Abstract
In mammalian primordial germ cells (PGCs), DNA demethylation is indispensible for parental imprint erasure, which is a reprogramming process essential for normal developmental potential. Thus, it is important to elucidate how DNA demethylation occurs in each imprinted region in PGCs and to determine which DNA demethylation pathway, passive or active, essentially contributes to the erasure of the imprint. Here, we report that active DNA demethylation via a putative Poly(ADP-ribose) polymerase (PARP) pathway is involved in H19-DMR imprint erasure in PGCs, as shown by an in vivo small molecule inhibitor assay. To the best of our knowledge, this is the first direct demonstration of a DNA replication-independent active DNA demethylation pathway in the erasure process of genomic imprinting in PGCs in vivo. The data also suggest that active DNA demethylation plays a significant role in the complete erasure of paternal imprinting in the female germ line.
Genomic imprinting is an epigenetic mechanism that results in functional differences between paternal and maternal genomes by regulating the expression of paternally and maternally expressed genes, and is indispensible for mammalian development, growth and behavior1,2,3,4. Genomic imprinting undergoes certain specific reprogramming processes, including erasure and reestablishment, via DNA demethylation and de novo DNA methylation of differentially methylated regions (DMRs) in each imprinted region containing a cluster of imprinted genes, respectively5. Its erasure in primordial germ cells (PGCs) starts between embryonic day (E) 9.5 and E10.5, and is completed by E12.56,7, and it is then reestablished based on gender during oocyte maturation and fetal gonocyte development8,9,10,11. Given that the reprogramming of PGCs is essential for acquiring totipotency, it is critically important to elucidate how the DNA demethylation of DMRs actually proceeds. The two possible pathways are passive and active DNA demethylation. The former depends on DNA replication, while the latter is carried out via enzymatic reactions which remove 5-methylcytosine (5mC) residues and replace them with cytosine residues, presumably via DNA repair mechanisms, such as base excision repair (BER). Recently, it was proposed that 5-hydroxymethylcytosine (5hmC) and possibly other Tet-converted bases (5-formylcytosine, 5fC, and 5-carboxycytosine, 5caC) are intermediates of the active DNA demethylation pathway in the BER system12. Conversely, the most recent report showed that although the erasure of DNA methylation in PGCs includes conversion from 5mC to 5hmC by Tet enzymes, the DNA demethylation itself may proceed via a DNA replication-coupled dilution mechanism, suggesting a major role of passive DNA demethylation in PGCs13,14. However, the mosaic-like DNA methylation pattern observed during the erasure in DMRs strongly suggests the direct involvement of active DNA demethylation during this process6. It was also reported that active DNA demethylation is involved in the reprogramming of genomic imprinting in PGCs through an organ culture of aorta gonad-mesonephros regions (AGMs)15.
In this study, we examined the temporal changes in the DNA methylation status of DMRs in various imprinted regions in PGCs. We also investigated the contribution of the DNA replication-dependent and -independent DNA demethylation pathways by inhibiting each of them using the small molecular inhibitors aphidicolin and 3-aminobenzamide (3-AB). The findings clearly demonstrate the existence of the DNA replication-independent, active DNA demethylation pathway in the erasure of genomic imprinting in PGCs in vivo. These results provide important insight into the active DNA demethylation pathway in mammalian reproduction.
Results
DNA demethylation of H19-DMR starts before E10.0 and proceeds in a step-by-step manner
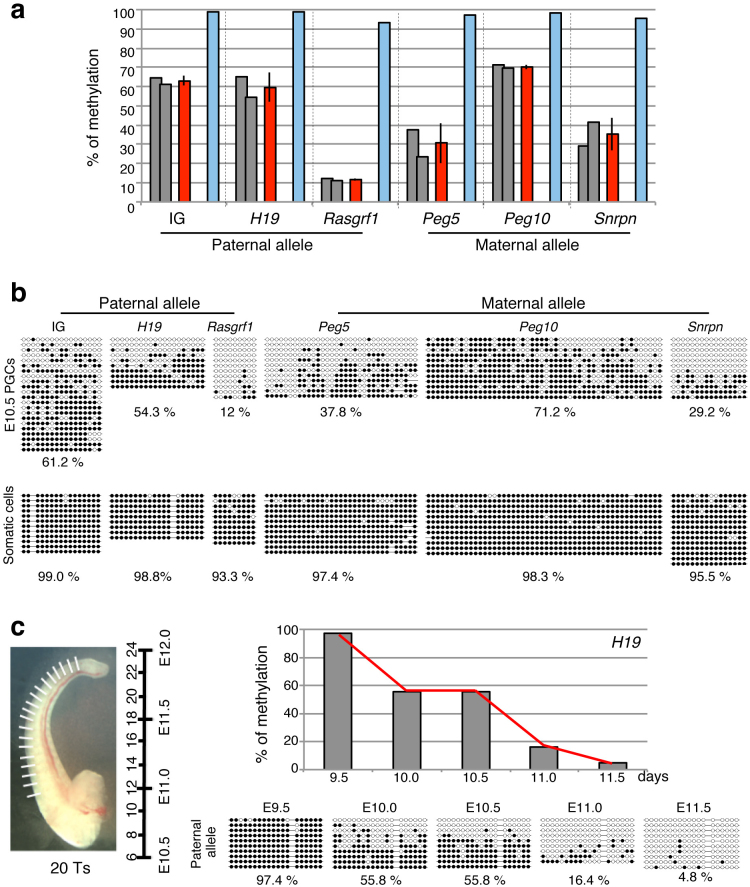
To elucidate the DNA demethylation pathway of DMRs in the germline, we extensively investigated the temporal changes in the DNA methylation status in PGCs. The DNA methylation status of 3 paternally imprinted regions (IG-DMR, H19-DMR and Rasgrf1-DMR) in which the paternal alleles are fully methylated and the maternal alleles are not methylated in PGCs before being erased as well as embryonic somatic cells, were analyzed in E10.5 PGCs and somatic cells (Fig. 1a, b). The methylation status was also examined in 3 maternally imprinted regions (Peg5-DMR, Peg10-DMR and Snrpn-DMR). In this experiment, the parental alleles were distinguished by DNA polymorphisms and the methylation patterns of somatic cells were used for indicating the fully methylate state as the control (Fig. 1a, b). Full methylation in one of the parental alleles was expected unless any DMR DNA demethylation had occurred in PGCs. Relatively higher DNA methylaton levels were detected in IG-DMR (63.0%), H19-DMR (59.7%) and Peg10-DMR (70.3%). However, hypomethylation was more frequently observed in Rasgrf1-DMR (11.6%), Peg5-DMR (30.5%) and Snrpn-DMR (35.2%) (Fig. 1a, b), clearly indicating that the DNA demethylation of the DMRs had already started before E10.5 in the PGCs, perhaps in the migrating stage, and that DNA demethylation proceeds in a region-specific manner. Especially in the case of Rasgrf1-DMR, the DNA demethylation process was almost finished by E10.5.
Figure 1. DNA methylation analysis of imprinted DMRs in PGCs.
(a), (b) DNA methylation level and status of DMRs in E10.5 PGCs and somatic cells. The gray and red bars represent each litter's and the average DNA methylation level of PGCs, respectively and the blue bars represent the DNA methylation level of somatic cells (a). Open circles indicate the unmethylated sites and closed circles indicate the methylated sites (b). (c) Dynamic DNA demethylation of H19-DMR in PGCs. PGCs were isolated from each stage in a manner precisely determined based on the number of tail somites (left). The bar represents DNA methylation level of the paternal allele (right, top) based on the result of bisulfite sequencing (right, bottom). Photograph by Kawasaki et al.
Next, the precise time course of the DNA demethylation of H19-DMR in PGCs was analyzed during E9.5-11.5 at half-day intervals (Fig. 1c). Full methylation was detected in E9.5 PGCs, the first DNA demethylation was observed by E10.0 (55.8%) the second between E10.5 (55.8%) and E11.0 (16.4%) and then the level went further down, to 4.8%, by E11.5. This result indicates that DNA demethylation is not continuous, but rather stepwise, over limited time periods, for example, from E9.5 to E10.0, and from E10.5 to E11.0 in the case of H19-DMR. It should also be noted that DMR demethylation always occurred in a mosaic-like pattern: i.e. methylated and unmethylated CpGs were mixed in the same strand, as reported previously6,7 (Fig. 1b, c). Along with the report by Ciccarone, et al.15, this mosaicism strongly suggests that the active DNA demethylation pathway is somehow involved in the DMR demethylation in PGCs, because random replacement of 5mC with C results in this pattern, regardless of the existence of DNA replication.
In PGCs in vivo, DNA demethylation of H19-DMR is disturbed by both a DNA polymerase inhibitor and PARP inhibitor
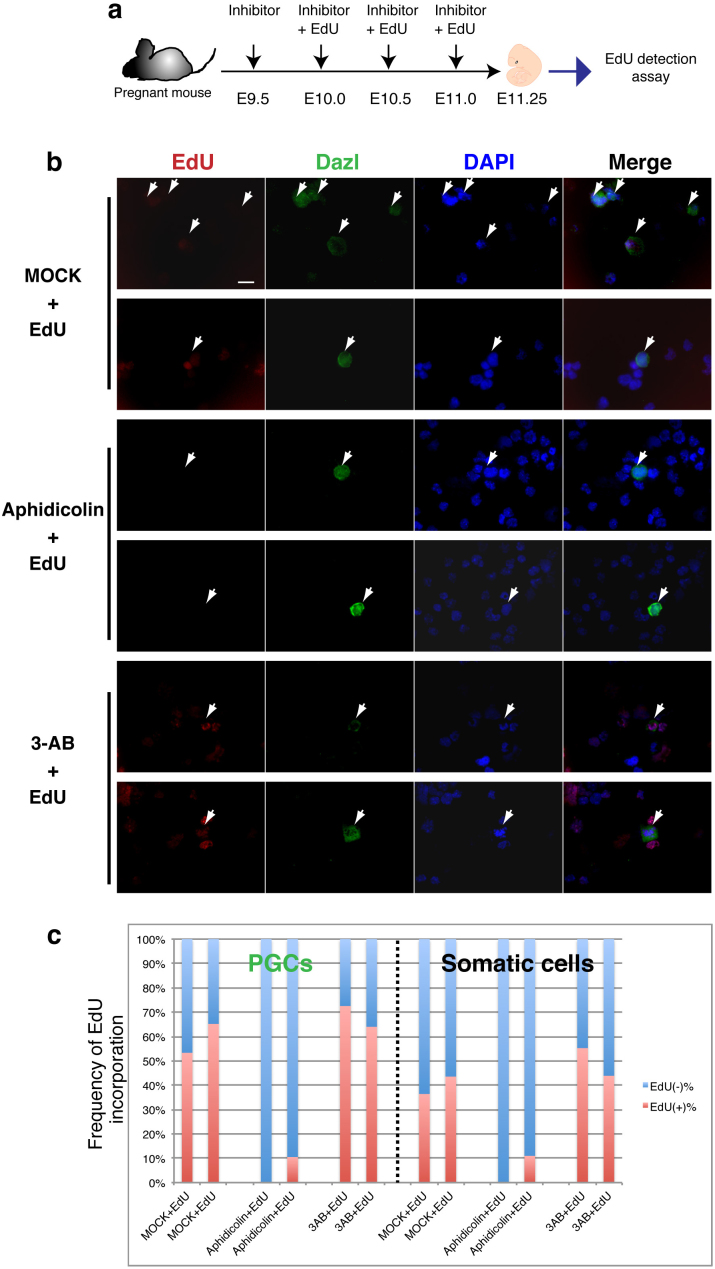
In order to detect DNA replication-independent DNA demethylation, we examined the in vivo effects of several small molecule inhibitors that would be expected to disturb the enzymatic reactions in either the passive or active DNA demethylation pathway, such as the DNA polymerase inhibitor aphidicolin or the PARP inhibitor 3-AB. PARP is known to be a nuclear protein that plays roles in DNA repair and apoptosis by the recognition of single strand breaks (SSBs) in genomic DNA16,17,18, and to therefore be a good target protein in the active DNA demethylation mediated by BER12,15. These inhibitors were administered 4 times by intraperitoneal injection into pregnant mice at half-day intervals from E9.5 (Fig. 2a), because full methylation of H19-DMR was detected at E9.5 PGCs (Fig. 1c). Also, DNA replication (the S phase of the cell cycle) in the PGCs was analyzed by the incorporation of 5-ethynyl-2′-deoxyuridine (EdU). Pregnant mice were administered either of the inhibitors from E9.5 and EdU was injected from E10.0 at the same intervals, and EdU-treated fetuses were analyzed for EdU incorporation on E11.25. EdU incorporation was found in the 3-AB-treated or MOCK fetal genomic DNA, but not in the aphidicolin-treated fetal genomic DNA of Dazl (deleted in azoospermia-like)19,20 positive PGCs or somatic cells (Fig. 2b, c). Dazl is a germ cell marker and Dazl-positive PGCs at E11.25 incorporated more EdU (indicating the S phase of cell cycle) than Dazl negative somatic cells (Fig. 2c), which was similar to the report in a previous study14. Also, immunostaining with Ddx4 (DEAD box polypeptide 4; Mvh), which is also a germ cell marker that expresses in PGCs from E10.520,21 (Supplementary Fig. S1), showed EdU incorporation in the 3-AB-treated and MOCK PGCs. These results indicated that aphidicolin almost completely inhibited DNA replication at the concentrations used, as expected, but 3-AB did not, indicating 3-AB is a useful drug for detecting DNA replication-independent DNA demethylation reactions.
Figure 2. Small molecule inhibitor assay in the fetus during embryonic development.
(a) Experimental scheme of the inhibitor assay (drawings by Kawasaki et al.). (b) EdU detection in PGCs and somatic cells in the genital ridges from an inhibitor-treated fetus. The incorporated EdU (red) was detected in PGCs and somatic cells. DAPI was used for nuclear staining (blue). For the identification of PGCs, gonad cells were stained with Dazl (green; arrows indicate PGCs). Scale bar = 20 μm. EdU was incorporated in the 3-AB-treated or MOCK fetal genomic DNA, but not in the aphidicolin-treated fetal genomic DNA. (c) Percentage of EdU (−) and EdU (+) cells of each group of PGCs and somatic cells was presented.
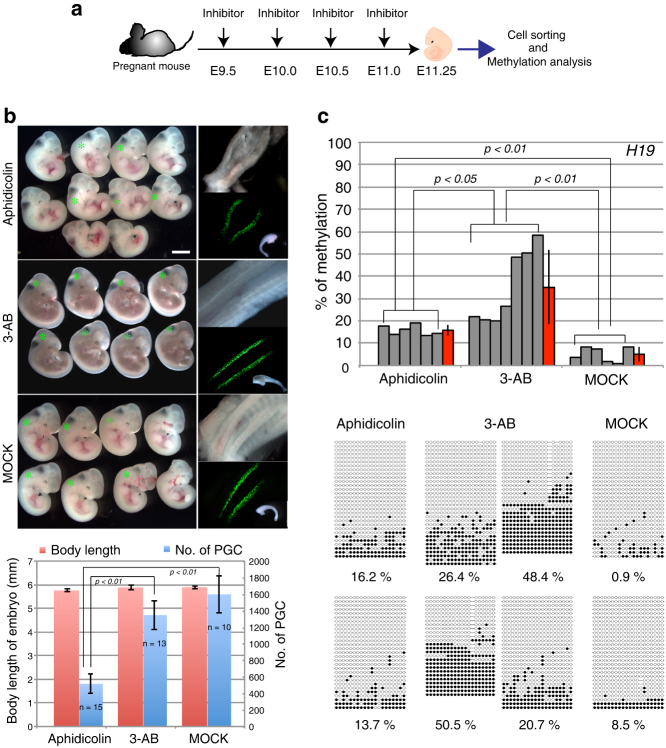
We next investigated the DNA methylation status of H19-DMR in PGCs that had been isolated from aphidicolin or 3-AB treated mice on E11.25 (Fig. 3a). To confirm whether the treatment with aphidicolin indeed blocks DNA replication in PGCs without growth and/or developmental retardation of the embryos, and that 3-AB does not block DNA replication without developmental retardation, we examined the developmental stage of embryos treated with inhibitors, measured the length of body in each embryo and counted the number of PGCs through a sorting of EGFP-positive PGCs by FACS. Although the developmental stage and body length of each embryo was not different among the aphidicolin-, 3-AB-treated and MOCK experiments, the number of PGCs decreased significantly in only the aphidicolin-treated embryos (Fig. 3b). Because paternal and maternal alleles could not be distinguished from each other in this experiment, the degree of methylation before the demethylation was represented as 50%, which is equivalent to full (100%) methylation of one parental allele (Fig. 1c). As shown in Figure 3c, the average percentage of DNA methylation in H19-DMR was reduced to 5.0% in the MOCK PGCs. However, the DNA methylation in the aphidicolin-treated PGCs remained 15.8%, indicating it had been inhibited due to the evident lack of DNA replication, but it should be noted that the DNA methylation level was significantly decreased regardless of DNA replication, suggesting the contribution of DNA replication-independent, active DNA demethylation during this period. Importantly, the average percentage of DNA methylation in the 3-AB-treated PGCs was 35.1%, significantly higher than that in the MOCK control and aphidicolin-treated samples, and there was reduced mosaicism of methylation patterns shown in the 3-AB treated PGCs (Fig. 3c), suggesting a PARP-dependent DNA demethylation pathway. Although the percentage loss of methylation was 68.3% in the aphidicolin-, 29.7% in 3-AB-treated, and 89.9% (similar to the predicted value of E11.25 PGCs from Fig. 1c) in MOCK PGCs, suggesting 21.6% passive DNA methylation and 60.2% active demethylation in PGCs from these inhibitor assays (Supplementary Fig. S2), this may be an overestimation because replication-coupled, passive DNA demethylation is theoretically a major event in PGC reprogramming in this period given the rapid proliferation of PGCs during E10.5-13.513,14. Therefore, it is not a simple matter to demonstrate the involvement of active DNA demethylation under normal developmental conditions13,14. Notwithstanding this difficulty, with the help of specific inhibitors, we have demonstrated that the active demethylation pathway, presumably mediated by PARP, actually contributes to the DNA demethylation of the DMRs in PGCs in vivo.
Figure 3. DNA methylation analysis of H19-DMRs in inhibitor-treated PGCs in vivo.
(a) Experimental scheme (drawings by Kawasaki et al.). After treatment of inhibitors, PGCs in the E11.25 fetus were isolated by sorting EGFP-positive cells and used for DNA methylation analysis. (b) E11.25 fetus and the dissected gonads after inhibitor treatment (top; * EGFP positive fetus). The body size of each fetus was measured (red bars) and the number of EGFP positive PGCs was counted by FACS (blue bars), as shown in the graph (bottom). Scale bar = 2mm. (c) DNA methylation level (top) and status (bottom) of H19-DMR in the inhibitor-treated PGCs, as shown by bisulfite sequencing. p value by t-test.
Discussion
Both the present and previous results clearly show that the DNA demethylation of DMRs in PGCs proceeds step-by-step and in a mosaic-like manner, indicating the involvement of the active DNA demethylation pathway in this process. By means of an experiment using the two inhibitors aphidicolin and 3-AB, which disturb the passive and active demethylation pathways, respectively, it was revealed that both inhibitors affected the DNA demethylation of H19-DMR, indicating that both the passive and active pathway are required for the DNA demethylation of DMR in PGCs. Notably, our findings indicated that presumable PARP-dependent active DNA demethylation pathway was contributed to the demethylation of H19-DMR in the PGCs in vivo. However, it is still unclear how PARP contributes to the DNA demethylation on the biochemical level, further experiments using other inhibitors or genetically modified mice will be required.
PARP plays a role in transcriptional regulation, histone modification and other epigenetic functions, such as the regulation of insulator activity in addition to DNA repair22,23,24,25,26,27,28. Thus, it is possible that PARP contributes to the active DNA demethylation via histone modification and/or transcriptional regulation in the PGCs. We also tried an AP endonuclease 1 (Ape1) inhibitor, which is known to be a component of BER, and another possible target of active DNA demethylation if a DNA repair mechanism is involved in this process. However, the Ape1 inhibitor did not show any effects on the DNA demethylation of H19-DMR, although it is unclear whether this drug actually functioned in vivo (data not shown). Because BER multi-protein complex also includes DNA polymerase β, XRCC1 and DNA ligase III29, further examination of inhibitor usage, such as the dosage and method of administration, will be necessary to reveal the precise mechanism of DMR demethylation30,31,32.
The origin of PGCs is the epiblast. PGCs are observed in a cluster at the allantois of the ectoplacental cone as early as E7.25 and migrate into the genital ridge until E10.533,34. According to a previous report, it was suggested that the demethylation of DMRs was triggered by the settlement of PGCs in the genital ridge6. However, our present results clearly show that DNA demethylation in six independent DMRs had already started by E10.5, and in the case of Rasgrf1-DMR, it had almost finished by then. We have confirmed the full DNA methylation of the H19-DMR in E9.5 PGCs but it was reduced to 55.8% in E10.0 PGCs, suggesting that the DNA demethylation of DMRs starts in the migrating PGCs after E9.5. Thus, further investigation of migrating PGCs will be necessary to address the effect of the settlement of the PGCs in the genital ridge and also the interaction between the PGCs and immediately surrounding cells (microenvironment).
As described above, DNA demethylation of Rasgrf1-DMR proceeds earlier than in the other DMRs, suggesting that a specific regulation mechanism exists in Rasgrf1-DMR. It is known that Rasgrf1-DMR contains extensive repeat sequences and that de novo DNA methylation is dependent on Dnmt3b, not Dnmt3a, as in the case of retrotransposons and endogenous retroviruses (ERVs). Therefore, it is likely that it behaves in like manner in these repeat sequences, although it was also reported that intracisternal A-particles (IAPs) and LTR-ERV1 retroelements are resistant to global DNA methylation erasure in PGCs35.
Why active demethylation functional in the DMR demethylation that takes place in PGCs, and is this essential in mammalian development? Active demethylation may play an essential role in female germ cells acquiring normal developmental potential. In the male germ line, the spermatogonial stem cells (SSCs) derived from gonocytes36 maintain cell division by self-renewal, while female germ cells stop cell division on E12.5 and enter into meiosis on E13.537. On the assumption that the PGC cell cycle is 16 hours13, it is expected that female germ cells can at most divide 3-4 times from the time of the initiation of DNA demethylation to the entry into meiosis. Therefore, if there is no active demethylation pathway, the female PGCs would theoretically retain 1/8–1/16 of the DNA methylation level, even when meiosis started after fertilization. Maternal-specific imprints are established during maturation of the oocyte, but no DNA methylation changes occur in the paternally imprinted regions8,9,10. Therefore, the incomplete erasure (i.e. DNA demethylation) of the paternally imprinted region in the oocytes would lead to the repression of H19 and Meg3 in the maternal alleles in association with an overexpression of Igf2, Dlk1 and Peg11/Rlt1, due to their biallelic expression, and ultimately result in abnormal embryonic development38,39,40 Consistent with this idea, Tet1 knock-out (KO) mice clearly exhibited female infertility, such as a reduced fertilization rate and increased fetal lethality at various developmental stages, although no abnormalities were observed in male KO mice41.
Rapid loss of 5mC is observed in paternal pronuclei just after fertilization in vivo, but this seems to be due to a simple conversion of 5mC to 5hmC by Tet3 enzyme and no changes from 5mC to cytosine have been confirmed yet42. This work also demonstrates that the PARP-dependent active demethylation pathway has the capacity to complete the removal of remaining 5mCs during as well as after the passive demethylation process in vivo. There has been previous evidence of active DNA demethylation in PGCs in vitro15, however, this is the direct demonstration active DNA demethylation is at work in PGCs in vivo, thus providing insight into active DNA demethylation in female germ cells that is required for normal mammalian reproduction.
Methods
Animals
The Institutional Animal Care and Use Committee of Tokyo Medical and Dental University approved all of the animal experimentation protocols. TgN(deGFP)18Imeg (RBRC00821), transgenic mice carrying EGFP cDNA under the control of the Oct4 distal promoter were transferred from RIKEN BRC (Japan). The mouse strain on the JF1 genetic background that possesses the TgN(deGFP)18Imeg transgene was created by 4 backcrosses to JF1. Transgenic mice with both the B6 and JF1 genetic background were used for this experiment. Male transgenic mice and female B6 mice were crossed and the fetuses were recovered in the period E9.5-E11.5 for the isolation of PGCs. In these experiments, the tail somites were counted to determine the correct embryonic stage: the tail somite number of 5–7 (5–7 ts) was used for E10.5, 11–13 ts for E11.0 and 17–19 ts for E11.5 (Fig. 1c). For the inhibitor experiments, pregnant mice were treated with small molecule inhibitors and the effect on the DMR demethylation in the PGCs in the fetuses were analyzed.
Isolation of PGCs
The embryonic fragments containing PGCs (at E9.5–E10.5) or genital ridges (at E11.0–E11.5) were dissected at each fetal stage under stereomicroscopy, dispersed into single cells by Trypsin-EDTA treatment and finally EGFP positive cells (PGCs) were isolated by FACS (MoFlo or FACS AriaII). Approximately 300 PGCs per fetus on E10.5 and 1000 PGCs on E11.5 were isolated by this procedure.
Bisulfite sequencing
Genomic DNA from PGCs was extracted using the Allprep DNA/RNA micro kit (QIAGEN) according to the manufacturer's instructions and eluted with elution buffer. After denaturation with NaOH, genomic DNA was treated with bisulfite solution (2.5 M sodium bisulfite, 0.3 M NaOH, 12.5 mM hydroquinone, pH 5.0) and subsequently purified. Bisulfite PCR was performed as follows. For denaturation, 96°C for 45 seconds; for amplification, 38 cycles, 96°C for 15 seconds, 60°C for 30 seconds and 72°C for 60 seconds, followed by a final extension at 72°C for 1 minute. The PCR primers used were
H19-DMR forward: 5′-GATTTATAAGGGTTATGGGGTGG-3′,
H19-DMR reverse: 5′-TTAAACCCCAACCTCTACTTTTATAAC-3′,
IG-DMR forward: 5′-GGTTTGGTATATATGGATGTATTGTAATATAGG-3′,
IG-DMR reverse: 5′-ATAAAACACCAAATCTATACCAAAATATACC-3′,
Rasgrf1-DMR forward:
5′-GGGATTTAAAATGTTTTTTTTTGGTTATTAGGGAT-3′,
Rasgrf1-DMR reverse: 5′-ACATTCTCAACAAAAACAATAACCTACCTA-3′,
Peg5-DMR forward: 5′-GAGGATATAAGTTTTATTTTGAAATTAGAAG-3′,
Peg5-DMR reverse: 5′-TACCTTAAATACCCTCTTACCACCTAAA-3′,
Peg10-DMR forward: 5′-GTAAAGTGATTGGTTTTGTATTTTTAAGTG-3′,
Peg10-DMR reverse: 5′-TTAATTACTCTCCTACAACTTTCCAAATT-3′,
Snrpn-DMR forward: 5′-AATTTGTGTGATGTTTGTAATTATTTGG-3′,
Snrpn-DMR reverse: 5′-ATAAAATACACTTTCACTACTAAAATCCACAA-3′.
Bisulfite PCR products were subcloned using the pGEM-T Easy Vector System (Promega) and sequenced.
Inhibitor assays in vivo
3-AB (A0788, Sigma) and aphidicolin (011-09811, Wako) were prepared as a 2.5 g/ml and 10 mg/ml stock solution with DMSO, respectively. 4 μl of the 3-AB stock solution or 2 μl of the aphidicolin stock solution were diluted with 500 μl of PBS and administered 4 times per a half day from E9.5 to pregnant mice by intraperitoneal injection. Then, each inhibitor-treated pregnant mouse was dissected on E11.25 (14–16 ts) and PGCs were isolated as described above. The length of body in each embryo was measured, and the number of PGCs was counted by FACS. Inhibition of DNA replication by aphidicolin was monitored with the EdU detection assay, as described below.
EdU administration and detection
For analysis of DNA replication in the fetus, EdU was administered to pregnant mice and incorporated into fetal genomic DNA. 500 μl of 10 mM EdU (A10044, Invitrogen) in PBS was administered by intraperitoneal injection 3 times per half day from E10.0 into pregnant mice. After the EdU-treated fetus at E11.25 was dissected, PGCs and somatic cells from genital ridges were dispersed into single cells, as described above, and were concentrated onto poly-L-lysine coated slides using CYTOSPIN 4. For the detection of EdU, slides were stained using the Click-It EdU Imaging kit (Invitrogen) according to the manufacturer's instructions. For the identification of PGCs, gonad cells were stained with Dazl (germ cell marker) or Mvh (germ cell marker) as a primary antibody (Abcam) and subsequently stained with Alexa fluor 488- goat anti rabbit IgG antibody (Molecular probes). DAPI was used for nuclear staining.
Author Contributions
T.K.-I. and F.I. conceived of the study and J.L., T.K., T.K.-I. and F.I. participated in the experimental design and data analysis. Y.K., J.L. and A.M. performed most analyses. J.L., T.K.-I. and F.I. wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was supported by grants from the Global Center of Excellence (GCOE) Program for International Research Center for Molecular Science in Tooth and Bone Diseases from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (J.L. and F.I.), the JST PRESTO program in Japan (J.L.), Joint Usage/Research Program of Medical Research Institute Tokyo Medical and Dental University (F.I., T.K. and T.K.-I.), Funding Program for Next Generation World-Leading Researchers (NEXT Program) from JSPS to T.K.-I., Creative Science Research from the Japan Society for the Promotion of Science (JSPS) and Grants-in-Aid for Scientific Research (S) from MEXT to F.I.
References
- Surani M. A., Barton S. C. & Noris M. L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550 (1984). [DOI] [PubMed] [Google Scholar]
- McGrath J. & Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984). [DOI] [PubMed] [Google Scholar]
- Cattanach B. M. & Kirk M. Differential activity of maternally and paternally derived chromosome regions in mouse. Nature 315, 496–498 (1985). [DOI] [PubMed] [Google Scholar]
- Barlow D. P. & Bartolomei M. S. Genomic imprinting in mammals: Epigenetics (Cold Spring Harbor Press, 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Dean W. & Walter J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001). [DOI] [PubMed] [Google Scholar]
- Lee J. et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development 129, 1807–1817 (2002). [DOI] [PubMed] [Google Scholar]
- Hajkova P. et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117, 15–23 (2002). [DOI] [PubMed] [Google Scholar]
- Davis T. L., Yang G. J., McCarrey J. R. & Bartolomei M. S. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9, 2885–2894 (2000). [DOI] [PubMed] [Google Scholar]
- Ueda T. et al. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5, 649–659 (2000). [DOI] [PubMed] [Google Scholar]
- Li J. Y., Lees-Murdock D. J., Xu G. L. & Walsh C. P. Timing of establishment of parental methylation imprints in the mouse. Genomics 84, 952–960 (2004). [DOI] [PubMed] [Google Scholar]
- Obata Y. & Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 277, 5285–5289 (2002). [DOI] [PubMed] [Google Scholar]
- Maiti A. & Drohat A. C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286, 35334–35338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J. A. et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S. et al. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32, 340–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone F. et al. Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One 7, e46927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D., Desnoyers S., D'Silva I. & Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342, 249–68 (1999). [PMC free article] [PubMed] [Google Scholar]
- de Murcia G. & Ménissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci 19, 172–176 (1994). [DOI] [PubMed] [Google Scholar]
- de Murcia J. M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A 94, 7303–7307 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke H. J., Lee M., Kerr S. & Ruggiu M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet 5, 513–516 (1996). [DOI] [PubMed] [Google Scholar]
- Maatouk D. M. et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 133, 3411–3418 (2006). [DOI] [PubMed] [Google Scholar]
- Toyooka Y. et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 93, 139–149 (2000). [DOI] [PubMed] [Google Scholar]
- Hassa P. O. et al. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem 280, 40450–40464 (2005). [DOI] [PubMed] [Google Scholar]
- Pavri R. et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18, 83–96 (2005). [DOI] [PubMed] [Google Scholar]
- Schreiber V., Dantzer F., Ame J. C. & de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7, 517–28 (2006). [DOI] [PubMed] [Google Scholar]
- Hassa P. O. & Hottiger M. O. An epigenetic code for DNA damage repair pathways? Biochem. Cell Biol 83, 270–285 (2005). [DOI] [PubMed] [Google Scholar]
- Caiafa P. & Zlatanova J. CCCTC-binding factor meets poly(ADP-ribose) polymerase-1. J Cell Physiol. 219, 265–70 (2009). [DOI] [PubMed] [Google Scholar]
- Zampieri M. et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One 4, e4717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar D. et al. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol 30, 1199–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein C., Berstein H., Payne C. M. & Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathway: fail-safe protection against carcinogenesis. Mutat Res 511, 145–178 (2002). [DOI] [PubMed] [Google Scholar]
- Loizou J. I. et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117, 17–28 (2004). [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A. et al. CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy: mechanistic rationale for drug combination therapy. Mol Cancer Ther 11, 994–1005 (2012). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 68, 3169–3177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclaren A. Primordial germ cells in the mouse. Dev Biol 262, 1–15 (2003). [DOI] [PubMed] [Google Scholar]
- Gisburg M., Snow M. H. L. & Mclaren A. Primordial germ cells in the mouse embryo during gastrulation. Development 110, 521–528 (1990). [DOI] [PubMed] [Google Scholar]
- Guibert S., Forné T. & Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22, 633–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooji D. G. Stem cells in the testis. Int J Exp Pathol 79, 67–80 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. & Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol 187, 107–113 (1997). [DOI] [PubMed] [Google Scholar]
- Leighton P. A., Ingram R. S., Eggenschwiler J., Efstratiadis A. & Tilghman S. M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34–39 (1995). [DOI] [PubMed] [Google Scholar]
- Georgiades P., Watkins M., Surani M. A. & Ferguson-Smith A. C. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127, 4719–4728 (2000). [DOI] [PubMed] [Google Scholar]
- Georgiades P., Watkins M., Burton G. J. & Ferguson-Smith A. C. Roles for genomic imprinting and the zygotic genome in placental development. Proc Natl Acad Sci U S A 98, 4522–4527 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K., Jin S. G., Pfeifer G. P. & Szabó P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A 108, 3642–3647 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures