Abstract
Summary: The characterization of the complex phenomenon of cell differentiation is a key goal of both systems and computational biology. GeStoDifferent is a Cytoscape plugin aimed at the generation and the identification of gene regulatory networks (GRNs) describing an arbitrary stochastic cell differentiation process. The (dynamical) model adopted to describe general GRNs is that of noisy random Boolean networks (NRBNs), with a specific focus on their emergent dynamical behavior. GeStoDifferent explores the space of GRNs by filtering the NRBN instances inconsistent with a stochastic lineage differentiation tree representing the cell lineages that can be obtained by following the fate of a stem cell descendant. Matched networks can then be analyzed by Cytoscape network analysis algorithms or, for instance, used to define (multiscale) models of cellular dynamics.
Availability: Freely available at http://bimib.disco.unimib.it/index.php/Retronet#GESTODifferent or at the Cytoscape App Store http://apps.cytoscape.org/.
Contact: marco.antoniotti@unimib.it
1 INTRODUCTION
Cell differentiation is the process by which the progeny of toti- and multi-potent stem cells progressively develops in more specialized cell types, characterized by particular properties and functions. The characterization of this complex and still mostly undeciphered phenomenon is of key importance for both computational and systems biology.
To address this problem, we here introduce GeStoDifferent, a plugin for Cytoscape (Smoot et al., 2011) that builds on the dynamical model of cell differentiation introduced in Villani et al. (2011). The key hypothesis of the model is that cell types are characterized by coherent gene activation patterns of the underlying gene regulatory networks (GRNs), and that the degree of differentiation is correlated with the stability of these patterns to noise and perturbations (e.g. mutations). In particular, more differentiated cells would show more sophisticated control mechanisms against noise and mutations (compare Hoffman et al., 2008 and references therein). In Villani et al. (2011), this is achieved by considering noisy random Boolean networks (NRBNs) as a GRN model and by focusing on the dynamical behavior that emerges from the interaction of a large number of genes. The model is general (i.e. it does not refer to any specific organism) and was proven to reproduce some key properties of the differentiation process, namely, (i) the presence of different degrees of differentiation, i.e. from toti-/multi-potent stem cells to intermediate states, to fully differentiated cells; (ii) the stochastic differentiation process, according to which a population of toti-/multi-potent cells generates progenies of distinct types by means of a stochastic process; and (iii) the induced pluripotency phenomenon, according to which fully differentiated cells can achieve a pluripotent stage through the perturbation of some key genes (Yamanaka, 2009).
Within this framework, GeStoDifferent can generate—according to some specific (user-defined) features—and select GRNs that display an appropriate emergent behavior matching an input cell lineage differentiation tree (e.g. that of intestinal crypts or hematopoietic cells). The Cytoscape environment then allows for further network-specific analysis and matching against well-known biological databases.
Our group is working on meshing different simulation levels for intestinal crypt development in conjunction with colorectal cancer development. GeStoDifferent has been used to identify GRNs describing the lineage commitment tree of cell populations in colonic crypts in De Matteis et al. (submitted for publication). Matched networks have been subsequently used within a multicellular and multiscale simulation model (Graudenzi et al., 2012), which we used to study some key cellular processes, such as cell growth, migration, sorting, division and lineage commitment; the selected NRBNs are consistent with the expected behaviour and are amenable to perturbation studies. Finally, new matching filters could in principle be devised to consider epigenetic factors pertaining to the cell differentiation process.
2 THE MODEL
GeStoDifferent is based on a dynamical model of GRN for cell differentiation, described by NRBNs (Villani et al., 2011), an extension of RBNs (Kauffman, 1969) accounting for noise.
An RBN is composed of  Boolean nodes, each of them associated with a Boolean variable
Boolean nodes, each of them associated with a Boolean variable 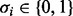 representing the expression of a gene: if
representing the expression of a gene: if 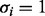 the
the  -th gene synthesizes its product (i.e. proteins or RNAs), otherwise it is inactive. Hence, states of an RBN are Boolean vectors. Each node is connected to
-th gene synthesizes its product (i.e. proteins or RNAs), otherwise it is inactive. Hence, states of an RBN are Boolean vectors. Each node is connected to 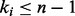 input nodes, and the values of the variables change at discrete time steps, according to the deterministic time evolution of the RBN. The value of the node
input nodes, and the values of the variables change at discrete time steps, according to the deterministic time evolution of the RBN. The value of the node 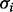 at time
at time 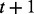 is determined according to a specific Boolean function
is determined according to a specific Boolean function 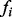 and the input nodes at time
and the input nodes at time  ,
, 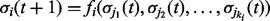 . Here,
. Here, 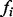 is the Boolean function associated with
is the Boolean function associated with 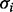 . The evolution of the system is determined by the parallel update of all the elements, at any discrete time step. An RBN is synchronous and deterministic; thus, its evolution will eventually cycle through some states, called attractor. Varying the structural features of the networks, different dynamical regimes appear, ranging from ordered to disordered behaviors.
. The evolution of the system is determined by the parallel update of all the elements, at any discrete time step. An RBN is synchronous and deterministic; thus, its evolution will eventually cycle through some states, called attractor. Varying the structural features of the networks, different dynamical regimes appear, ranging from ordered to disordered behaviors.
Introducing noise in RBNs makes the model more pliable; one of the effects of noise on an RBN is to allow for transitions between attractors, as resulting from a gene’s random ‘flips’. This allows for the evaluation of an attractors transition network (ATN), i.e. a stability matrix between attractors. In turn, by pruning ‘rare’ transitions from an ATN (as determined by introducing a noise threshold), NRBNs can model GRN noise-resistance. By defining multiple thresholds and by identifying strongly connected components within pruned ATNs, it is possible to define (hierarchical) threshold ergodic sets (TES) (i.e. noise dependent gene activation patterns) (see Fig. 1). In Villani et al. (2011), TESs are mapped to cell types, and less differentiated cells are associated to lower thresholds (i.e. those yielding larger TESs) because of less refined noise resistance mechanisms, hence, allowing the GRNs to roam in a wider portion of their state space (and vice versa).
Fig. 1.
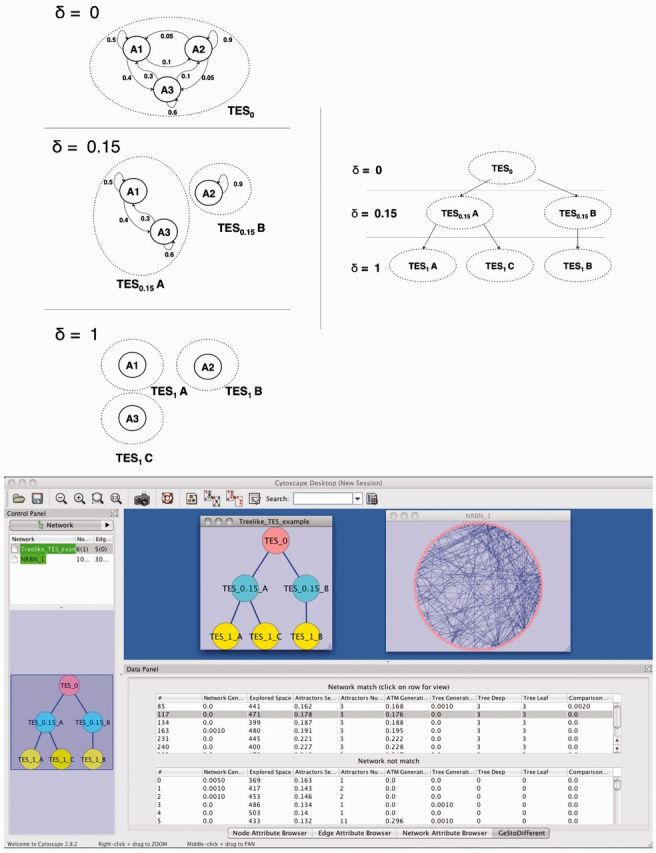
(Top) Threshold-dependent ATNs and the matched stochastic lineage differentiation tree defined by the TESs landscape of a NRBN. (Bottom) Screenshot of the GeStoDifferent plugin showing the input differentiation tree and a randomly generated NRBN matching such a tree
3 THE PLUGIN
GeStoDifferent is coded in Java and works with Cytoscape version 2.8 (Smoot et al., 2011). Information about download and installation can be found at the plugin website (see Availability).
GeStoDifferent sessions are user-defined batch computations. Parameters are specified by a step-by-step wizard. These include: a differentiation tree (in the.sif Cytoscape formats), the NRBNs structure (number of genes, type of Boolean functions and network topology) and the analysis accuracy (exhaustive or sampling-based approach, recommended for large NRBNs). Two topologies are supported: (i) random and (ii) scale-free, with further restrictions, such as fixed incoming/outgoing connections. Three types of Boolean functions are available: (i) standard ‘and/or’, (ii) random and (iii) canalyzing.
Each computational task is tracked by a progress bar. Once a task is finished, (un)matched GRNs are shown in two tables within the Cytoscape Data Panel. By clicking on a network, it gets visualized within Cytoscape, so that it can be further analyzed (see Fig. 1). Also, all the matching networks are exported as textual files, so that they can be used in simulation environments external to Cytoscape, [e.g. CompuCell3D or the simulator described in Graudenzi et al. (2012)]. An illustration of GeStoDifferent performance for different sized networks is available of the plugin website (see Availability).
Funding: Regione Lombardia of Italy under the research projects RetroNet through the ASTIL [12-4-5148000-40]; U.A 053 and NEDD Project [ID14546A Rif SAL-7] Fondo Accordi Istituzionali 2009.
Conflict of Interest: none declared.
REFERENCES
- Graudenzi A, et al. Proceedings of the Italian Workshop on Artificial Life and Evolutionary Computation, WIVACE 2012. 2012. A multiscale model of intestinal crypts dynamics. number ISBN: 978-88-903581-2-8. [Google Scholar]
- Hoffman M, et al. Noise driven stem cell and progenitor population dynamics. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002922. e2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Smoot M, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani M, et al. A dynamical model of genetic networks for cell differentiation. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0017703. e17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]


