants typically experience multiple environmental stresses simultaneously, such as cold combined with high light or salinity combined with water deficiency. Therefore, while a subset of stress-inducible genes show specific expression in response to a certain stress, others show similar patterns of expression in response to different types of stresses, resulting in common downstream events involving signaling cascades that lead to the increased production or accumulation of ROS or the activation of mitogen-activated protein kinases.
ROS, including the hydroperoxyl radical, superoxide anion, hydrogen peroxide (H2O2), and the hydroxyl radical, are unavoidably generated during any metabolic process that disrupts the electron transport chain (Apel and Hert, 2004; Møller et al., 2007). Because ROS are highly reactive oxidants of cellular components, including amino acids, lipids, and DNA, and cause widespread damage in plants and animals, they are considered to be toxic (Møller et al., 2007). However, many reports indicate that living organisms seem to react differently to ROS in a dose-dependent manner. A redox-dependent signaling pathway regulates cyclinD1 expression to control the induction of cell division in animal systems (Burch and Heintz, 2005). Low levels of H2O2 have also been reported to regulate cell cycle progression (Reichheld et al., 1999) or to function during the onset of secondary cell wall differentiation in plants (Potikha et al., 1999; Schopfer et al., 2002). In addition, numerous reports have provided evidence that oxidative ROS bursts are common phenomena under biotic and abiotic stresses. The slight alteration in intracellular ROS levels that occurs following exposure to various stresses often triggers the signaling cascade leading to increased tolerance to a specific stress. Plants overexpressing RCI3, which is a cold-inducible gene encoding an active cationic peroxidase, showed increased tolerances to dehydration and salt (Llorente et al., 2002). Genes that are induced by H2O2, such as OXI1 and AtNDPK2, have been shown to play important roles in the activation of MPK3 and MPK6 (Moon et al., 2003; Rentel et al., 2004), which act as crucial links in the MAPK cascade during ROS signaling (Teige et al, 2004; Xing et al., 2008; Yuasa et al., 2001). A transgenic poplar plant overexpressing AtNDPK2 under the control of an oxidative stress-inducible promoter was recently reported to have a higher tolerance to oxidative stress conditions due to the enhanced activities of various antioxidant enzymes (Kim et al., 2011). Therefore, the fine tuning of the ROS steady-state levels between generations and the scavenging of these ROS is critical for multiple cellular processes. Plants have developed efficient antioxidant systems to handle the excessive accumulation of ROS. The formation of the most toxic ROS, the hydroxyl radical is prevented by the action of superoxide dismutase, which converts superoxide to hydrogen peroxide. Although it is less toxic than superoxide, the resulting hydrogen peroxide (H2O2), a relatively long-lived ROS, should also be eliminated (Mittler et al., 2004; Møller and Sweetlove, 2010).
Peroxidases are a large family of enzymes that play critical roles in the detoxification of H2O2 (Tognolli et al., 2002; Valério et al., 2004). There are three different classes of peroxidases containing a heme group (Edward et al., 1993; Smulevich et al., 2006). The intracellular class I consists of ascorbate peroxidases, cytochrome c peroxidases, and catalase-peroxidase, which are found in intracellular organelles, including the chloroplast, peroxisome, and mitochondria. Because class I peroxidases are found in most living organisms, the other two classes are thought to have originated from them. The class II peroxidases, including manganese peroxidase, lignin peroxidase, and versatile peroxidase are mainly found in fungi. All land plants possess multigene families of class III peroxidases that are usually secreted into the apoplastic space, where they catalyze the reduction of hydrogen peroxide with various electron donors, including phenolic compounds, lignin precursors, and various secondary metabolites (Cosio and Dunand, 2009). In Arabidopsis, 73 genes encoding class III peroxidases have been reported. Most of these genes (66 out of 73) are expressed in 6-week-old plants (Valério et al., 2004), while the others are likely to be expressed in specific stress conditions or developmental stages (Irshad et al., 2008; Swanson et al., 2005).
Brassinosteroids (BR) are well-known plant hormones that promote various aspects of plant growth and development. The binding of BR to the plasma membrane-localized receptor brassinosteroid-Insensitive 1 (BRI1) triggers the BR signaling cascade. Therefore, mutant alleles that are defective in BRI1 display severely growth-inhibited dwarf phenotypes, whereas BRI1-overexpressing transgenic plants appear as constitutive BR-treated phenotypes with longer and narrower growth in their leaves and petioles (Kinoshita et al., 2005; Li and Chory, 1997). In addition to their roles in plant growth, BRs are thought to confer tolerance to a variety of environmental stresses. Pretreatment with 24-epibrassinolide (EBR) resulted in increased heat shock protein synthesis following heat stress (Dhaubhadel et al., 2002), the prevention of chlorophyll loss in cold-stressed plants (He et al., 1991), and the enhancement of photosynthesis in chilled plants (Yu et al., 2002). However, the mechanisms underlying the regulation of stress tolerance by exogenous treatment with EBR have not been elucidated.
We previously reported that although overall growth was greatly inhibited in bri1-9 mutants, the leaves were not damaged by cold stress and the plant showed much higher cold tolerances than wild type. In contrast, BRI1-overexpressing transgenic plants that had been transformed with a BRI1-GFP construct showed more elongated petioles and leaves than the wild type plants and were the least tolerant of the same stress conditions. We found that this phenomenon was partly attributable to the constitutive activation of not only the stress-inducible genes but also the genes encoding the transcription factors that are responsible for stress-inducible gene expression in bri1-9 even in normal conditions (Kim et al., 2010). In this report, we further focused on a group of genes, specifically peroxidases that are involved in ROS metabolism, and investigated their functional relationships with cold stress in bri1-9 and BRI1-GFP plants compared with wild type.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana Columbia-0 (Col-0) was used as the wild type, and bri1-9 mutant and BRI1-GFP transgenic plants from the same ecotype background were also used. T-DNA insertional AtPrx knock-out lines for this study (atprx1: SALK_ 060130, atprx22: SALK_144487, atprx39: SALK_096147, and atprx69: SALK_137991) were obtained from the ABRC stock center. For the observation of gross morphologies, seeds were sown directly onto Sunshine #5 soil. Otherwise, they were sterilized with 75% ethanol containing 0.05% Tween-20 for 15 min, and then washed twice with 95% ethanol, and germinated in a 1/2 MS (Duchefa) plate containing 0.8% PhytoAgar. All plants were grown at 22°C under long-day conditions (16 h L/8 h D).
Cold treatment, leaf damage, and ion leakage measurements
The seedlings were grown on plates containing 1/2 MS for 10 days and then exposed to a temperature of 4°C in a growth chamber. To investigate the effects of SHAM treatment, seedlings were grown on 1/2 MS plates containing 100 μM SHAM (Sigma-Aldrich) for 10 days. To measure electrolyte leakage, we first measured the electrical conductivities (EC1) of the 10 seedlings from each line that had been treated with or without cold after incubating them in 10 ml distilled deionized water at room temperature for 2 h. Using a conductivity meter (SevenGo proTM conductivity meter SG3, Mettler Toledo), electrical conductivities (EC2) were measured again of the samples after autoclaving to release all electrolytes and cooling to room temperature. The ratio of initial electrolyte leakage to 100% electrolyte leakage was calculated using the following formula: [(EC1/EC2) × 100] (Kim et al., 2010). To determine the ratio of cold-damaged leaves, cold-treated plates were transferred back to normal conditions and grown for an additional 2 days. The numbers of total leaves and damaged leaves were counted for all seedlings, and the proportion was calculated. Experiments were repeated three times and showed similar results.
Determination of H2O2 concentrations
To visualize H2O2 concentrations, the seedlings grown for 10 days under normal conditions and then treated with or without cold were incubated in a staining solution containing 1 mg/ml diaminobenzidine (DAB, Sigma D-8001) for 5 to 8 h at 25°C. The stained seedlings were boiled in 96% EtOH for 10 min, and cooled to room temperature. To measure the H2O2 level quantitatively, we used the Amplex Red Peroxidase/H2O2 Assay Kit (Invitrogen). To serve as H2O2-containing biological sources, crude extracts were obtained from the 10-day-old seedlings in extraction buffer [50 mM sodium phosphate pH 7.4, 100 mM NaCl, 1 mM EDTA, protease inhibitor cocktail (Roche)]. Reaction mixtures containing horseradish peroxidase (HRP, 0.2 U/ ml) were made according to the manufacturer’s protocol. After incubation for 30 min. at room temperature, absorbances were detected at a wavelength of 571 nm using a spectrophotometer (Bekman Coulter DU730).
Determination of DNA damage
Total DNA was isolated from 10-day-old seedlings treated with or without cold for 6 h using the DNeasy Plant Mini Kit (Qiagen). The formation of 8-hydroxydeoxyguanosine (8-OHdG) was detected by an anti-8-OHdG monoclonal antibody followed by an HRP-conjugated secondary antibody using the OxiSelect™ Oxidative DNA Damage ELISA Kit (CELL BIOLABS). Absorbance was measured at a wavelength of 450 nm as per the manufacturer’s recommendations.
Semi-quantitative RT-PCR analyses
The RNA samples purified from the seedlings grown for 10 days were treated with RNase-free RQ1 DNase (Promega) and used for first-strand cDNA synthesis with the SuperscriptIII-MMLV reverse transcriptase (Invitrogen) using oligo d(T15) as the primer. Second-strand synthesis was performed using the same aliquot of first-strand cDNA as the template. The expression of each gene was normalized to β-Tubulin. All of the primer sequences that were used in this study are indicated in Supplementary Table 1.
Generation of transgenic plants
To generate the 35S-CaMV constitutive promoter-driven overexpression constructs of AtPrx1, AtPrx22, and AtPrx69, open-reading frames of each AtPrx were amplified by PCR using wild-type genomic DNA as a template and inserted into pCHF1 vectors. All of the resulting constructs were transformed into the BRI1-GFP transgenic plants by floral dipping methods through the transformation of Agrobacterium tumefaciens (GV3101). Twenty to thirty transgenic plants transformed with each AtPrx constructs were selected using gentamycin (50 μg/ml) in the T1 generation. We then selected the transgenic plants containing a single-copy of the transgene in the T2 generation. Three, four, and five independent homozygous lines were obtained from AtPrx1OE, AtPrx22OE and AtPrx69OE transgenic plants, respectively, in the T3 generation.
Microarray analysis
Microarray construction was previously performed as described with details in Kim et al. (2010) using the Arabidopsis thaliana ESTs obtained from the KAZUSA DNA Research Institute (Japan, http://www.kazusa.or.jp). Total RNA from 10-day-old Arabidopsis seedlings was labeled using a 3 DNA Array 50 kit (Genisphere, USA). Following the hybridization, arrays were scanned using a ScanArray Lite (GSI Lumonics, USA) in both Cy3 and Cy5 channels. Four replicates were analyzed.
RESULTS
Cold-resistant bri1-9 plants contain higher levels of ROS
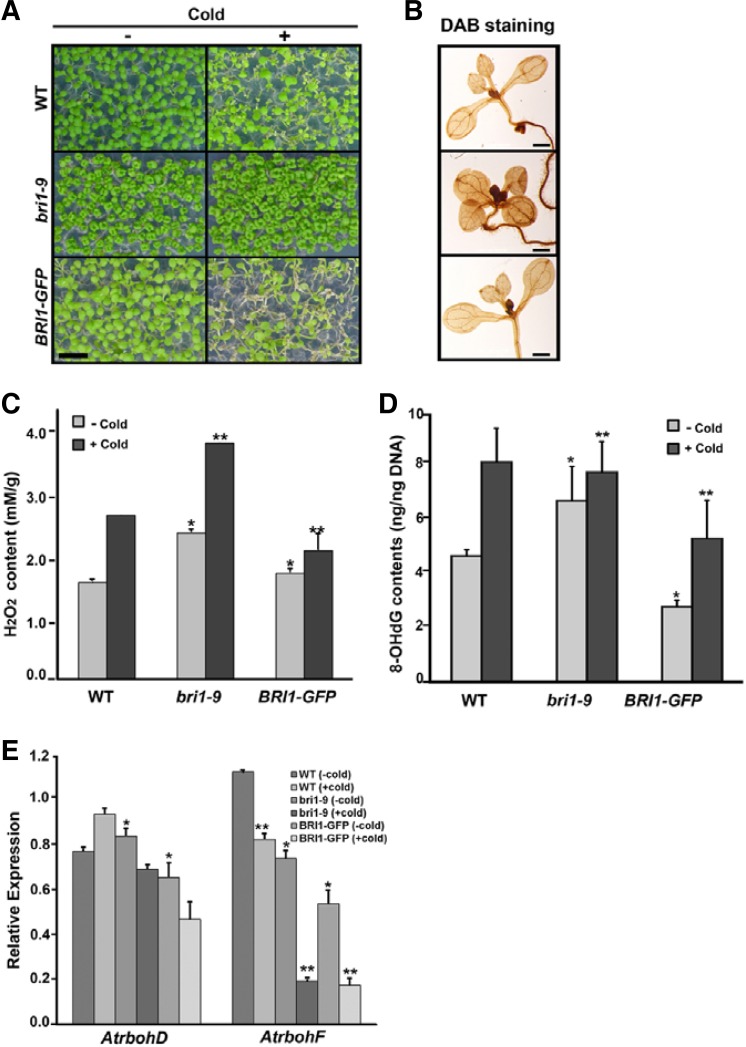
The opposite responses to low temperature in bri1-9 and BRI1-GFP plants were always observed in our conditions, with higher tolerance in bri1-9 and reduced tolerance in BRI1-GFP compared with wild type (Fig. 1A), as we previously showed (Kim et al., 2010). We predicted that the increases in leaf damage and ion leakage that were observed in BRI1-GFP plants following the cold treatment were indicators of unbalanced ROS production and subsequent oxidative stress. This hypothesis was supported by DAB-staining analyses, in which the bri1-9 mutant showed darker brownish-staining patterns than the wild type under normal conditions, indicating that higher levels of ROS were accumulated (Fig. 1B). Therefore, we directly measured the levels of hydrogen peroxide and observed that bri1-9 plants contained higher concentrations of endogenous H2O2 than the wild type and BRI1-GFP plants and that cold treatment caused increases in the H2O2 concentrations of 40% in the wild type and bri1-9 plants and 10% in the BRI1-GFP plants (Fig. 1C). The formation of 8-hydroxydeoxyguanosine (8-OHdG), a ubiquitous marker of oxidative damage of DNA bases (Møller et al., 2007), was detected more frequently in bri1-9 plants than in wild type plants at basal conditions. Cold treatment induced the increased formation of 8-OHdG to a lesser degree in bri1-9 plants than in wild type (Fig. 1D). These results indicated that bri1-9 plants contain higher levels of ROS in normal and cold-stressed conditions.
Fig. 1.
Brassinosteroid-signaling mutant contains higher ROS levels. (A) Cold-stressed phenotypes of bri1-9 and BRI1-GFP seedlings Pictures were taken 48 h after cold treatment. Scale bars denote 5 mm. (B) DAB-staining pattern showing accumulation of ROS in seedlings. Bar indicates 1 mm. Hydrogen peroxide concentrations (C) and formation of 8-OHdG (D) were determined from whole seedlings that were treated with or without cold. Bar denotes standard error. ANOVA statistical analyses were performed, and significant differences are indicated by P values [*P ≤ 0.001 and **P ≤ 0.0001 in (C); and *P ≤ 0.001 and **P ≤ 0.05 in (D)]. (E) The expression of AtrbohD and AtrbohF genes in bri1-9 and BRI1-GFP seedlings compared with that of wild type with or without cold (*P ≤ 0.05, **P ≤ 0.001).
To examine whether the higher ROS production in bri1-9 was occurred via the NADPH oxidase complex, we analyzed the expression of AtrbohD and AtrbohF genes, which encode catalytic subunits of NADPH oxidase (Torres et al., 2002). We observed that the expression levels of AtrbohD and AtrbohF in bri1-9 were similar to those of wild type and BRI1-GFP plants in normal conditions. In cold-stressed, the expression level of AtrbohD was not changed in bri1-9 and BRI1-GFP plants, though it was slightly increased in wild type. The expression level of AtrbohF was reduced in bri1-9 and BRI1-GFP plants compared with that of wild type (Fig. 1E). This result implied that higher levels of ROS in bri1-9 were produced by sources other than the NADPH oxidase complex. ROS are known to act as versatile signaling molecules that are generated under various stress conditions and affect plant growth and development in a dose-dependent manner. Therefore, our results suggest two possibilities, namely that higher basal levels of ROS in bri1-9 may be beneficial for coping with cold stress by inducing subsequent signaling or that bri1-9 has unique mechanisms to rapidly detoxify ROS.
Genes encoding class III plant peroxidases are more highly expressed in bri1-9 plants
To investigate the relationship between the increased accumulation of ROS and the higher cold tolerance of bri1-9, we analyzed the previous bri1-9 and BRI1-GFP transgenic plants microarray data and compared the data with those of wild type to evaluate the expression of genes, including those encoding peroxidase, catalase, superoxide dismutase, and thioredoxin. In this filtering process, we disregarded the two-fold cut off criterion, because more than 80% of BR-responsive genes are known to be transcriptionally regulated by less than two-fold upon BL treatment (Vert et al., 2005). Most of the genes related to ROS-metabolism showed less than two-fold fluctuation in bri1-9 and BRI1-GFP plants compared with those of wild type. Among 12,544 cDNA clones from the Arabidopsis library (Kim et al., 2010), 129 spots containing ROS-related cDNAs were differentially regulated in the bri1-9 and BRI1-GFP transgenic plants compared with the wild type. There were many different spots containing the same cDNA that showed similar trends in gene expression, indicating the consistency of the microarray analysis. From this analysis, we could summarize thirty-nine genes encoding peroxidases, three superoxide dismutase genes, nine glutathione S-transferase, and eighteen thioredoxin genes. The thirty-nine genes encoding peroxidases could be divided into three groups: ascorbate peroxidase (five genes), glutathione peroxidase (four genes), and peroxidases that belonged to the secreted class III peroxidases (thirty genes) (Table 1). Twentyeight out of the thirty genes encoding the class III peroxidases were up-regulated in bri1-9 compared with wild type, and interestingly, more than half of these genes (15 genes) were oppositely down-regulated in the BRI1-GFP transgenic plants.
Table 1.
Differential expression of genes encoding peroxidases in bri1-9 and BRI1-GFP transgenic plants compared with those of wild type plants
| Spot no | AGI | Gene description | log2 (bri1-9/WT)a | log2 (BRI1-GFP/WT)a |
|---|---|---|---|---|
| 5977 | At5g64120 | Peroxidase, AtPrx71 | 1.005 | 0.330 |
| 555 | At5g39580 | Peroxidase, AtPrx62 | 0.888 | 0.327 |
| 5165 | At5g67400 | Peroxidase, AtPrx63 | 0.793 | −0.419 |
| 8244 | At4g11290 | Peroxidase, AtPrx39 | 0.742 | −0.133 |
| 7509 | At4g37520 | Peroxidase, AtPrx50 | 0.725 | 0.213 |
| 341 | At3g49960 | Peroxidase, AtPrx35 | 0.723 | −0.636 |
| 8273 | At1g05240 | Peroxidase, AtPrx1 | 0.693 | −0.612 |
| 7038 | At3g32980 | Peroxidase, AtPrx32 | 0.661 | −0.123 |
| 4405 | At4g26010 | Peroxidase, AtPrx44 | 0.655 | −0.401 |
| 377 | At2g38390 | Peroxidase, AtPrx23 | 0.643 | −0.123 |
| 10701 | At4g08770 | Peroxidase, AtPrx37 | 0.625 | −0.154 |
| 1252 | At4g30170 | Peroxidase, AtPrx45 | 0.618 | 0.104 |
| 4289 | At2g38380 | Peroxidase, AtPrx22 | 0.606 | −0.033 |
| 1240 | At5g64100 | Peroxidase, AtPrx69 | 0.585 | 0.039 |
| 6267 | At3g49110 | Peroxidase, AtPrx33 | 0.558 | 0.433 |
| 7457 | At5g17820 | Peroxidase, AtPrx57 | 0.494 | −0.273 |
| 2253 | At3g49120 | Peroxidase, AtPrx34 | 0.463 | 0.293 |
| 6384 | At1g05260 | Peroxidase, AtPrx3 | 0.346 | −0.325 |
| 4258 | At1g71695 | Peroxidase, AtPx12 | 0.342 | 0.172 |
| 5111 | At4g21960 | Peroxidase, AtPrx42 | 0.237 | −0.095 |
| 10603 | At5g42180 | Peroxidase, AtPrx64 | 0.193 | −0.094 |
| 1201 | At3g21770 | Peroxidase, AtPrx30 | 0.190 | −0.008 |
| 12298 | At2g37130 | Putative peroxidase | 0.170 | −0.013 |
| 1259 | At5g66390 | Peroxidase, AtPrx72 | 0.163 | 0.230 |
| 2039 | At2g18980 | Peroxidase, AtPrx16 | 0.137 | 0.086 |
| 5897 | At5g06720 | Peroxidase, AtPrx51 | 0.090 | 0.085 |
| 3796 | At5g40150 | Peroxidase, AtPrx63 | 0.060 | 0.109 |
| 10731 | At5g06730 | Peroxidase, AtPrx55 | 0.014 | 0.089 |
| 5417 | At5g51890 | Peroxidase, AtPrx66 | −0.191 | 0.257 |
| 8267 | At3g28200 | Peroxidase, AtPrx31 | −0.223 | −0.179 |
| 523 | At1g07890 | L-ascorbate peroxidase, APX1 | 0.030 | 0.032 |
| 9998 | At4g08390 | Stromal ascorbate peroxidase | −0.016 | −0.120 |
| 1877 | At4g32320 | L-ascorbate peroxidase-like protein | −0.127 | 0.099 |
| 10903 | At1g77490 | Thylakoid-bound ascorbate peroxidase | −0.210 | 0.165 |
| 8225 | At4g35000 | L-ascorbate peroxidase | −0.258 | 0.073 |
| 10320 | At1g63460 | Glutathione peroxidase, GPX8 | 0.397 | 0.044 |
| 8427 | At2g43350 | Putative glutathione peroxidase | 0.381 | 0.149 |
| 2292 | At2g25080 | Glutathione peroxidase, GPX1 | −0.023 | 0.063 |
| 11897 | At2g31570 | Putative glutathione peroxidase | −0.098 | −0.153 |
Average fold ratio of replicated experiments
Peroxidase inhibitor, SHAM, reduced cold resistance of bri1-9 mutant
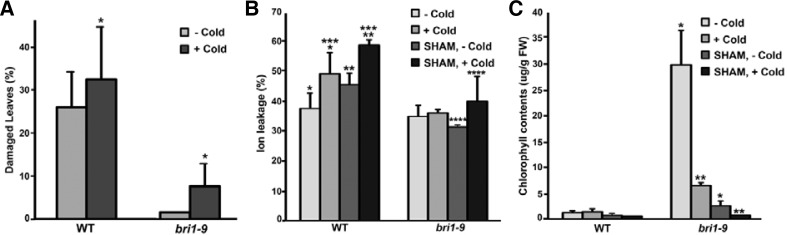
If the appropriate expression of a particular subset of AtPrx genes and the resulting higher levels of ROS production are pre-requisites for the resistance to cold phenotype, we predicted that the cold response would be altered by the inactivation of peroxidases. We used a salicylhydroxamic acid (SHAM), a pharmacological inhibitor of peroxidase (Brouwer et al., 1986), to examine whether the cold response would be altered by the inactivation of peroxidases. We first grew the wild type and bri1-9 on SHAM-containing 1/2 MS media for 10 days. Following cold treatment, we observed more damaged leaves in the SHAM-treated seedlings. Even in the bri1-9, we clearly observed the bleached whitish leaves in the SHAM-and cold-treated seedlings (Fig. 2A). When combined with cold, treatment with SHAM also enhanced cold-induced ion leakage both in wild type and bri1-9 (Fig. 2B). In addition, we observed pale green leaves, particularly in bri1-9, during the growth period with SHAM treatment. Because of this observation, we measured seedling chlorophyll contents. The chlorophyll content of bri1-9 under normal condition was much higher than that of wild type. Treatment with either cold or SHAM greatly reduced the chlorophyll content, and combined treatment led to the additive reduction of chlorophyll in bri1-9, to levels close to those of wild type subjected to the same treatment (Fig. 2C). These results suggest that bri1-9 plants lose their cold tolerance via the inactivation of peroxidase activity.
Fig. 2.
Cold-resistant phenotypes of bri1-9 mutants were reduced by treatment with SHAM, a peroxidase inhibitor. (A) Effects of SHAM combined with cold on the ratio of damaged leaves in bri1-9 compared with that of the wild type. The unpaired t-test was performed for the corresponding line without cold (*P = 0.0034). (B, C) Measurement of ion leakage and chlorophyll content from the bri1-9 and wild type seedlings treated with cold, SHAM, or both. Bar denotes the standard error. The unpaired t-test was performed and obtained two-tailed P values as follows: *P = 0.02, **P = 0.008, ***P = 0.22, ****P = 0.01.
Overexpression of certain AtPrx genes increases cold tolerance in BRI1-GFP plants
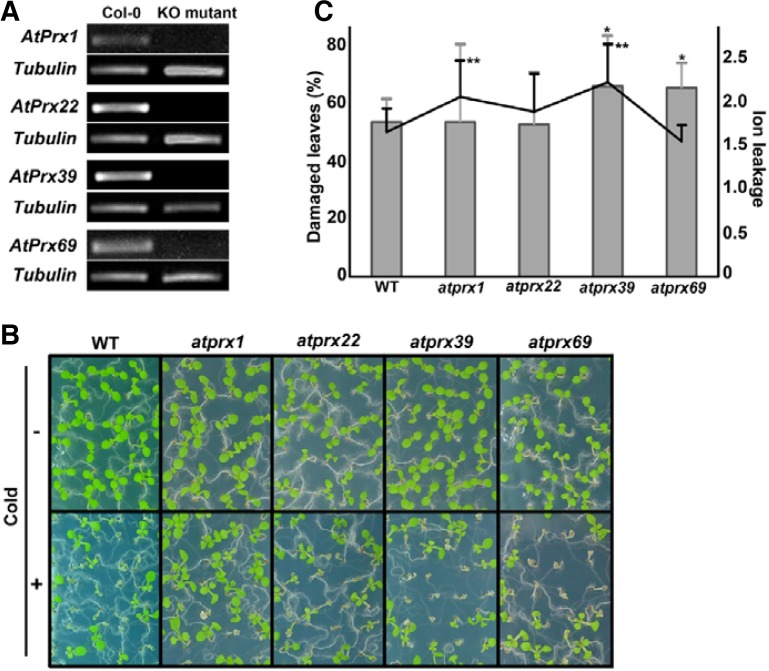
To identify the AtPrx genes involved in the response to cold, we screened the specific AtPrx genes shown in Table 1 based on the following categories: opposite expression patterns; higher expression levels in bri1-9 and lower expression in BRI1-GFP plants compared with wild type; and more than 0.5 difference of two-fold cut-off value. We also considered the availability of T-DNA insertional knock-out lines for each gene. We finally chose four AtPrx genes (AtPrx1, AtPrx22, AtPrx39, and AtPrx69). First, to verify the results from the microarray analysis, we performed RT-PCR using RNA from wild type, bri1-9 and BRI1-GFP plants. Consistent with the microarray data, the expression levels of AtPrx22, AtPrx39 and AtPrx69 in bri1-9 plants were higher than those in wild type plants. The expressions levels of these AtPrxs in BRI1-GFP plants were slightly reduced compared with those in the wild type. However, AtPrx1 showed an opposite expression pattern in the microarray and RT-PCR analyses (Supplementary Fig. 1). We obtained the homozygous mutant of each gene (Fig. 3A). When investigating the gross morphologies of each knock-out AtPrx mutant, we could not detect significant phenotypic changes in the aerial parts during the entire growth period (Supplementary Fig. 2A). The hydrogen peroxide concentrations, which were measured from 10-day-old whole seedlings of each atprx mutant, showed that there were no significant differences in the H2O2 levels compared with those of wild type (Supplementary Fig. 2B). These results are likely attributable to the functional redundancy of the AtPrx genes; here the lack of any single AtPrx gene did not seem to affect normal developmental processes. However, following cold treatment for 24 h, a greater number of leaves in the knock-out mutants remained bleached and wilted. The damaged leaf ratios and ion leakages in the mutants were greater in the knock-out plants compared with the wild type (Figs. 3B and 3C).
Fig. 3.
Knockout AtPrx mutants showed increased susceptibilities to cold stress compared with wild type. (A) Confirmation of lack of transcription of AtPrx1, AtPrx22, AtPrx39, and AtPrx69 in each corresponding mutant. (B) Cold-treated AtPrx seedlings displayed increasingly damaged phenotypes after a 2-day recovery period. (C) Quantitative analysis of cold response as determined by the ratio of damaged leaves following cold treatment (bar graph, right) and electrolyte leakage (line graph, left). ANOVA statistical analyses were performed, and significant differences are indicated by P values (*P ≤ 0.6 for damaged leaves, **P ≤ 0.3 for ion leakage). Bar denotes standard deviation.
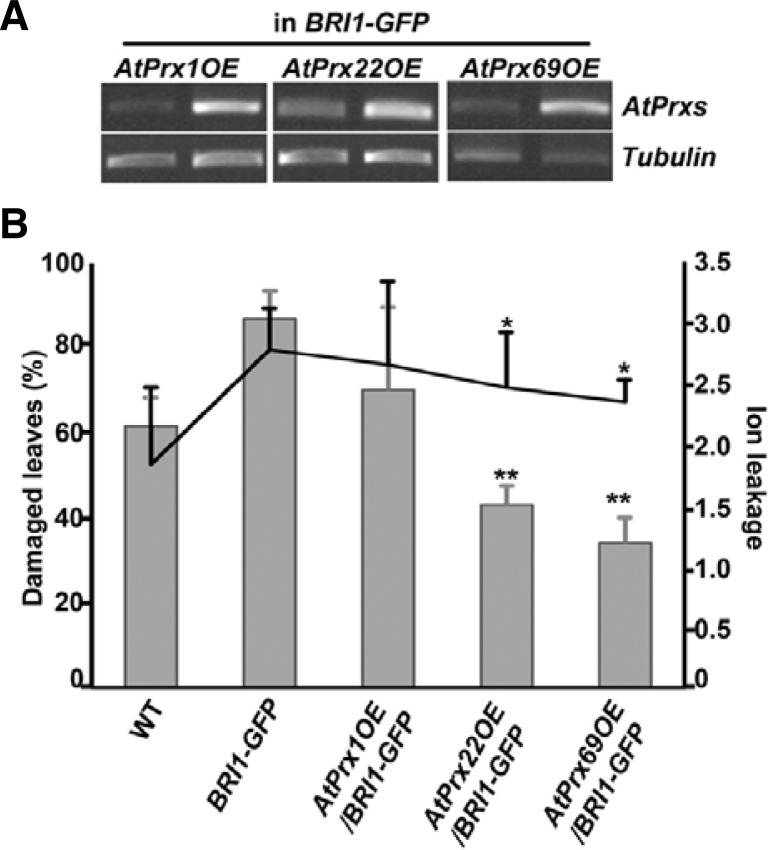
We further examined whether the constitutive overexpression of these genes would increase the tolerances of the least cold-tolerant BRI1-GFP transgenic plants. We cloned full-length AtPrx1, AtPrx22, and AtPrx69 cDNAs into binary vectors driven by the 35S-CaMV constitutive promoter and generated transgenic BRI-GFP to overexpress them (Fig. 4A). None of these AtPrx genes affected the overall growth of the BRI1-GFP plants when they were expressed at high levels. We then performed the DAB staining analysis to determine whether the overexpression of certain AtPrxs would cause accumulation of ROS in the transgenic plants. Under normal conditions, the intensity of DAB-staining in the transgenic seedlings was not significantly different from that of un-transformed BRI1-GFP (Supplementary Fig. 3). To further explore the cold response, we exposed BRI1-GFP plants overexpressing AtPrx genes to cold and monitored the presence of leaf damage and ion leakage (Fig. 4B). The BRI1-GFP plants that had been transformed with AtPrx22 and AtPrx69 showed clear reductions in leaf damage. The overexpression of AtPrx genes in BRI1-GFP also reduced ion leakage to levels that were comparable to those of the wild type. These results suggest that cold sensitive BRI1-GFP plants acquired the capacity to cope with cold stress due to the overexpression of subsets of AtPrx genes. The overexpression of AtPrx69 conferred the highest tolerance to the BRI1-GFP plants.
Fig. 4.
Cold-susceptible BRI1-overexpressing transgenic plants showed increased cold resistance via AtPrx overexpression. (A) Confirmation of AtPrx overexpression in BRI1-GFP transgenic plants. (B) Quantitative analysis of cold response as determined by the ratio of damaged leaves following cold treatment (bar graph) and electrolyte leakage (line graph) in AtPrx-overexpressing BRI1-GFP plants compared with wild type and untransformed BRI1-GFP plants. Homozygous lines in the T3 generation were used. Bar denotes standard deviation. ANOVA statistical analyses were performed and significant differences are indicated by P values (*P ≤ 0.04, **P ≤ 0.006).
DISCUSSION
Recently, extensive analyses of physiological and molecular regulation in response to various environmental stresses in plants have elucidated the manner in which plants merge and coordinate their needs for basal growth and abiotic stress responses during times of acute stress. Considering that ROS play roles in the positive and/or negative regulation of cellular functions in plants, plants may use ROS to balance their growth and stress responses during times of stress (Swanson and Gilroy, 2010). In this context, our initial findings that higher levels of ROS accumulation and the increased expression of the genes encoding ROS-metabolizing enzymes occurred simultaneously in bri1-9 mutant were very interesting (Fig. 1 and Table 1). Because bri1-9 exhibited a cold-resistant phenotype (Fig. 1) and the knock-out mutants for a subset of class III peroxidases, whose expressions are up-regulated in bri1-9 compared with wild type plants, were more susceptible to cold stress (Fig. 3), it can be inferred that greater accumulation of ROS due to the peroxidase activities in bri1-9 is significant with regard to the higher tolerance of bri1-9 to cold stress. This notion was further confirmed by two findings: ROS production in bri1-9 did not seem to occur through the activation of the NADPH complex (Fig. 1E) and the inhibition of peroxidase activity by the pharmacological inhibitor SHAM resulted in decreased cold tolerance in bri1-9 (Fig. 2).
BR affect ROS production and ROS-mediated abiotic stress responses
Overall, our results indicated that a BR signaling defective mutant, bri1-9, showed a higher tolerance to cold stress, despite its severely retarded growth. A similar growth-retarded mutant caused by the failure of active BR biosynthesis, det2, was reported to have higher endogenous SOD activity than the wild type and exhibited a higher tolerance to oxidative stress (Cao et al., 2005). Together with these results and considering that endogenous BR accumulate in BR-signaling mutants (Noguchi et al., 1999), our data indicate that tolerance to abiotic stress seems to be determined by a combination of complex cellular responses rather than by absolute levels of BR.
However, there have been many reports regarding the effects of BR on ROS-mediated cellular responses and ROS-induced oxidative stress, although the results are somewhat contradictory. The exogenous application of 24-epiBL to a salt-sensitive rice variety resulted in an increased tolerance to the subsequent salt stress and enhanced catalase (CAT), superoxide dismutase (SOD), and glutathione reductase (GR) activities (Özdemir et al., 2004). Water-stressed maize seedlings that had been treated with 24-epiBL showed an increase in the SOD, CAT, and ascorbate peroxidase (APX) activities (Li et al., 1998). In contrast, reductions in Prx and APX activities were reported in osmotically-stressed sorghum following BR treatment (Vardhini and Rao, 2003). Although the effects of exogenous BR treatment were not always consistent with the activities of ROS-metabolic enzymes, ROS production is enhanced by BR in other ways. In cucumber plants, the increased expression of the RBOH gene and the accompanying increase in NADPH oxidase activity were detected in seedlings that had been treated with 24-epiBL. The BR-induced elevation of H2O2 accumulation led to tolerance of photo-oxidative damage and cold stress and resistance to the cucumber mosaic virus (Xia et al., 2009).
A subset of genes encoding class III peroxidases, including AtPrx1, AtPrx22, AtPrx39, and AtPrx69, plays a role in cold response
Class III peroxidases are secreted into the apoplastic space; one of the first symptom of stress is disintegration of cell shape. Therefore, apoplastic peroxidases play a critical role in the production of ROS that act on the peroxidative cross-linking of the cell wall, conferring rigidity to stressed-plants so that they can maintain cellular integrity as a first line of defense. However, few reports have exclusively investigated the distinct functions of the individual genes that encode the proteins that are involved in ROS metabolism. We identified twenty-eight genes that encode the class III peroxidases that were up-regulated in bri1-9 plants compared with wild type plants via microarray analysis (Table 1) and also observed higher levels of ROS accumulation in bri1-9. However, when we directly measured the activity of peroxidase in bri1-9 and BRI1-GFP plants compared with that of the wild type, we did not detect increased or decreased peroxidase activities in the bri1-9 or BRI1-GFP plants respectively (data not shown). Because the peroxidase activity assay used high concentration of H2O2 as a substrate in a reaction mixture to determine the degree of detoxification of H2O2, it may be possible that in vitro peroxidase activity might not represent a specific peroxidase activity or reflect the relevant physiological conditions.
To confirm the microarray data, we performed RT-PCR analyses to investigate the expression of the AtPrx genes that were selected for further investigation using different batches of RNAs. We found that the expression pattern of AtPrx1 was not correlated with the previous result from microarray analysis. However, as the cold-inducible expression pattern of AtPrx was evident and its knock-out mutant was available, we decided to include AtPrx1 together with AtPrx22, AtPrx39 and AtPrx69 for further experiments. The AtPrx1, AtPrx22, AtPrx39, and AtPrx69 genes had been previously identified only by large-scale microarrays or proteomics studies and were included among thousands of genes. AtPrx22 was identified as a protein induced by potassium deficiency (Kang et al., 2004). AtPrx69 was known to be induced by Pseudomonas syringae infection (Mohr and Cahill, 2007) and deficiencies of various ions, such as phosphate, sulfur, and aluminum (Hammond et al., 2003; Kumari et al., 2008; Nikiforova et al., 2003). Although a variety of physiological roles for AtPrxs have been reported (Cosio and Dunand, 2009; Tognolli et al., 2002; Valério et al., 2004), the identification of unique functions of individual AtPrx has been difficult due to the presence of many homologous AtPrx genes. Their functional redundancies can mask the effects of a single-gene knock-out, as was shown by the shoot growth and whole cellular concentrations of H2O2 (Supplementary Fig. 2). Few studies have reported specific functions of individual AtPrx gene. Recently, AtPrx39 was shown to be expressed only in the transition zone of the root, affecting root development (Tsukagoshi et al., 2010). In this study, we also provided evidences to support the observation that the homozygous mutants of atprx1, atprx22, atprx39, and atprx69 exhibited noticeable changes only when exposed to cold (Fig. 3), although the growth pattern and total H2O2 content of each mutant were not changed. Generation of multiple knock-out mutants with different combination of single atprx mutant will help us realize the distinctive or combinatorial functions contributed by each AtPrx gene.
Manipulation of certain AtPrx genes provides a way to generate plants with better tolerance to various stresses without sacrificing growth
Compared with microarray analyses of plants in various environmental conditions, few reports have been published describing transgenic approaches to identify the functions of specific peroxidases because alterations in single-gene expression may be masked, resulting in a lack of visual phenotypic alterations. The physiological substrates of specific peroxidases may also be limiting factors, although they can react to various plant compounds in vitro (Cosio and Dunand, 2009). In a few cases, the overexpression of AtPrx62 in wild type plant led to an increase in the resistance to Botrytis cinerea of over 80% (Chassot et al., 2007), similar to AtPrx21 and Atprx71. The expression of AtPrx62 and AtPrx71 was induced by phosphate starvation (Hammond et al., 2003) and low oxygen concentrations (Klok et al., 2002). These results suggest that the manipulation of the expression of a certain group of AtPrx genes may provide practical tools for the development of plants with increased stress tolerance. Here, we showed that the overexpression of AtPrx22 and AtPrx69 in BRI1-GFP plants clearly improved cold tolerance (Fig. 4). In addition to showing less damaged leaf morphologies and reduced ion leakage following the cold treatment, the BRI1-GFP transgenic plants overexpressing these AtPrx genes did not display growth inhibition compared with the un-transformed BRI1-GFP plants. This observation may be critical when considering the agricultural improvement of crop plants. The naturally constitutive freezing-tolerant mutant eskimo1 (esk1) has been shown to exhibit growth retardation (Xin and Browse, 1998). Additionally, many studies have reported that transgenic plants overexpressing CBF/DREB1 genes acquire cold tolerance compared with untransformed plants. The overexpression of the CBF/DREB1 genes resulted in increased freezing tolerances in non-acclimated plants, accompanied by the constitutive overexpression of downstream Cor genes and increased proline and soluble sugar concentrations (Gilmore et al., 2000; Kasuga et al., 1999; Liu et al., 1998). However, the growth of these transgenic plants was severely retarded. Moreover, feedback repression among the CBF/DREB1 genes was also reported. In the cbf2-null mutant, the increased expression of CBF1 and CBF3 were observed (Novillo et al., 2004), while in the ice1 mutant, the expression levels of CBF3 and CBF2 showed opposite trends (Chinnusamy et al., 2003). The expression levels of CBF/DREB1 genes were also affected by the upstream transcription factor MYB15, which is a negative regulator of CBFs (Agarwal et al., 2006). Therefore, the manipulation of CBF regulons can induce pleiotropic effects in an attempt to control a specific stress. In comparison, the modulation of a subset of AtPrx genes showed improved cold tolerance without growth inhibition.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, grant #: PJ008003), Rural Development Administration, Republic of Korea, and the SRC Research Center for Women’s Diseases of Sookmyung Women’s University (2009) (grant # 3-0903-0022 to K.H.N.).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Agarwal M., Hao Y., Kapoor A., Dong C., Fujii H., Zheng X., Zhu J. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Apel K., Hert H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Brouwer K.S., van Valen T., Day D.A., Lambers H. Hydroxamate-stimulated O2 uptake in roots of Pisum sativum and Zea mays, mediated by a peroxidase. Plant Physiol. 1986;82:236–240. doi: 10.1104/pp.82.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch P.M., Heintz N.H. Redox-regulation of cell-cycle re-entry: cyclin D1 as a primary target for the mitogenic effects of reactive oxygen and nitrogen species. Antioxid. Redox Signal. 2005;7:741–751. doi: 10.1089/ars.2005.7.741. [DOI] [PubMed] [Google Scholar]
- Cao S., Xu Q., Cao Y., Qian K., An K., Zhu Y., Binzeng H., Zhao H., Kua B. Loss-of function mutations in DET2 gene lead to an enhanced resistance to oxidative stress in Arabidopsis. Physiol Plant. 2005;123:57–66. [Google Scholar]
- Chassot S., Nawarth C., Metraux J.P. Cuticular defects leads to full immunity to a major plant pathogen. Plant J. 2007;49:972–980. doi: 10.1111/j.1365-313X.2006.03017.x. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B., Hong X., Agarwal M., Zhu J. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio C., Dunand C. Specific functions of individual classIII peroxidases genes. J. Exp. Bot. 2009;62:391–408. doi: 10.1093/jxb/ern318. [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S., Browning K.S., Gallie D.R., Krishna P. Brassinosteroid functions to protect the translational machinery and heat-shock protein synthesis following thermal stress. Plant J. 2002;29:681–691. doi: 10.1046/j.1365-313x.2002.01257.x. [DOI] [PubMed] [Google Scholar]
- Edward S.L., Raag R., Wariishi H., Gold M.H., Poulos T.L. Crystal structure of lignin peroxidase. Proc. Natl. Acad. Sci USA. 1993;90:750–754. doi: 10.1073/pnas.90.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.P., Bennett M.J., Bowen H.C., Broadly M.R., Eastwood D.C., May S.T., Rahn C., Swarup R., Woolaway K.E., Whote P.J. Changes in gene expression in Arabidopsis shoot during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R.Y., Wang G.Y., Wang S.X. Effect of brassinolide on growth and chilling resistance of maize seedlings. In: Cutler H.G., Yokoda T., Adam G., editors. Brassinosteroids: Chemistry, Bioactivity and Applications. American Chemical Society; Washington DC: 1991. pp. 220–230. [Google Scholar]
- Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol. 2008;8:94. doi: 10.1186/1471-2229-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.G., Pyo Y.J., Cho J.W., Cho M.H. Comparative proteome analysis of differentially expressed proteins induced by K+ deficiency in Arabidopsis thaliana. Proteomics. 2004;4:3549–3559. doi: 10.1002/pmic.200400898. [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Kim B.H., Lim C.J., Lim C.O., Nam K.H. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol. Plant. 2010;138:191–204. doi: 10.1111/j.1399-3054.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Kim Y.H., Kim M.D., Choi Y.I., Park S.C., Yun D.J., Noh E.W., Lee H.S., Kwak S.S. Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotech. J. 2011;9:334–347. doi: 10.1111/j.1467-7652.2010.00551.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Kolk E.J., Wilson I.W., Wilson D., Champman S.C., Ewing R.M., Somerville S.C., Peacock W.J., Dolferus R., Dennis E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M., Tayor G.J., Deyholos M.K. Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol. Genet Genomics. 2008;279:339–357. doi: 10.1007/s00438-007-0316-z. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li L., van Staden J., Jäger A.K. Effects of plant growth regulators on the antioxidant system in seedlings of two maize cultivars subjected to water stress. Plant Growth Regul. 1998;25:81–87. [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F., Lópes-Cobollo R.M., Catalá R., Martínes-Zapater J., Salinas J. A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 2002;32:13–24. doi: 10.1046/j.1365-313x.2002.01398.x. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Breusengem F.V. Reactive oxygen species gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mohr P.G., Cahill D.M. Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct. Integr Genomics. 2007;7:181–191. doi: 10.1007/s10142-006-0041-4. [DOI] [PubMed] [Google Scholar]
- Møller I.M., Sweetlove L.J. ROS signaling-specificity is required. Trends Plant Sci. 2010;15:370–374. doi: 10.1016/j.tplants.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Møller I.M., Jensen P.E., Hansson A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Moon H., Lee B., Choi G., Shin D., Prasad D.T., Lee O., Kwak S.S., Kim D.H., Nam J., Bahk J., et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci USA. 2003;100:358–363. doi: 10.1073/pnas.252641899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgan R. Transcriptome analysis of sulfur depletion on Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Tuan H., Feldmann K.A., Tax F.E. Brassinostroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Alonso J.M., Ecker J.R., Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdemir F., Bor M., Demiral T., Türkan I. Effects of 24-epibrassinolide on seed germination, seedling growth, lipid peroxidation, proline content and antioxidative system of rice (Oryza sativa L.) under salinity stress. Plant Growth Regul. 2004;42:203–211. [Google Scholar]
- Potikha T.S., Collins C.C., Johnson D.I., Delmer D.P., Levine A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 1999;119:849–858. doi: 10.1104/pp.119.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.P., Vernoux T., Lardon F., Montagu M.V., Inze D. Specific checkpoints regulate plant cell cycle progresssion in response to oxidative stress. Plant J. 1999;17:647–656. [Google Scholar]
- Rentel M.C., Lecourieux D., Ouaked F., Usher S.L., Petersen L., Okamoto H., Knight H., Peck S.C., Grierson C.S., Hirt H., et al. OXI1 kinase is necessary for oxidative burst-mediated signaling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- Schopfer P., Liszkay A., Bechtold M., Frahry G., Wagner A. Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta. 2002;214:821–828. doi: 10.1007/s00425-001-0699-8. [DOI] [PubMed] [Google Scholar]
- Smulevich G., jakopitsch C., Droghetti E., Obinger C. Probing the structure and bifunctionality of catalase-peroxidase (KatG) J. Inorg Biochem. 2006;100:568–585. doi: 10.1016/j.jinorgbio.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Swanson S., Gilroy S. ROS in plant development. Physiol Plant. 2010;138:384–392. doi: 10.1111/j.1399-3054.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- Swanson R., Clark T., Preuss D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 2005;18:173–171. [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Do’czi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. Analysis and expression of the class III peroxidases large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/s0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dnagl J.L., Jones J.D.G. Arabidposis gp91phox homologs AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Valério L., Meyer M.D., Penel C., Dunand C. Expression analysis of the Arabidopsis peroxidase multigene family. Phytochemistry. 2004;65:1331–1342. doi: 10.1016/j.phytochem.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Vardhini B.V., Rao S.S.R. Amelioration of osmotic stress by brassinosteroids on seed germination and seedling growth of three varieties of sorghum. Plant Growth Regul. 2003;41:25–31. [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- Xia X.J., Wang Y.J., Zhou Y.H., Tao Y., Mao W.H., Shi K., Asami T., Chen Z., Yu J.Q. Reactive oxygen species are involved in brassinosteroid-Induced stress tolerance in cucumber. Plant Physiol. 2009;150:801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Browse J. Eskimo1 mutants of Arabidopsis are constitutively freezing tolerant. Proc. Natl. Acad. Sci USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Xing Y., Jia W., Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–521. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Yu J.Q., Zhou Y.H., Ye S.F., Huang L.F. 24-Epibrassinolide and abscisic acid protect cucumber seedlings from chilling injury. J. Hortic. Sci. Biotechnol. 2002;77:470–473. [Google Scholar]
- Yuasa T., Ichimura K., Mizoguchi T., Shinozaki K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 2001;42:1012–1016. doi: 10.1093/pcp/pce123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.