Abstract
Human mesenchymal stem cells (hMSCs) remodel or regenerate various tissues through several mechanisms. Here, we identified the hMSC-secreted protein SCRG1 and its receptor BST1 as a positive regulator of self-renewal, migration, and osteogenic differentiation. SCRG1 and BST1 gene expression decreased during osteogenic differentiation of hMSCs. Intriguingly, SCRG1 maintained stem cell marker expression (Oct-4 and CD271/LNGFR) and the potentials of self-renewal, migration, and osteogenic differentiation, even at high passage numbers. Thus, the novel SCRG1/BST1 axis determines the fate of hMSCs by regulating their kinetic and differentiation potentials. Our findings provide a new perspective on methods for ex vivo expansion of hMSCs that maintain native stem cell potentials for bone-forming cell therapy.
Mesenchymal stem cells (MSCs) are non-hematopoietic stromal cells, which retain the ability to self-renew and differentiate into mesenchymal cells such as osteoblasts (OBs), adipocytes (APs), chondrocytes (CCs), and skeletal muscle cells1. Therefore, MSCs are strong candidates for use in regenerative medicine. Cell therapy with adult stem cells such as bone marrow-derived MSCs involves expansion of isolated stem cells in vitro, followed by transplantation back into the body at the site of injury to initiate regeneration2. The criteria for expanded human MSCs (hMSCs) have been defined by the International Society for Cell Therapy as follows: (1) adherence to plastic cell culture plates; (2) positive for CD73, CD90, and CD105 expression; (3) negative for CD34, CD45, HLA-DR, CD14, CD11b, CD79a, and CD19 expression; and (4) in vitro differentiation into OBs, APs, and CCs3. However, loss of self-renewal and multilineage differentiation potentials occurs at high numbers of cell doublings4. Effective stem cell therapies with hMSCs require the establishment of new techniques that preserve MSC multipotency after lengthy expansion.
MSCs can be identified by their ability to form colony-forming unit fibroblasts (CFU-Fs) in vitro5. Mabuchi et al. recently demonstrated that clones of hMSCs retaining high rates of CFU-Fs expressing surface markers CD271/LNGFR, Thy-1, and VCAM-1, exhibited robust multilineage differentiation and self-renewal6. Thus, the combination marker LNGFR+THY-1+VCAM-1high+ (LTV) could be used to isolate potent hMSCs. In-depth investigation of MSC markers has made it possible to identify and purify MSCs; for instance, an anti-CD49a antibody is useful for identifying hMSCs7,8. Intriguingly, rapidly expanding clones of hMSCs express abundant CD49a and VCAM-1 and are highly migratory6. In addition, LTV cells showed 2-fold higher CFU-F formation in comparison to LNGFR+THY-1+VCAM-1− or LNGFR+THY-1+VCAM-1low+ cells. These results suggest that VCAM-1 can be used as a marker for enriching migratory, multipotent, and proliferative cells from culture-expanded hMSCs. However, it is unclear which ligand-receptor signals regulate expression of LNGFR, Thy-1, and VCAM-1, nor is it clear how each marker is associated with proliferation, migration, or differentiation.
The capacity for self-renewal is a key feature of MSCs: self-renewal is the ability to divide while preserving multipotency, which is a prerequisite for sustaining the stem cell pool. In addition, an increased proliferation rate is necessary for efficient use of MSCs in regenerative therapies. After a long period of expansion, MSCs become large and flatten, and lose their ability to divide. Tsai et al. demonstrated the importance of octamer-binding transcription factor 4 (Oct-4) and Nanog in maintaining MSC proliferation activity and differentiation potential, and inhibited spontaneous differentiation9. Oct-4 and Nanog induce expression of DNA (cytosine-5-)-methyltransferase 1 via direct promoter binding, thereby leading to repression of p16, p21, and genes associated with development and lineage differentiation.
Scrapie responsive gene 1 (SCRG1) was identified by Dron et al. in 1998 for its increased expression in the brains of mice infected with scrapie10; the gene is associated with the neurodegenerative changes observed in transmissible spongiform encephalopathies (TSE). In a recent study, Dron et al. reported induction of SCRG1 in the neurons of scrapie-infected mice and the presence of SCRG1 in autophagic vacuoles in terminal-stage disease11. The major studies by Dron and collaborators have shown that SCRG1 is induced in TSE and brain injuries, and is associated with autophagy12. The SCRG1 gene encodes a 98-amino acid, cytokine-like peptide with an N-terminal signal peptide13,14. The predicted protein is highly conserved in mammals and has no significant homology with any other known protein14,15. SCRG1 is preferentially expressed in the central nervous system; SCRG1 transcript levels are similar in primary cultures of neurons and in whole brain, indicating that SCRG1 expression is predominant in neurons in vivo14. However, the SCRG1 receptor and its intracellular signal transduction remain an important but unsolved mystery.
Bone marrow stromal cell antigen 1 (BST1), also known as CD157, is a member of the CD38 gene family and a glycosyl phosphatidylinositol (GPI)-anchored member of the NADase/ADP-ribosyl cyclase family; it is an ectoenzyme that cleaves extracellular nicotinamide adenine dinucleotide (NAD+), generating ADP ribose (ADPR) and cyclic ADPR (cADPR)16,17. BST1 also establishes functional and structural interactions with other transmembrane molecules and thereby gains the ability to transduce intracellular signals18,19,20. BST1 was initially characterized as a stromal21 and myeloid surface glycoprotein22 that mediates control of cell migration and diapedesis23. The receptor and signaling features of BST1 have also been investigated using agonistic monoclonal antibodies to mimic putative ligand(s). These methods have demonstrated that BST1 ligation induces tyrosine phosphorylation of a 130-kDa protein identified as focal adhesion kinase (FAK) in myeloid cell lines24,25; BST1 engagement regulates Ca2+ homeostasis and mediates superoxide production in human myelomonocytic U937 cells16. Accumulating evidence indicates that BST1 is a key player in the control of leukocyte adhesion, migration and diapedesis26,27. In this context, BST1 behaves as a receptor by establishing lateral interactions with other transmembrane molecules, thus overcoming its structural limitation (i.e., of being a GPI-anchored molecule) and acquiring the ability to transduce signals17.
Upon activation by tissue damage, MSCs contribute to tissue-repair processes through a multitude of properties such as self-renewal, migration, and differentiation. In order to identify the key genes that control these MSC properties, we compared gene expression profiles in undifferentiated and OB-differentiated hMSCs. We identified a novel SCRG1/BST1 ligand-receptor combination that maintains hMSC expressions of stem cell markers Oct-4 and CD271/LNGFR, as well as self-renewal, migration, and osteogenic differentiation potentials during ex vivo expansion.
Results
SCRG1 synthesis and secretion are downregulated after osteogenic commitment
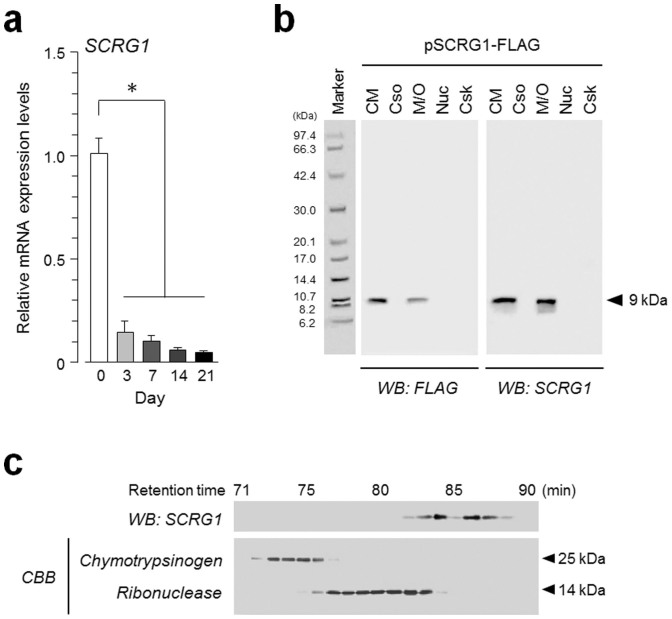
To identify genes that modulate the migration, self-renewal, and multipotency of hMSCs, we used DNA microarrays to characterize the expression profiles of undifferentiated and osteogenically differentiated hMSCs at various time points (supplementary Fig. S1). Genes that were downregulated more than 5-fold 21 days after osteogenic induction are listed in Table S1. We focused on SCRG1, because the precise function of SCRG1, which is generally known as a secreted protein, has not been investigated in hMSCs. SCRG1 gene expression was downregulated more than 20-fold, suggesting its importance in the undifferentiated stage of hMSCs. This result was confirmed by qRT-PCR (Fig. 1a). SCRG1 transcription decreased rapidly from day 3 after osteogenic induction, falling to 4.7% on day 21. Next, we investigated the subcellular distribution of SCRG1 by using western blotting. The SCRG1-FLAG fusion protein was overexpressed in HEK293 cells; the 9-kDa protein was detected in the membrane/organelle fraction and in the conditioned medium, indicating that SCRG1 is secreted from hMSCs (Fig. 1b). The molecular mass of secreted SCRG1 was confirmed by western blotting of the gel-filtrated fraction of the conditioned medium (Fig. 1c).
Figure 1. Synthesis and secretion of SCRG1 are downregulated in hMSCs after osteogenic commitment.
(a) hMSCs were cultured in osteogenic differentiation medium for 0, 3, 7, 14, and 21-days. qRT-PCR was performed with specific oligonucleotide primers for SCRG1. Transcript expression of SCRG1 was normalized to GAPDH and results are indicated as fold-decrease relative to the control (day 0). Data are presented as mean ± SD. *p < 0.05 was considered significant. (b) To examine the subcellular localization of SCRG1, pSCRG1-FLAG was transfected into HEK293 cells. After 48 h, the cells and conditioned medium were collected. The cells were fractionated using the ProteoExtract Subcellular Proteome Extraction Kit. Five-fold concentrated conditioned medium (CM), cytosol (Cso), membrane/organelle (M/O), nucleus (Nuc), and cytoskeleton (Csk) fractions were analyzed by western blotting with anti-SCRG1 or anti-FLAG antibody. (c) hMSC cultured medium was applied to a Superdex 75 pg column equilibrated with 10 mM Tris-HCl, pH 7.5, containing 0.5 M KCl and chromatographed at 1.0 mL/min with the same buffer. Collected fractions were analyzed by western blotting with anti-SCRG1 antibody (upper panel). Chymotrypsin and ribonuclease were used as molecular mass standards (lower panel). Although cropped blot/gel were used, the gels were run under the same experimental conditions.
SCRG1 stimulates ERK, JNK, and PI3K signaling in hMSCs
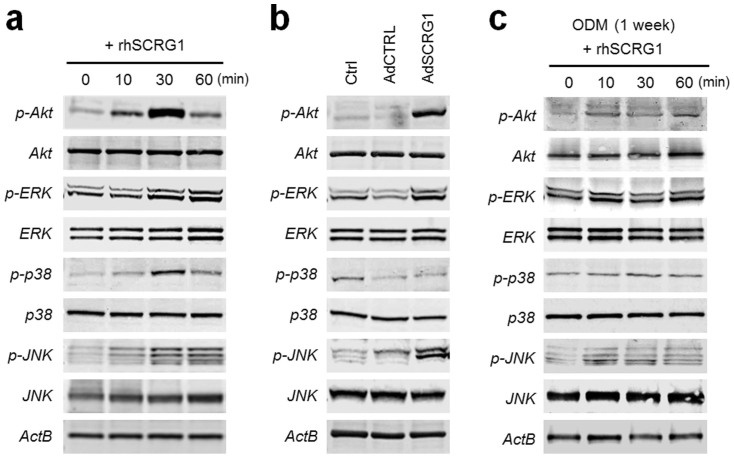
Intracellular signals mediated by extracellular signal-regulated kinase (ERK), c-jun N-terminal kinase (JNK), and phosphoinositide 3-kinase (PI3K)/Akt are crucial for the migratory activity of MSCs28. To examine the effect of SCRG1 on the activities of PI3K/Akt and mitogen-activated protein kinases (MAPKs), the phosphorylation status of these molecules was examined by western blotting; the phosphorylation levels of Akt, ERK, p38 MAPK, and JNK were upregulated within 10–30 min after the stimulation with rhSCRG1 in human bone marrow-derived UE7T-13 MSCs (Fig. 2a). Overexpression of SCRG1-FLAG with pAdSCRG1-FLAG upregulated the phosphorylation levels of Akt, ERK, and JNK, but not p38 MAPK (Fig. 2b), suggesting that SCRG1 mediates autocrine or paracrine stimulation of Akt, ERK, and JNK in hMSCs. Intriguingly, activation of these kinases by rhSCRG1 was not detected in osteogenically induced hMSCs (Fig. 2c), suggesting the SCRG1-receptors that activate Akt, ERK, and JNK are not dominantly expressed in osteogenically induced hMSCs. As the receptor for SCRG1 had not been identified, we sought to identify it in our list of genes that are inhibited during osteogenic differentiation of hMSCs (supplementary Table S1).
Figure 2. SCRG1 stimulates ERK, JNK, and PI3K pathways in hMSCs.
(a) UE7T-13 cells were serum-starved overnight and stimulated with 500 ng/mL rhSCRG1. (b) AdSCRG1-FLAG–transfected UE7T-13 cells were cultured with serum-reduced DMEM for 1 week. (c) UE7T-13 cells cultured in osteogenic differentiation medium (ODM) for 1 week were serum-starved overnight and stimulated with 500 ng/mL rhSCRG1. Cells in (a–c) were washed twice with ice-cold PBS and lysed in RIPA buffer. Samples were separated by SDS-PAGE and analyzed by western blotting. Although cropped blots were used, the gels were run under the same experimental conditions.
The SCRG1 receptor BST1 relays ERK and PI3K signals in hMSCs
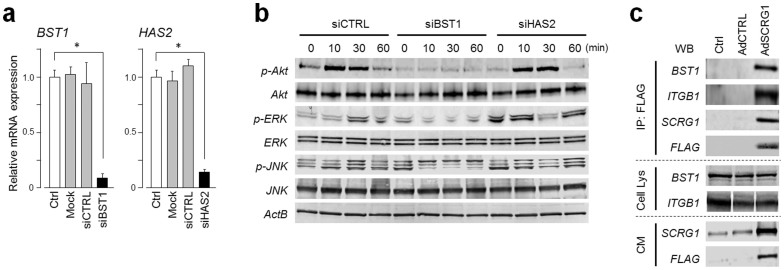
Membrane proteins BST1 and hyaluronan synthase 2 (HAS2), expression of which was >5-fold downregulated in 21 days after osteogenic induction (supplementary Table S1), were chosen as candidates SCRG1 receptors. To evaluate these membrane molecules as functional receptors for SCRG1, BST1 and HAS2 were knocked down in UE7T-13 cells culture. siRNAs against BST1 and HAS2 clearly knocked down the expression of these genes (Fig. 3a and supplementary Fig. S2), but control siRNA did not. Intriguingly, knockdown of BST1 with siBST1 clearly suppressed the rhSCRG1-induced phosphorylation of Akt and ERK, but siHAS2 did not, strongly suggesting that BST1 transduces the SCRG1-induced signals that stimulate Akt and ERK (Fig. 3b). Buono et al. demonstrated that BST1 recruits β1 and β2 integrins, and the multimolecular complex activates ERK and PI3K/Akt, thereby activating migratory activity in human monocytes20. Here, we investigated whether BST1 forms a complex with SCRG1, β1, and β2 integrins. SCRG1-FLAG was overexpressed and immunoprecipitated with anti-FLAG antibody in UE7T-13 cells, then the immunoprecipitate was examined by western blotting. The immunoprecipitate indeed contained SCRG1-FLAG, β1 integrin, and BST1 (Fig. 3c), suggesting BST1 and β1 integrin form a receptor complex for SCRG1. In contrast, UE7T-13 did not express β2 integrin (supplementary Fig. S3).
Figure 3. SCRG1 receptor BST1 relays ERK and PI3K signals in hMSCs.
(a) UE7T13 cells were transfected with siRNA for BST1 (siBST1) or HAS2 (siHAS2). mRNA expression of BST1 (left panel) and HAS2 (right panel) was measured by qRT-PCR and normalized to GAPDH; results are expressed as fold increase or decrease relative to the control (Ctrl). Data are presented as mean ± SD. *p < 0.05 was considered significant. (b) UE7T-13 cells were transfected with siBST1 or siHAS2. After transfection for 48 h, the cells were serum-starved overnight and then stimulated with 500 ng/mL rhSCRG1. The cells were washed twice with ice-cold PBS and lysed in RIPA buffer. Samples were separated by SDS-PAGE and analyzed by western blotting. Although cropped blots were used, the gels were run under the same experimental conditions. (c) UE7T-13 cells were transfected with pAdSCRG1-FLAG. After 1 week, the cells were treated with 0.5 mM dithiobis-sulfosuccinimidylpropionate and then lysed in ice-cold RIPA buffer. Lysates were incubated with anti-FLAG M2 Agarose Affinity Gel and proteins were eluted by adding Laemmli sample buffer (IP: FLAG). Eluted proteins were analyzed by western blotting. Total cell lysate (Cell Lys) and 5-fold concentrated conditioned medium (CM) were used as loading controls. Although cropped blots were used, the gels were run under the same experimental conditions.
The SCRG1/BST1 axis stimulates migratory activity of hMSCs in a FAK/PI3K-dependent manner, even after ex vivo expansion
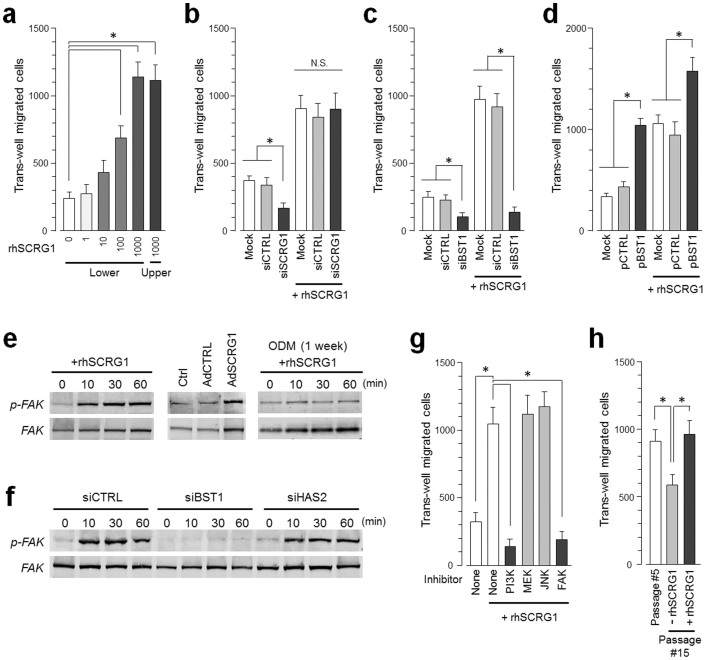
rhSCRG1 stimulated the dose-dependent migratory activity of UE7T-13 cells in a Boyden chamber assay (Fig. 4a). In addition, UE7T-13 migratory activity was suppressed by siRNA-mediated down-regulation of endogenous SCRG1, which was restored by the addition of exogenous rhSCRG1 (Fig. 4b and supplementary Fig. S4). This exogenous rhSCRG1-induced migratory activity was clearly suppressed by siBST1 transfection (Fig. 4c). Next, we examined how BST1 overexpression affects rhSCRG1-induced migratory activity. Bicistronic vector pCMV-BST1-IRES-AcGFP simultaneously expresses BST1 and AcGFP in the same transcript. Therefore, BST1-overexpressing cells transfected with pCMV-BST1-IRES-AcGFP were detected as GFP-positive cells. As shown in supplemental Fig. S5, the transfection efficiencies of pCMV-BST1-IRES-AcGFP and pCMV-null-IRES-AcGFP into UE7T-13 cells were 60.5% (34.4% + 26.1%) and 54.3% (4.1% + 50.2%), respectively. In addition, the percentage of BST1-overexpressing pCMV-BST1-IRES-AcGFP-transfectants was 60.1% (25.7% + 34.4%) and in pCMV-null-IRES-AcGFP -transfectants was 13.0% (8.9% + 4.1%). Thus, pCMV-BST1-IRES-AcGFP potentiates the expression of BST1 in UE7T-13 cells, whereas pCMV-null-IRES-AcGFP did not. Moreover, BST1 overexpression in UE7T-13 cells enhanced rhSCRG1-induced migratory activity (Fig. 4d).
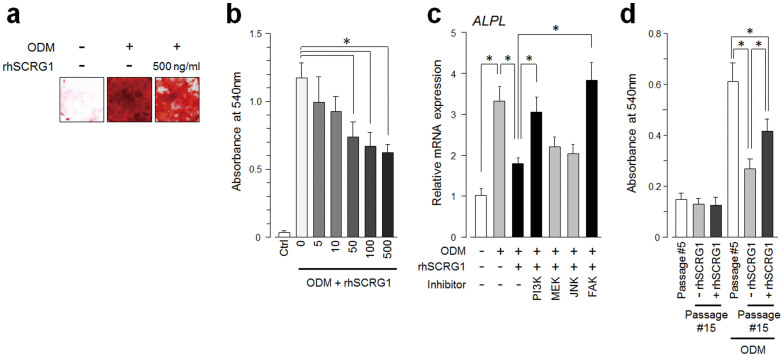
Figure 4. SCRG1/BST1 stimulates FAK/PI3K-dependent hMSC migration and preserves migratory activity after ex vivo expansion.
(a) Migration of UE7T-13 cells was investigated as described in the Methods. rhSCRG1 was added at various concentrations (1–1000 ng/mL). After incubation for 15 h, the number of cells that had migrated to the lower side was counted. (b and c) UE7T-13 cells were transfected with siSCRG1 (b) or siBST1 (c). rhSCRG1 (500 ng/mL) was added to the lower well of the Transwell plate and trans-well migration was analyzed as described in (a). (d) UE7T-13 cells were transfected with pCMV-null-IRES-AcGFP (pCTRL) or pCMV-BST1-IRES-AcGFP (pBST1). rhSCRG1 (500 ng/mL) was added to the lower well of the Transwell plate and trans-well migration was analyzed as described in (a). (e) Phosphorylation of FAK was detected in UE7T-13 as in Figs. 2a–c. Although cropped blots were used, the gels were run under the same experimental conditions. (f) Phosphorylation of FAK was detected in UE7T-13 as in Fig. 3b. Although cropped blots were used, the gels were run under the same experimental conditions. (g) UE7T-13 cells were treated with 1 μM kinase inhibitors trans-well migration was analyzed as in (a). PI3K inhibitor LY294002 (PI3K), MEK inhibitor U0126 (MEK), JNK inhibitor SP600125 (JNK), and FAK inhibitor I (FAK) were added to the lower and upper wells. (h) Primary cultured hMSCs (passage number #5) were subcultured ten times in the presence (passage number #15, +rhSCRG1) or absence (passage number #15, −rhSCRG1) of 500 ng/mL rhSCRG1. Trans-well migration was analyzed as in (a). In a–d, g, and h, data are presented as mean ± SD. *p < 0.05 was considered significant.
In general, cell adhesion receptors such as integrins modulate signal transduction cascades29. PI3K or FAK are cytoplasmic signaling molecules that interface with adhesion receptors at the cell surface to induce migration. Therefore, we investigated SCRG1 activation of these signaling molecules and migratory activity in UE7T-13 cells. rhSCRG1 treatment or SCRG1-FLAG overexpression upregulated FAK phosphorylation in UE7T-13 cells; however, rhSCRG1-induced FAK phosphorylation was not detected in osteogenically differentiated cells (Fig. 4e). rhSCRG1-induced FAK phosphorylation was suppressed by siBST1, but not by control siRNA and siHAS2 (Fig. 4f). In addition, rhSCRG1-induced migration was suppressed by PI3K inhibitor LY294002 and FAK inhibitor I (Fig. 4g). In contrast, ERK kinase (MEK) inhibitor U0126 and JNK inhibitor SP600125 did not affect rhSCRG1-induced migration. After ex vivo expansion (ten passages), hMSC migration was downregulated (Fig. 4h, lane 2), but rhSCRG1 restored migratory activity (lane 3).
SCRG1/BST1 preserves osteogenic differentiation of hMSCs even after ex vivo expansion
SCRG1 and BST1 were strongly expressed in undifferentiated hMSCs (supplementary Table S1); however, expression was dramatically reduced during osteogenic differentiation, suggesting the SCRG1/BST1 axis is important in the undifferentiated stage. Self-renewal of stem cells, preserving their ability to differentiate into specialized cell types, is essential for their therapeutic utility. Alizarin red staining revealed that rhSCRG1 caused dose-dependent inhibition of ECM mineral deposition by osteogenically differentiated UE7T-13 cells (Fig. 5a and b). In addition, qRT-PCR revealed that upregulation of alkaline phosphatase (ALPL) by osteogenic differentiation medium was significantly but incompletely inhibited by rhSCRG1; this inhibition was suppressed by PI3K inhibitor LY294002 and FAK inhibitor I, but not by MEK inhibitor U0126 or JNK inhibitor SP600125 (Fig. 5c). rhSCRG1 preserved the ability of hMSCs to differentiate into osteoblasts even after ex vivo expansion (Fig. 5d), but did not influence their ability to differentiate into adipocytes (supplementary Fig. S6 and S7).
Figure 5. SCRG1/BST1 preserves osteogenic differentiation of hMSCs after ex vivo expansion.
(a and b) UE7T-13 cells were cultured in osteogenic differentiation medium (ODM) containing 500 ng/mL (a), or various concentrations of rhSCRG1 (b, 5–500 ng/mL). After 2 weeks, the cells were evaluated for extra cellular matrix mineralization by alizarin red staining (a). Alizarin red was extracted with 10% cetylpyridinium chloride and absorbance was measured at 540 nm (b). (c) UE7T-13 cells were cultured in ODM with 500 ng/mL rhSCRG1 and 1 μM PI3K inhibitor LY294002 (PI3K), MEK inhibitor U0126 (MEK), JNK inhibitor SP600125 (JNK), and FAK inhibitor I (FAK) for 1 week. Total RNAs were extracted and qRT-PCR was performed with tissue non-specific (liver/bone/kidney) alkaline phosphatase (ALPL)-specific oligonucleotide primers. mRNA expression was normalized to GAPDH and results are expressed as fold-increase or decrease relative to the control. (d) Primary cultured hMSCs (passage number #5) were subcultured ten times in the presence (passage number #15, +rhSCRG1) or absence (passage number #15, −rhSCRG1) of 500 ng/mL rhSCRG1. The cells were evaluated for extra cellular matrix mineralization as in (b). In b–d, data are presented as mean ± SD. * p < 0.05 was considered significant.
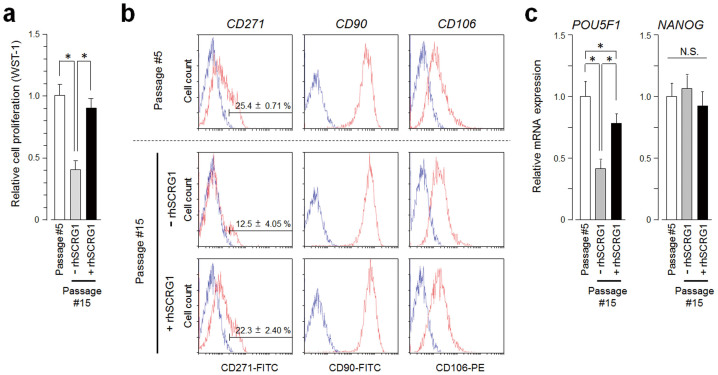
SCRG1/BST1 preserves self-renewal potential and stem cell marker expression in hMSCs after ex vivo expansion
Lengthy ex vivo expansion of hMSCs is necessary to produce sufficient cell volumes for transplantation; thus, we investigated the effect of SCRG1 on proliferation activity. In UE7T-13 cells, proliferation activity was unaffected by addition of rhSCRG1 (supplementary Fig. S8). On the other hand, rhSCRG1 maintained the proliferative activity of hMSCs even after the cells were subcultured ten times (Fig. 6a). As LNGFR+THY-1+VCAM-1high+ (CD271+CD90+CD106high+) cells exhibit high self-renewal, migration, and multipotency potentials, we investigated whether rhSCRG1 affects expression of these stem cell markers in hMSCs Indeed, rhSCRG1 maintained CD271/LNGFR expression even after 15 passages; however, THY-1/CD90 and VCAM-1/CD106 expression was unchanged between passages 5 and 15 (Fig. 6b). rhSCRG1 had no effect on the expression of these MSC markers. Expression of hMSC markers Stro-1, MSCA-1, CD73, CD105, and CD146 were unchanged between passages 5 and 15 (Fig. S9) and their expression was unaffected by rhSCRG1. Churchman et al. previously demonstrated that CD271/LNGFR-positive hMSCs exhibit strong transcription activity, particularly of Wnt-related genes such as embryonic stem cell (ES cell) markers NANOG and POU5F130. Therefore, we investigated the influence of the SCRG1/BST1 axis on expression of these ES cell marker genes. Interestingly, rhSCRG1 preserved the expression of POU5F1, a gene product of Oct-4, even after 15 passages (Fig. 6c, left). There was no difference in NANOG expression between passages 5 and 15, and expression was unaffected by rhSCRG1 (Fig. 6c, right).
Figure 6. SCRG1/BST1 preserves self-renewal potential and the expression of stem cell markers in hMSCs even after long-term culture.
(a–c) Primary cultured hMSCs (passage number #5) were subcultured ten times in the presence (passage number #15, +rhSCRG1) or absence (passage number #15, −rhSCRG1) of 500 ng/mL rhSCRG1. These primary hMSCs under long-term culture conditions were used for the experiments in a–c. (a) The cells were cultured in growth medium on 96-well culture plates for 5 days. After incubation, proliferation was evaluated by WST-1 assay. (b) Expression of cell surface CD271/LNGFR (left column), CD90/THY-1 (middle column), and CD106/VCAM1 (right column) was analyzed by flow cytometry with FITC- or PE-conjugated specific antibodies. Specific antibody (red) and isotype control IgG (blue) are shown. (c) mRNA expression of POU5F1 (left panel) and NANOG (right panel) were analyzed by qRT-PCR and normalized to GAPDH; results are expressed as fold-increase or decrease relative to the control. In a–d, data are presented as mean ± SD. *p < 0.05 was considered significant.
Discussion
MSCs contribute to tissue-repair processes through a multitude of properties such as cell proliferation, migration, and differentiation. In this study, we demonstrated that a novel ligand-receptor complex SCRG1/BST1 and its signaling pathways facilitate hMSC migration, and preserve CD271/LNGFR expression and osteogenic differentiation potential even after in vitro expansion.
BST1 interacts with β1 and β2 integrins, and antibody-induced cross-linking of BST1 promotes relocation of these complexes into detergent-resistant membrane domains19. Moreover, BST1 contributes to the integrin-driven signaling network, which is critical during leukocyte transmigration, leading to optimal phosphorylation of tyrosine kinase receptors and activation of PI3K and MAPK signaling cascades20. However, the native ligand of BST1 in vivo and the functions of BST1 in MSCs are not clear. We found that SCRG1 secreted from hMSCs forms a complex with BST1 and β1 integrin (Fig. 3); it also activates MEK/ERK and PI3K/Akt, consistent with Buono's report that BST1/integrin activates PI3K and MAPK. FAK is a cytoplasmic tyrosine kinase that plays critical roles in integrin-mediated signal transduction and signaling by other cell surface receptors to induce cytoskeletal reorganization during migration29. As shown in Fig. 4g, rhSCRG1-induced migration was inhibited by FAK inhibitor I, suggesting β1 integrin and FAK play important roles in SCRG1/BST1-induced hMSC migration. hMSCs cultured with rhSCRG1 exhibited strong migratory activity even after long-term culture, whereas hMSCs cultured without rhSCRG1 did not (Fig. 4h). These results suggest the SCRG1/BST1 axis is important to the tissue-forming ability of hMSCs by stimulating and maintaining their migratory activity.
Because of their rarity in vivo, hMSCs could be used after expansion for therapies in regenerative medicine. Ex vivo expanded hMSCs have been used for bone regeneration31. However, in vitro expansion of MSCs is associated with gradual accumulation of senescent cells32, telomere erosion33, and changing phenotypes34,35. Thus, ex vivo expansion of hMSCs seems to degrade multipotency; it is thus important to establish novel hMSC expansion techniques that do not sacrifice multipotency even after long-term culture.
CD271/LNGFR-positive hMSCs exhibit strong transcription activity, particularly with respect to the Wnt-related genes30, as shown by bone-related expression profiles of CD271/LNGFR-positive native and culture-expanded hMSCs (15 population doublings), in which CD271/LNGFR expression was downregulated. In addition, expression of Wnt target genes NANOG and OCT4 were higher in CD271/LNGFR-positive hMSCs than in culture-expanded cells. These Wnt-related molecules play important roles in skeletal homeostasis in health and disease36. We demonstrated that rhSCRG1 preserved expression of CD271/LNGFR (Fig. 6b) and POU5F1 (Fig. 6c) in hMSCs, even after ten passages. In addition, rhSCRG1 preserved the osteogenic activity of MSCs even after the ten times subculture in hMSCs (Fig. 5d). Therefore, the SCRG1/BST1 axis may maintain the osteogenic activity of hMSCs during ex vivo expansion through the upregulation of POU5F1 expression. POU5F1 is an EC marker and a critical transcription factor that maintains pluripotency and the undifferentiated state of stem cells37,38.
As shown in Fig. 5b, rhSCRG1 suppressed the osteoblastic differentiation of immortalized hMSC cell line UE7T-13 cells, which exhibit consistent osteoblastic potential even after long-term culture (data not shown). However, it is unclear whether rhSCRG1 abrogates the ability of MSCs to differentiate into osteoblasts or attenuates osteoblast commitment. Attenuation of osteoblast commitment is not equivalent to inhibiting the ability of hMSCs to differentiate; thus, these cells differentiate into osteoblasts following expansion. Yew et al. demonstrated that primary cultured hMSCs show decreased expression of stemness markers such as Oct-4 and NANOG and reduced osteogenic potential in later passages39. Therefore, we evaluated the effects of rhSCRG1 on osteogenic potential in late-passage human bone marrow-derived primarily cultured MSCs. As shown in Fig. 5d, rhSCRG1 preserved the ability of primary hMSCs to differentiate into osteoblasts even after ex vivo expansion. We concluded that rhSCRG1 attenuates osteoblast commitment in hMSCs while preserving their osteoblastic potential. On the other hand, rhSCRG1 did not preserve the adipogenic activity of hMSCs even after ten passages (supplementary Fig. S7). Osteogenic differentiation in MSCs is inhibited with hypoxia inducible factor 1α (HIF1α) stabilization40,41,42, which also produces an increase in adipogenic markers43. This may be mediated by inhibition of the E2A-p21 axis by the HIF1α-TWIST pathway44. After p21 inhibition by overexpression of TWIST, the proliferation rate and differentiation potential also increase39. Oct-4, gene product of POU5F1, also suppresses expression of p219. We predict that osteogenic differentiation in hMSCs is restricted with p21 repression through maintenance of POU5F1 expression by rhSCRG1; thus, the osteogenic differentiation potential is preserved. Osteoblast and adipocyte differentiation mechanisms in MSCs are opposite-directed commitments. Generally, once MSCs are committed to the osteoblastic lineage, they do not transdifferentiate into adipocytes. Here, we demonstrated that SCRG1 preserved the ability of MSCs to differentiate into osteoblasts, but not adipocytes. Therefore, SCRG1 may attenuate the differentiation of MSCs at some late stage of osteoblast commitment.
MSCs contribute to tissue-repair processes through a multitude of properties such as cell proliferation, migration, and differentiation. The clinical use of adult organ-derived MSCs depends on culture expansion to obtain sufficient cell populations for transplantation. The novel SCRG1/BST1 axis maintained the proliferative activity of hMSCs even after long-term culture. Thus, this ligand/receptor complex maintained the self-renewal, migration, and osteogenic differentiation potentials of hMSCs, even at high passage numbers.
This is the first report to show that the SCRG1/BST1 axis positively regulates the self-renewal, migration, and osteogenic differentiation potentials of hMSCs. Our findings provide a novel perspective for methods for ex vivo expansion that maintain native stem cell potentials for bone-forming cell therapy.
Methods
Cells
Primary human mesenchymal stem cells (hMSCs) were purchased from Lonza (Walkersville, PT-2501). hMSCs were cultured in MSCGM™ BulletKit™ (Lonza, PT-3001). Human bone marrow-derived MSCs, UE7T-13, the life span of which was prolonged by a retrovirus encoding human papillomavirus E7 and hTERT45,46, were purchased from Health Science Research Resources Bank (JCRB No. 1154, Japan Health Sciences Foundation). UE7T-13 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma, D6046) supplemented with 10% fetal bovine serum (FBS, Hyclone, CC3008-504). All cell cultures were maintained at 37°C in humidified 5% CO2. Cultured hMSCs were used to identify candidate genes for migration, self-renewal, and multipotency, and for investigations of stem cell marker expression and the preservation of stem cell potential. All other experiments employed hMSC line UE7T-13, which is an immortalized line suitable for transfection and long-term culture to evaluate differentiation ability. However, UE7T-13 cells were not suitable for the evaluation of reduced stemness after expansion, because these cells were immortalized and exhibit consistent osteoblastic potential even after vigorous expansion. Therefore, primary cultured hMSCs were used to evaluate stemness after expansion.
Reagents
FAK inhibitor I (1,2,4,5-benzenetetramine, 4HCl, 324877) was provided by Merck Millipore. PI3K inhibitor LY294002 (440202), MEK inhibitor U0126 (662005), and JNK inhibitor II SP600125 (420119) were purchased from Calbiochem. Protease inhibitor cocktail for use with mammalian cell and tissue extracts (13786) and phosphatase inhibitor cocktails 1 (P0044) and 2 (P5726) were purchased from Sigma. All other reagents were of analytical grade.
DNA microarray
Whole-genome expression was analyzed after 0, 3, 7, 14, and 21 days culture of hMSCs in osteogenic differentiation medium. Total RNA was extracted using ISOGEN reagent (Nippon Gene, 311-02501). Kurabo performed the DNA microarray analyses, including reverse transcription labeling, microarray hybridization, scanning, and raw data analyses (GeneSpring, GX, Agilent Technologies). For hybridization, five human 8.5 K Genome Focus GeneChips (Affymetrix) were used. Of the 8,755 genes except the control probe, 189 up- and 171 downregulated genes were identified during osteogenic differentiation. Osteogenic differentiation-related genes were defined as those that varied more than 5-fold on day 21 in comparison to day 0. These analyses were conducted by the Research Institute of Bio-System Informatics, Tohoku Chemical Co., Ltd. (Morioka, Iwate, Japan).
SCRG1 polyclonal antibody
N-terminal His-tagged mature SCRG1 with the signal peptide (Met21-Gln98) removed was synthesized in the Escherichia coli expression system pCold I (Takara, 3361) according to the manufacturer's instructions. The recombinant protein was purified to homogeneity by chromatography on HisTrap HP (GE Healthcare, 17-5248). The purified recombinant protein was emulsified with Freund's incomplete adjuvant and injected subcutaneously into rabbits. Blood was collected after two additional booster injections administered at 14-day intervals. The IgG fraction of the immunized rabbit sera was purified by Protein-A Sepharose (GE Healthcare, 17-0780) column chromatography.
Constructs and cell transfection
pSCRG1-FLAG was constructed by using the pFLAG-CMV 5b mammalian transient expression system (Sigma, E3762) according to the manufacturer's instructions. pSCRG1-FLAG was transfected into cells with Lipofectamine LTX reagent (Invitrogen, 15338-100). pAdSCRG1-FLAG provided stronger SCRG1-FLAG expression and was constructed with the pAd/CMV/V5-DEST Gateway Vector Kit (Invitrogen, V493-20). IRES-containing bicistronic vectors pCMV-BST1-IRES-AcGFP allowed simultaneous expression of BST1 and AcGFP from the same RNA transcript and was constructed from the bicistronic IRES vectors, pIRES2-AcGFP1 (Clontech, 632435). Electroporation of pCMV-BST1-IRES-AcGFP into cells was performed with a Super Electroporator NEPA21 (Nepagene).
Recombinant human SCRG1 (rhSCRG1)
C-terminal FLAG-tagged recombinant human SCRG1 (rhSCRG1) was produced by using a pSCRG1-FLAG vector and FreeStyle MAX 293 Expression System (Invitrogen, K9000-10) according to the manufacturer's instructions. rhSCRG1 secreted into the culture medium of HEK293F cells was purified by anti-FLAG M2 Agarose Affinity Gel (Sigma, A2220). The purified rhSCRG1 produced a single band after SDS-PAGE and CBB staining (supplementary Fig. S10).
siRNA transfection
Transcriptional knockdown was performed by transfection with siRNA oligonucleotide duplexes at a final concentration of 20 nM in DMEM using Lipofectamine RNAiMAX (Invitrogen, 13778-150) for 48 h. Sequences of the siRNA oligonucleotide duplexes were as follows: BST1 (5′-GUUCCAGCACAGCUGUAUUTT-3′; 5′-AAUACAGCUGUACUGGAACTT-3′), HAS2 (5′-CCAGUAUCAGUUUGGUUUATT-3′; 5′-UAAACCAAACUGAUACUGGTT-3′), and SCRG1 (5′-UCUGUGUCAGGUCAGCUACUCCUUC-3′; 5′-GAAGGAGUAGCUGACCUGACACAGA-3′).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted with ISOGEN reagent (Nippon Gene, 311-02501). cDNA was synthesized with the PrimeScript RT reagent kit (Takara, RR036A) according to the manufacturer's instructions. Fluorescence real-time RT-PCR was performed on a Thermal Cycler Dice Real Time System (Takara) using the SYBR Premix Ex Taq II (Takara, RR820). Primer pairs for SCRG1 (sense, 5′-CCCAGTGAGTGTGAGCATTTAAGAA-3′; antisense, 5′-AGCAAAGTTAGCCCAATGGTGA-3′), ALPL (sense, 5′-GGACCATTCCCACGTCTTCAC-3′; antisense, 5′-CCTTGTAGCCAGGCCCATTG-3′), NANOG (sense, 5′-CAACATCCTGAACCTCAGCTACAA-3′; antisense, 5′-GGCATCCCTGGTGGTAGGAA-3′), POU5F1 (sense, 5′-GTGCCGTGAAGCTGGAGAA-3′; antisense, 5′-TGGTCGTTTGGCTGAATACCTT-3′), and the control GAPDH (sense, 5′-GCACCGTCAAGGCTGAGAAC-3′; antisense, 5′-ATGGTGGTGAAGACGCCAGT-3′) were used to detect target gene transcripts. mRNA levels were normalized to GAPDH.
Western blotting
Cells were washed twice with ice cold PBS and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing protease and phosphatase inhibitor cocktails (Sigma). Protein content was measured with BCA reagent (Pierce, 23225). Equivalent protein samples were separated by 10–20% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore, IPVH00010). After blocking with 5% nonfat dry milk in T-TBS (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, and 0.1% Tween-20), the membrane was incubated with primary anti-SCRG1, anti-FLAG M2 (Sigma, F3165), anti-Akt (Cell Signaling, 9272), anti-phospho-Akt (Ser473) (p-Akt, Cell Signaling, 9271), anti-p44/42 MAPK (ERK, Cell Signaling, 9102), anti-phospho-p44/42 MAPK (Thr202/Tyr204) (p-ERK, Cell Signaling, 9101), anti-p38 MAPK (p38, Cell Signaling, 9212), anti-phospho-p38 MAPK (T180/Y182) (p-p38, Cell Signaling, 9211), anti-SAPK/JNK (JNK, Cell Signaling, 9252), anti-phospho-SAPK/JNK (Thr183/Tyr185) (p-JNK, Cell Signaling, 9251), anti-BST1 (eBioscience, 14-1579-82), anti-β1 integrin (ITGB1, Santa Cruz, sc-8978), anti-FAK (Cell Signaling, 3285), anti-phospho-FAK (Y397) (p-FAK, Cell Signaling, 3283), and anti-β-actin (clone C4, Santa Cruz, sc-4778) antibody as a loading control for normalization. The blots were incubated with alkaline phosphatase-conjugated secondary antibody and developed using the BCIP/NBT membrane phosphatase substrate system (KPL, 50-81-00).
Subcellular localization of SCRG1
To examine the subcellular localization of SCRG1, pSCRG1-FLAG was transfected into HEK293 cells. After 48 h, the cells and conditioned medium were collected. The cells were fractionated using the ProteoExtract Subcellular Proteome Extraction Kit (Calbiochem, 539790) according to the manufacturer's instructions. The conditioned medium and fractionated samples were analyzed by western blotting using anti-SCRG1 and anti-FLAG M2 antibody (Sigma, F3165).
Immunoprecipitation
UE7T13 cells were transfected with adenovirus expression vector, pAdSCRG1-FLAG. After 1 week, the cells were treated with 0.5 mM membrane-impermeable cross-linker dithiobis-sulfosuccinimidylpropionate (Pierce, 21578) for 30 min at room temperature. The reaction was stopped with 20 mM Tris-HCl, pH7.5, for 15 min, and then cells were lysed in ice-cold RIPA buffer. Cell lysates were centrifuged at 12,000 × g for 30 min, and then incubated overnight at 4°C with Anti-FLAG M2 Agarose Affinity Gel (Sigma, A2220). The gels were washed with PBS and proteins were eluted by adding Laemmli sample buffer and boiling for 5 min. Eluted proteins were analyzed by western blotting.
Gel filtration chromatography
hMSCs cultured medium was collected at 1 week and concentrated 5-fold in a Centricon (Amicon, 4241). The concentrated medium was applied to a Superdex 75 pg column (GE Healthcare, 28-9893-33) equilibrated with 10 mM Tris-HCl, pH 7.5, containing 0.5 M KCl and chromatographed at 1.0 mL/min with the same buffer. Fractions were collected once per minute and analyzed by western blotting with anti-SCRG1 antibody. Chymotrypsin and ribonuclease were used as molecular mass standards.
Flow cytometry
Cells were suspended in PBS containing 0.5% FBS and 2 mM EDTA at (1.0 × 105) and incubated with phycoerythrin (PE)-conjugated anti-BST1 (clone: eBioSY11B5, eBioscience, 12-1579-4), fluorescent isothiocyanate (FITC)-conjugated anti-CD271 (clone: ME204, BioLegend, 345103), FITC-conjugated anti-CD90 (clone: 5E10, BioLegend, 328107), PE-conjugated anti-CD106 (clone: STA, BioLegend, 305805), FITC-conjugated anti-Stro-1 (BioLegend, 340105), PE-conjugated anti-MSCA-1 (clone: W8B2, BioLegend, 327305), FITC-conjugated anti-CD105 (clone: 43A3, BioLegend, 323203), FITC-conjugated anti-CD73 (clone: AD2, BioLegend, 344015), or PE-conjugated anti-CD146 (clone: SHM-57, BioLegend, 342003) antibody for 1 h at 4°C in the dark. Acquisition was performed with an EPICS XL ADC System (Beckman Coulter).
Cell migration assay
Migration was determined with Transwell cell culture inserts (BD Falcon, 353504) that were 6.5 mm in diameter with 8-μm pore filters. The cells (2.0 × 104) were suspended in 350 μL serum-free DMEM containing 0.1% BSA (Sigma, A2153) and seeded into the upper well; 600 μL normal growth medium was placed in the lower well of the Transwell plate. After incubation for 6 h at 37°C, cells that had not migrated from the upper side of the filters were scraped off with a cotton swab, and filters were stained with the Three-Step Stain Set (Diff-Quik, Sysmex, 16920). The number of cells that had migrated to the lower side of the filter was counted under a light microscope in five high-power fields (×400). The experiment was performed in triplicate.
Cell proliferation assay
Cell proliferation was analyzed using a colorimetric assay for cleavage of the tetrazolium salt WST-1 (Roche, 5015944) by mitochondrial dehydrogenases in viable cells. The dye is quantified by spectrophotometry and is directly correlated to the number of metabolically active cells in culture. Cells were cultured in growth medium containing rhSCRG1 on 96-well plates (Nunc) for 5 days. After various time points, the cells were incubated for a further 1 h at 37°C with 100 μL medium containing 10 μL WST-1 reagent. The samples were shaken for 1 min and absorbance was measured at 450 nm on an MPR-A4i microplate reader (Tosoh).
In vitro differentiation
To induce adipogenic differentiation, cells were cultured to near confluence and cultured in adipogenic induction medium consisting of DMEM (Sigma) supplemented with 10% FBS, 10 μg/mL insulin (Sigma, I9278), 1 μM dexamethasone (Sigma, D4902), 0.5 μM isobutyl-methylxanthine (Sigma, 15879), and 100 μM indomethacin (Sigma, 17378) for 2 weeks. The induction medium was changed every 3 days. At the end of the differentiation period, cells were fixed with 10% formalin for 10 min and lipid droplets were stained with Oil Red O (Sigma, O0625). Oil Red O stain was quantified by extraction from lipid droplets with dimethyl sulfoxide (DMSO, Sigma, D8418) and absorbance was measured at 540 nm.
To induce osteogenic differentiation, confluent cells were incubated in osteogenic induction medium consisting of MEM alpha (Wako Pure Chemical, 135-15175) supplemented with 10% FBS, 0.1 μM dexamethasone (Sigma), 10 mM β-glycerol phosphate (Sigma, G9422), and 50 μM ascorbic acid (Sigma, A7506) for 2 weeks. The induction medium was changed every 3 days. Bone matrix mineralization was evaluated by Alizarin red S (Sigma, A5533) staining. Alizarin red S was extracted by adding 10% cetylpyridinium chloride (Sigma, C0732) in 8 mM Na2HPO4 (Merck) and 1.5 mM KH2PO4 (Merck) and absorbance was measured at 540 nm.
Statistics
All experiments were repeated at least three times with similar results. Representative images or data are shown. Data were presented as the mean ± standard deviation (SD). Differences between averages and percentages between control and tests were statistically analyzed using paired two-tailed Student's t-tests. P-values less than 0.05 were considered statistically significant.
Author Contributions
E.A. was responsible for the design, analysis, and writing of the manuscript. N.T., S.S., N.O., T.H. and M.T. managed data collection and analysis. H.M. helped with the analysis and discussion. A.I. and N.C. wrote the paper, contributed to the design, analysis, and discussion of the study and acknowledge all individuals who have contributed to this work.
Supplementary Material
Supplementaly infomation
Acknowledgments
This work was supported in part by JSPS KAKENHI Grant Numbers 25463224 to N.T. 24792149 to N.O. 25463013 to M.T. 23592896 to A.I. and 25463053 to N.C.; the Open Research Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan, 2008–2012; Grant-in-aid for Strategic Medical Science Research Centre from the Ministry of Education, Culture, Sports, Science and Technology of Japan, 2010–2014.
References
- Prockop D. J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276, 71–74 (1997). [DOI] [PubMed] [Google Scholar]
- Mosna F., Sensebé L. & Krampera M. Human bone marrow and adipose tissue mesenchymal stem cells. Stem Cells Dev. 19, 1449–1470 (2010). [DOI] [PubMed] [Google Scholar]
- Dominici M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317 (2006). [DOI] [PubMed] [Google Scholar]
- Muraglia A., Cancedda R. & Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J. Cell Sci. 113, 1161–1166 (2000). [DOI] [PubMed] [Google Scholar]
- Friedenstein A. J. et al. Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method. Exp. Hematol. 2, 83–92 (1974). [PubMed] [Google Scholar]
- Mabuchi Y. et al. LNGFR+THY-I+VCAM-Ihi+ cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 1, 152–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschaseaux F. et al. Direct selection of human bone marrow mesenchymal stem cells using an anti-CD49a antibody reveals their CD45med, low phenotype. Brit. J. Haematol. 122, 506–517 (2003). [DOI] [PubMed] [Google Scholar]
- Jones E. A. et al. Optimization of a flow cytometry-based protocol for detection and phenotypic characterization of multipotent mesenchymal stromal cells from human bone marrow. Cytometry Part. B. 70B, 391–399 (2006). [DOI] [PubMed] [Google Scholar]
- Tsai C. C. et al. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol. Cell. 47, 169–182 (2012). [DOI] [PubMed] [Google Scholar]
- Dandoy-Dron F. et al. Gene expression in scrapie. Cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J. Biol. Chem. 273, 7691–7697 (1998). [DOI] [PubMed] [Google Scholar]
- Dron M. et al. Scrg1 is induced in TSE and brain injuries, and associated with autophagy. Eur. J. Neurosci. 22, 133–146 (2005). [DOI] [PubMed] [Google Scholar]
- Dron M. –. SCRG1, a potential marker of autophagy in transmissible spongiform encephalopathies. Autophagy 2, 58–60 (2006). [DOI] [PubMed] [Google Scholar]
- Dron M. et al. Characterization of the human analogue of a Scrapie-responsive gene. J. Biol. Chem. 273, 18015–8 (1998). [DOI] [PubMed] [Google Scholar]
- Dron M. et al. Mouse scrapie responsive gene 1 (Scrg1): genomic organization, physical linkage to sap30, genetic mapping on chromosome 8, and expression in neuronal primary cell cultures. Genomics 70, 140–9 (2000). [DOI] [PubMed] [Google Scholar]
- Dandoy-Dron F., Griffond B., Mishal Z., Tovey M. G. & Dron M. Scrg1, a novel protein of the CNS is targeted to the large dense-core vesicles in neuronal cells. Eur. J. Neurosci. 18, 2449–59 (2003). [DOI] [PubMed] [Google Scholar]
- Ishihara K. & Hirano T. BST-1/CD157 regulates the humoral immune responses in vivo. Chem. Immunol. 75, 235–55 (2000). [DOI] [PubMed] [Google Scholar]
- Malavasi F. et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol. Rev. 88, 841–886 (2008). [DOI] [PubMed] [Google Scholar]
- Liang F., Qi R. Z. & Chang C. F. Signalling of GPI-anchored CD157 via focal adhesion kinase in MCA102 fibroblasts. FEBS. Lett. 506, 207–210 (2001). [DOI] [PubMed] [Google Scholar]
- Lavagno L., Ferrero E., Ortolan E., Malavasi F. & Funaro A. CD157 is part of a supramolecular complex with CD11b/CD18 on the human neutrophil cell surface. J. Biol. Regul. Homeost. 21, 5–11 (2007). [PubMed] [Google Scholar]
- Lo et al. The CD157-integrin partnership controls transendothelial migration and adhesion of human monocytes. J. Biol. Chem. 286, 18681–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisho T. et al. BST-1, a surface molecule of bone marrow stromal cell lines that facilitates pre-B-cell growth. Proc. Natl. Acad. Sci. U. S. A. 91, 5325–5329 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S. C. & Todd R. F. 3rd Structural and biosynthetic features of the Mo5 human myeloid differentiation antigen. Tissue Antigens. 41, 214–218 (1993). [DOI] [PubMed] [Google Scholar]
- Funaro A. et al. Ectoenzymes and innate immunity: the role of human CD157 in leukocyte trafficking. Front Biosci. 14, 929–943 (2009). [DOI] [PubMed] [Google Scholar]
- Hussain A. M., Lee H. C. & Chang C. F. Functional expression of secreted mouse BST-1 in yeast. Protein Expr. Purif. 12, 133–137 (1998). [DOI] [PubMed] [Google Scholar]
- Okuyama Y. et al. Human BST-1 expressed on myeloid cells functions as a receptor molecule. Biochem. Biophys. Res. Commun. 228, 838–845 (1996). [DOI] [PubMed] [Google Scholar]
- Funaro A. et al. CD157 is an important mediator of neutrophil adhesion and migration. Blood. 104, 4269–4278 (2004). [DOI] [PubMed] [Google Scholar]
- Ortolan E. et al. CD157 plays a pivotal role in neutrophil transendothelial migration. Blood. 108, 4214–4222 (2006). [DOI] [PubMed] [Google Scholar]
- Li L. & Jianxin J. Regulatory factors of mesenchymal stem cell migration into injured tissues and their signal transduction mechanisms. Front. Med. 5, 33–39 (2011). [DOI] [PubMed] [Google Scholar]
- Zhao X. & Guan J. L. Fokal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 63, 610–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman S. M. et al. Transcriptional profile of native CD271+ multipotential stromal cells. Evidence for multiple fates, with prominent osteogenic and Wnt pathway signaling activity. Arthr. Rheumat. 64, 2632–2643 (2012). [DOI] [PubMed] [Google Scholar]
- Deschaseaux F., Pontikoglou C. & Sensebe L. Bone regeneration: the stem/progenitor cells point of view. J. Cell. Mol. Med. 14, 103–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W. et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 3, e2213 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter M. A. et al. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells 2, 675–82 (2004). [DOI] [PubMed] [Google Scholar]
- Jones E. et al. Large-scale extraction and characterization of CD271+ multipotential stromal cells from trabecular bone in health and osteoarthritis: implications for bone regeneration strategies based on uncultured or minimally cultured multipotential stromal cells. Arthritis Rheum 62, 1944–1954 (2010). [DOI] [PubMed] [Google Scholar]
- Halfon S., Abramov N., Grinblat B. & Ginis I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells 20, 53–66 (2011). [DOI] [PubMed] [Google Scholar]
- Yang Y. Wnt signaling in development and disease. Cell Biosci. 2, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolf C. M., Cho E. & Tuan R. S. Mesenchymal stromal cells: Biology of adult mesenchymal stem cells: Regulation of niche, self-renewal and differentiation. Arthritis Res. Ther. 9, 204 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R., Ciesiolka T. & Gruss P. A nexus between Oct-4 and E1A: Implications for gene regulation in embryonic stem cells. Cell 66, 291–304 (1991). [DOI] [PubMed] [Google Scholar]
- Yew T. L. et al. Knockdown of p21(Cip1/Waf1) enhances proliferation, the expression of stemness markers, and osteogenic potential in human mesenchymal stem cells. Aging Cell 10, 349–361 (2011). [DOI] [PubMed] [Google Scholar]
- Fehrer C. et al. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell 6, 745–757 (2007). [DOI] [PubMed] [Google Scholar]
- D'Ippolito G., Diabira S., Howard G. A., Roos B. A. & Schiller P. C. Low oxygen tension inhibits osteogenic differentiation and enhances stemness of human MIAMI cells. Bone 239, 513–522 (2006). [DOI] [PubMed] [Google Scholar]
- Tamama K. et al. Differential roles of hypoxia inducible factor subunits in multipotential stromal cells under hypoxic condition. J Cell Biochem 112, 804–817 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin R., LaPres J. J., Kinser S. & McCabe L. R. Prolyl-hydroxylase inhibition and HIF activation in osteoblasts promotes an adipocytic phenotype. J Cell Biochem 100, 762–772 (2007). [DOI] [PubMed] [Google Scholar]
- Tsai C. C. et al. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood 117, 459–469 (2011). [DOI] [PubMed] [Google Scholar]
- Mori T. et al. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol. Cell. Biol. 25, 5183–5195 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura T. et al. Hepatic differentiation of human bone marrow-derived UE7T-13 cells: effects of cytokines and CCN family gene expression. Hepatol. Res. 37, 1068–1079 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementaly infomation