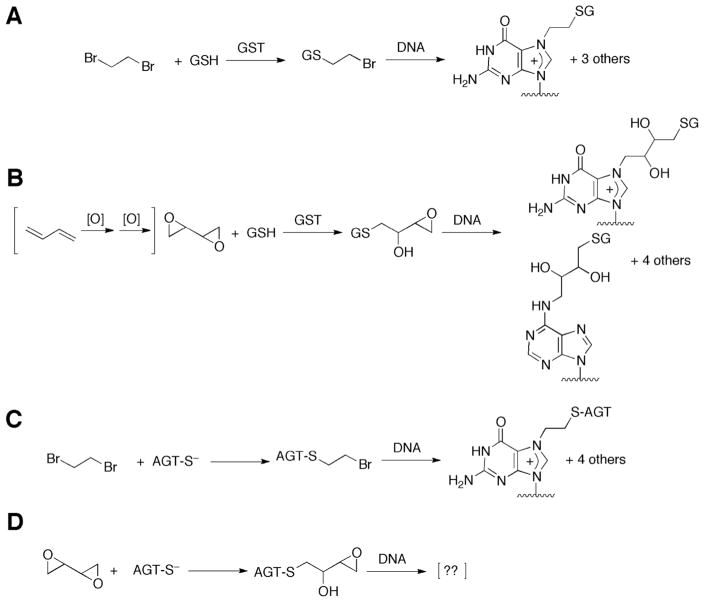
Abstract
Several studies with bacteria and in vitro mammalian systems have provided evidence for the roles of two thiol-based conjugation systems, glutathione (GSH) transferase and O6-alkylguanine DNA-alkyltransferase (AGT), in the bioactivation of the bis-electrophiles 1,2-dibromoethane and 1,2,3,4-diepoxybutane (DEB), the latter an oxidation product of 1,3-butadiene. The in vivo relevance of these conjugation reactions to biological activity in mammals has not been addressed, particularly with DEB. In the present work we used transgenic Big Blue® mice, utilizing the cII gene, to examine the effects of manipulation of conjugation pathways on liver mutations arising from dibromoethane and DEB in vivo. Treatment of the mice with butathionine sulfoxime (BSO) prior to dibromoethane lowered hepatic GSH levels, dibromoethane-GSH DNA adducts (N7-guanyl), and cII mutation frequency. Administration of O6-benzylguanine (O6-BzGua), an inhibitor of AGT, did not change the mutation frequency. Depletion of GSH (BSO) and AGT (O6-BzGua) both lowered the mutation frequency induced by DEB, and BSO lowered the levels of GSH-DEB N7-guanyl and N6-adenyl DNA adducts. Our results provide evidence that the GSH conjugation pathway is a major in vivo factor in dibromoethane genotoxicity; both GSH and AGT conjugation are major factors in the genotoxicity of DEB. The latter findings are considered to be of relevance to the carcinogenicity of 1,3-butadiene.
INTRODUCTION
1,2-Dibromoethane (ethylene dibromide) has been used extensively as a pesticide, but its industrial use was curtailed after demonstration of carcinogenicity.1–4 In rodents, dibromoethane produces mammary gland, spleen, adrenal, liver, kidney, and subcutaneous tissue tumors.1,2 This compound is classified as “Probably carcinogenic to humans” by the International Agency for Cancer Research (IARC).5
1,3-Butadiene is used in the synthetic rubber industry and its annual use in the United States is ~ 2 × 109 kg.6,7 It is carcinogenic in rodents (much more in mice than rats) and has been classified as “Carcinogenic to humans” by the IARC.8 There is also concern about exposure to humans from other sources, e.g. cigarette smoke.9,10
The mechanisms of action of both dibromoethane and 1,3-butadiene are both generally accepted to be genotoxic and involve metabolism. Dibromoethane is conjugated with glutathione (GSH) by GSH transferase (GST) and the resulting half-mustard (GSCH2CH2Br) reacts with DNA via the intermediacy of an episulfonium ion (Scheme 1).14–181,3-Butadiene is oxidized by P450s (P450 2E1, 2A6)19,20 to butadiene monoepoxide21 and then to 1,2,3,4-diepoxybutane (DEB). Of the known oxidative metabolites, DEB is the most toxic and mutagenic.22,23 The much higher level of DEB found in mice compared to rats is thought to explain the much greater carcinogenicity in mice relative to rats.24–28
Scheme 1. GSH (A, B) and AGT (C, D) Conjugation Pathways for Activation of Dibromoethane (A, C) and DEB (B,D).
For the identities of the other DNA adducts of dibromoethane (GSH),11 DEB (GSH),12 and dibromoethane (AGT)13 see the indicated references.
The major DNA adduct formed from dibromoethane is S-[2-(N7-guanyl)ethyl]GSH.11,16,29 Minor adducts (formed in vitro) include S-[2-(N2-guanyl)ethyl]GSH, S-[2-(O6-guanyl)ethyl]GSH, and S-[2-(N1-adenyl)ethyl]GSH.11 Of these, the N7-guanyl adduct has been found in vivo.18,30 Limited studies have been done on misincorporation opposite these adducts with DNA polymerases.31
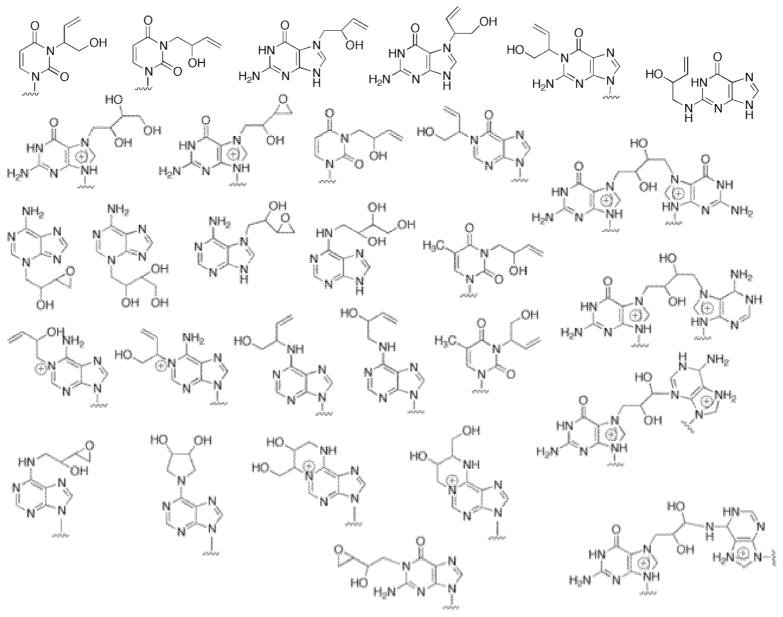
A myriad of DNA adducts have been identified from reactions of DEB with nucleosides and DNA, including those in Scheme 2.32–44 Only some of these have been identified in in vivo settings.12,32, 45–48 Four of these have been incorporated into oligonucleotides and found to be miscoding under some conditions: N1-(2-hydroxy-3-buten-1-yl)deoxyinosine,49 a DEB N6-N6-adenine-adenine cross-link,49 N3-(2-hydroxy-3-buten-1-yl)deoxycytidine,42 and 1,N6-(2-hydroxy-3-hydroxymethyl-2-deoxyadenosine.50 However, none of these studies provides direct evidence of in vivo mutagenicity or a role in carcinogenicity.
Scheme 2. DNA Adducts from Reaction of Oxidized Products of 1,3-Butadiene.
See the references.33–42 (Known stereoisomers of several of the adducts are not considered here.)
With dibromoethane, a strong case for the role of GSH conjugation can be made in toxicity. Bacterial mutagenesis of dibromoethane is highly dependent upon GST activity.51 Disulfiram increases both tumor incidence1,52 and in vivo levels of the DNA adduct S-[2-(N7-guanyl)ethyl]GSH,18 apparently by blocking the oxidative detoxication of dibromoethane (to 2-bromoacetaldehyde)53 (1,2-dichloroethane was used in ref.52).
With 1,3-butadiene and DEB, a role for GSH conjugation in mutagenicity and carcinogenesis has been proposed.54 Although DEB is a direct-acting mutagen, its bacterial mutagenicity is strongly enhanced by GSTs.54–56 The GSH-DEB conjugate S-(2-hydroxy-3,4-epoxybutyl)GSH is 20-fold more mutagenic than DEB in the Salmonella typhimurium TA1535 base pair tester system.56,57 In Escherichia coli TGR8, GST also increased the mutagenicity of DEB and for systems in which 1,3-butadiene was oxidized by P450s.57 In this test strain, the mutation spectra of GSH-enhanced systems differed from that obtained with DEB.57 The DNA adduct S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH was miscoding with human DNA polymerase κ (and bacteriophage DNA polymerase T7 exonuclease−) but not with other DNA polymerases.58 The DNA adducts S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH and S-[4-(N7-guanyl)-2,3-dihydroxybutyl]GSH have been detected and quantitated in vivo in livers of rats and mice.12
Another conjugation system that activates bis-electrophiles is the DNA repair protein O6-alkylguanine DNA-alkyltransferase (AGT). Expression of bacterial or human AGT in bacterial or human cells enhances the toxicity and the mutagenicity of DEB.59–63 The reactivity of AGT is related to the low pKa (4–5) of AGT Cys-145, rendering it a thiolate anion at physiological pH.64 The chemistry of the reaction is presumed to be similar to GSH activation (Scheme 1).65 The putative AGT-Cys-145-CH2CH2Br half-mustard reacts with DNA to form adducts at the guanine N7,65 N1, N2, and O6 atoms and the adenine N6 atom.13 The sites of DNA modification by the putative AGT Cys-145-DEB conjugate have not been determined.
All of the previous work established a case for thiol conjugation in the bioactivation of both dibromoethane and DEB. However, the in vivo biological relevance has not been established. In the present work we used transgenic Big Blue® mice, utilizing the cII gene, to examine the effects of manipulation of conjugation pathways on in vivo mutations arising from dibromoethane and DEB. Our results provide evidence that the GSH conjugation pathway is a major in vivo factor in dibromoethane genotoxicity, and both GSH and AGT conjugation are major factors in the genotoxicity of DEB and probably 1,3-butadiene.
EXPERIMENTAL PROCEDURES
Materials
1,2-Dibromoethane, (racemic) DEB, butathionine-S,R-sulfoximine (BSO), O6-benzylguanine (O6-BzGua), 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB), piperidine, polyethylene glycol 400, and enzymes for digestion were purchased from SigmaAldrich (St. Louis, MO). Phusion High-Fidelity DNA polymerase and uracil DNA glycosylase (UDG) were obtained from New England Biolabs (Ipswich, MA). The oligonucleotides were purchased from Midland Certified Reagents (Midland, TX) for AGT assays or from Integrated DNA Technologies (Coralville, IA) for sequence analysis of the cII mutants and were purified by the manufacturers using HPLC.
The three major DNA adducts formed by GSH conjugation with dibromoethane—S-[2-(N2-guanyl)ethyl]GSH, S-[2-(O6-guanyl)ethyl]GSH), and S-[2-(N6-adenyl)ethyl]GSH—and the internal standard S-[2-(N2-guanyl) [2H4]-ethyl]GSH were synthesized and purified as described previously.11.66 The six major DNA adducts induced by GSH conjugation with DEB—S-[4-(N3-adenyl)2,3-dihydroxybutyl)GSH (N3A-(OH)2butyl-GSH), S-[4-(N6-deoxyadenosinyl)2,3-dihydroxybutyl)GSH (N6dA-(OH)2butyl-GSH), S-[4-(N7-guanyl)2,3-dihydroxybutyl)GSH (N7G-(OH)2butyl-GSH), S-[4-(N1-deoxyguanosinyl)2,3-dihydroxybutyl)GSH (N1dG-(OH)2butyl-GSH), S-[4-(N4-deoxycytidinyl)2,3-dihydroxybutyl)GSH (N4dC-(OH)2butyl-GSH), S-[4-(N3-thymidinyl)2,3-dihydroxybutyl)GSH (N3dT-(OH)2butyl-GSH)—and internal standards (N6dA-(OH)2butyl-[glycine-13C2,15N]-GSH, N7G-(OH)2butyl-[glycine-13C2,15N]-GSH) were also synthesized and purified as described previously.12
Animals and Treatments
Seventy male Big Blue® transgenic mice (five mice per group × 7 treatment groups × 2 time points), age 8 weeks (Agilent/Taconic) were housed in plastic cages (with bedding), according to U. S. National Institutes of Health guidelines. All procedures involving the use of animals were approved by the LRRI Institutional Animal Care and Use Committee. Following an acclimation period of 7 days, Big Blue® transgenic mice were treated with vehicle (corn oil (n = 4), saline (n = 4), and 40% polyethylene glycol 400 in phosphate-buffered saline (n = 2); total n = 10), dibromoethane (30 mg/kg, ip, in corn oil) (n = 10), BSO (8 mg/kg, ip, in saline)/dibromoethane (30 mg/kg, ip, in corn oil) (n = 10), O6-BzGua (80 mg/kg, ip, in 40% polyethylene glycol 400 in phosphate-buffered saline (PBS, 10 mM potassium phosphate, pH 7.4, containing 0.9% NaCl, w/v)/dibromoethane (30 mg/kg, ip, in corn oil) (n = 10), DEB (25 mg/kg, ip, in corn oil) (n = 10), BSO (8 mg/kg, ip, in saline)/DEB (25 mg/kg, ip, in corn oil) (n = 10), or O6-BzGua (80 mg/kg, ip, in 40% polyethylene glycol 400 (v/v) in PBS)/DEB (25 mg/kg, ip, in corn oil) (n = 10). O6-BzGua was administered 1 h prior to treatment with dibromoethane or DEB and BSO was administered 2 h prior to treatment with dibromoethane or DEB. Mice were killed 6 h (five mice per group) or 24 h later (five mice per group). Livers were isolated, frozen, and stored at −80 °C until further analysis.
GSH Assay
The GSH assay was performed as previously described.67 Liver (100 mg) was homogenized in 8 mL of 20 mM EDTA, and trichloroacetic acid was added to a final concentration of 5% (w/v). GSH levels from the mouse liver were measured spectrophotometrically after the incubation of the trichloroacetic acid supernatant with DTNB.
AGT Assay
Mouse liver was homogenized in cold 70 mM HEPES buffer (pH 7.8) containing 1.6 mM EDTA, 1 mM dithiothreitol (DTT), and 5% glycerol (v/v). The homogenate was sonicated and centrifuged at 4 °C for 2 min (12,000 × g). The resulting supernatants were used for the AGT assay.
AGT assays were performed based on a previously described method68 by measuring the transfer of an alkyl group from DNA (O6-methylguanine (O6-MeGua)) to AGT using a 30-/36-mer duplex DNA substrate (5′-GCCTCGAGCCAGCCGCAGACGCAGCGAGGA-3′, 3′-CGGAGCTCGGTCGGCGTCTGCGUCXCTCCTGCGGCT-32P-5′ (X: O6-MeGua), with the 36-mer (with O6-MeGua) 5′-end-labeled with 32P58,68). Tissue extracts (0.5 mg of protein) were incubated with 60 nM DNA substrate in 50 mM Tris-HCl buffer (pH 7.5) containing 1.6 mM EDTA (to block nuclease action), 1.0 mM DTT, and 5% glycerol (v/v) at 37 °C for 1 h. The reactions were quenched with 0.3 M NaOH, followed by neutralization with 0.3 M HCl. DNA substrate/product was isolated by spin column separations. The resulting product was incubated with 3 units of UDG in 50 mM Tris-HCl buffer (pH 7.5) containing 1.0 mM DTT at 37 °C for 30 min, followed by 0.20 M piperidine treatment at 95 °C for 30 min. The resulting product was dried by lyophilization and redissolved in a mixture of H2O and formamide (1:3, v/v). Products were separated using 16% acrylamide (w/v) electrophoresis gels, and results were visualized using a phosphorimaging system (Bio-Rad Molecular Imager FX, BioRad, Hercules, CA).
Measurement of DNA Adducts
DNA from mouse liver was isolated as previously described,18 followed by thermal or enzymatic digestion.12 The reactions were filtered through MWCO Centricon filters (3 kDa cut-off, Millipore, Billerica, MA) and spiked with synthetic N6dA-(OH)2butyl-[glycine-13C2,15N]-GSH, N7G-(OH)2butyl-[glycine-13C2,15N]-GSH, and S-[2-(N2-guanyl) [2H4]-ethyl]GSH. The resulting reactions were analyzed by LC-MS/MS as previously described.12 LC-MS/MS analysis was performed using a Waters Acquity UPLC system (Waters, Milford, MA) interfaced to a Thermo-Finnigan LTQ mass spectrometer (ThermoElectron, Sunnyvale, CA) equipped with an electrospray ionization (ESI) source. Chromatographic separation was achieved with a Waters Acquity UPLC BEH C18 octadecylsilane column (2.1 mm × 100 mm, 1.7 μm). LC conditions were as follows: Solvent A was 0.1% CH3CO2H in H2O (v/v) and solvent B was 0.1% CH3CO2H in CH3CN (v/v). The following gradient program (v/v) was used with a flow rate of 300 μL min−1: the gradient started with 5% B (v/v), increased to 15% B (v/v) at 2 min, to 30% B (v/v) at 6 min, and held at 30% B (v/v) for 1 min. The column was re-equilibrated for 3 min with 5% B (v/v). The temperature of the column was maintained at 40 °C. The MS conditions were as follows: positive ion mode; ion spray voltage, 4.5 kV; capillary voltage, 20 V; capillary temperature, 350 °C; tube lens voltage, 40 V.
CII Mutation Assay
High molecular weight genomic DNA was extracted from mouse liver using a RecoverEase DNA Isolation Kit (Agilent/Stratagene, La Jolla, CA). The packaging of the phage, plating the packaged DNA samples, and determination of mutation frequencies were performed according to the manufacturer’s instructions for the λ Select-cII Mutation Detection System for Big Blue Rodents (Agilent/Stratagene).
Sequence Analysis of the CII Mutants
Single, well-isolated cII plaques were picked and suspended in 100 μL of sterile distilled H2O. These suspensions were heated at 100 °C for 5 min and centrifuged at 12,000 × g for 3 min. The supernatant (10 μL) was used as the DNA template in PCR. The cII gene was amplified by PCR using 5′-CCACACCTATGGTGTATG-3′ (forward primer), 5′-CCTCTGCCGAAGTTGAGTAT-3′ (reverse primer), and Phusion High-Fidelity DNA polymerase. The PCR cycling conditions were as follows: initial melting (95 °C, 3 min), 35 cycles of denaturation (95 °C, 30 s), annealing (60 °C, 1 min), and extension (72 °C, 1 min) followed by a last extension step at 72 °C for 10 min. The PCR products were purified using a QiaQuick PCR purification kit (Qiagen, Hilden, Germany) and submitted to the Vanderbilt DNA Sequencing Facility for nucleotide sequence analysis.
Statistical Analysis
All GSH, AGT, DNA adduct, and mutation frequency results are expressed as means ± SD, with five mice per group (except n = 10 in control (vehicle) group). Statistical significance was determined by Student’s t-test and Fisher’s exact test, with significance levels indicated in the figures and tables.
RESULTS
Rationale
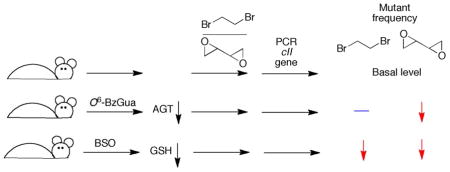
The object of this work was to determine if the GSH and AGT conjugation pathways are important in vivo for the biological effects of two bis-electrophiles, dibromoethane and DEB, for which there was considerable in vitro evidence. Cell culture models could be considered but these are not in vivo systems, and manipulation of GSH and AGT levels might not be relevant to in vivo situations. A mouse model with a reporter transgene was selected for ease in analysis of mutants. In principle, one approach would be to use animals in which a GST was deleted. However, previous studies showed that several GSTs are active toward dibromoethane51,69 and DEB.12 GSH depletion with BSO, an inhibitor of the γ-glutamylcysteine synthetase reaction in the synthesis of GSH, had previously been demonstrated to lower the level of dibromoethane-GSH-DNA adducts in rats and mice.18 Although AGT knockout mice (mgmt(−/−)) have been characterized, use of the established Big Blue® mutation system would require breeding. The AGT inhibitor O6-BzGua, in clinical development,70 has been shown to be a selective inhibitor of AGT.71 We utilized in vivo mutations as a biomarker for cancer. An actual chronic cancer bioassay might be difficult to interpret in the context of long term GSH depletion, which would be expected to have pleiotropic effects in light of the current understanding of the importance of thiol/redox regulatory systems.
Attenuation of GSH and AGT Levels
Preliminary trials were done with B6C3F1 strain mice (because of the expense of the Big Blue® animals) to define conditions that might be applicable for the transgenic animals (Supporting Information Figure S1). The results obtained in the studies with Big Blue® mice showed decreased liver GSH levels (to 30% basal) 6 h following treatment (Supporting Information Figure S2) with BSO and either DEB or dibromoethane (BSO administered 2 h prior to other chemicals). The level of GSH returned to ~ 70% of the basal level after 24 h.
Treatment with O6-BzGua (1 h prior to other chemicals) lowered the level of AGT activity to ~ 10% after 6 h (Supporting Information Figures S3, S4). After 24 h the activity returned to 30% of the basal level. Because AGT doe not recycle in the reaction, the results are representative of the amount of active AGT.
Effects of GSH and AGT Depletion on DNA Adducts
We had previously shown the effect of BSO treatment/GSH depletion on the levels of liver S-[2-(N7-guanyl)ethyl]GSH DNA adducts in B6C3F1, ICR, and A/J mice and in three strains of rats.18 This effect was observed again in the Big Blue® mice, with the adduct level attenuated to 45% at 6 h after treatment (Supporting Information Table S1). The level remained this low at 24 h. No N1- or N6-adenyl adducts were detected.
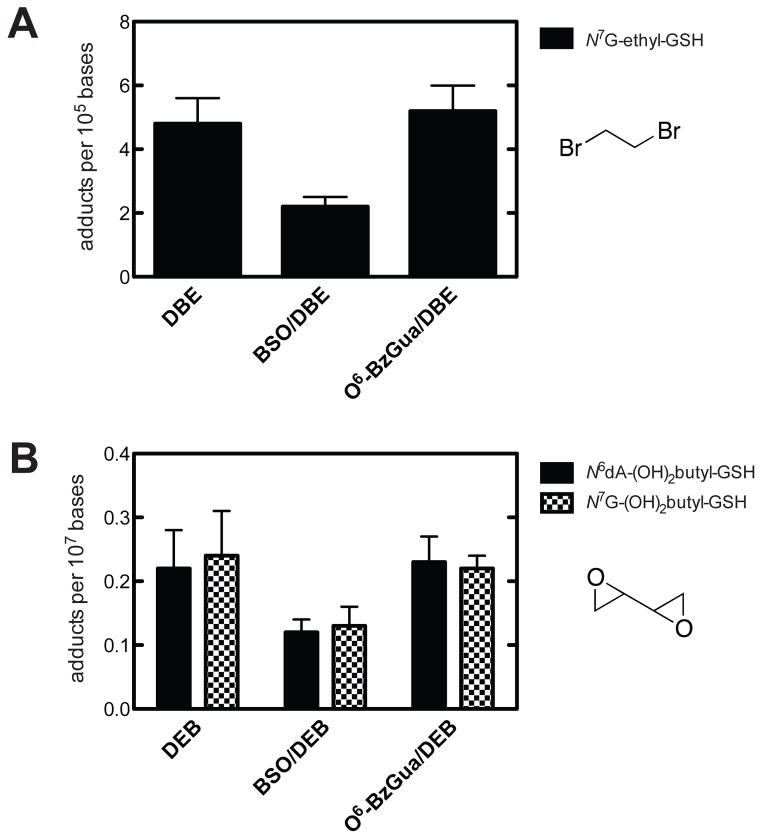
GSH-containing DEB DNA adducts were analyzed following treatment with BSO (Figure 1, Supporting Information Table S2). A similar pattern (~50% decrease) was seen with BSO treatment for the two adducts12 that were detected, the N7-guanyl and N6-adenyl GSH-containing adducts. These were the only GSH-containing DNA adducts detected in previous in vivo work in mice and rats.12 The levels also remained attenuated after 24 h.
Figure 1.
Quantitative analysis of DNA adducts in livers of Big Blue® transgenic mice 6 h after treatment with dibromoethane (DBE), BSO/dibromoethane, or O6-BzGua/dibromoethane (A) and DEB, BSO/DEB, or O6-BzGua/DEB (B). O6-BzGua (80 mg/kg, ip) was administered 1 h prior to treatment with dibromoethane (30 mg/kg, ip) or DEB (25 mg/kg, ip), and BSO (8 mg/kg, ip) was administered 2 h prior to treatment with dibromoethane (30 mg/kg, ip) or DEB (25 mg/kg, ip).
Effects of GSH and AGT Depletion on Mutations
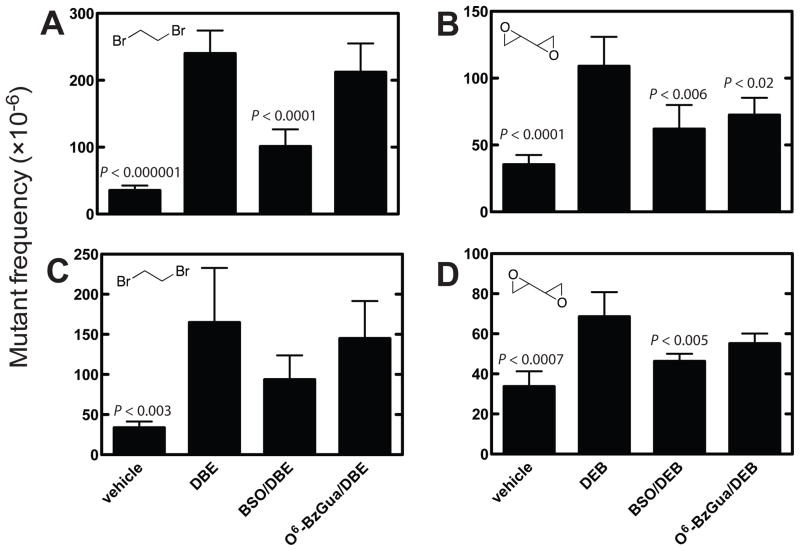
Dibromoethane treatment increased the cII mutant frequency ~ 6-fold at 6 h after treatment (Figure 2A). DEB also increased the mutant frequency, although only ~ ½ as much as dibromoethane did, at the 6 h time point (Figure 2B). At 6 h, BSO treatment (GSH depletion) decreased the dibromoethane-induced mutations by 70%, and both BSO and O6-BzGua treatment (AGT depletion) decreased the DEB-induced mutations by 50% (Figure 2A, 2B). The same patterns were seen 24 h after treatment (Figure 2C, 2D), although the mutation frequencies were attenuated less. However, the BSO and O6-BzGua inhibition patterns were similar.
Figure 2.
cII mutant frequencies in liver of Big Blue® transgenic mice at 6 (A, B) and 24 h (C, D) after treatment with vehicle, dibromoethane (DBE), BSO/dibromoethane, or O6-BzGua/dibromoethane (A, C) and vehicle, DEB, BSO/DEB, or O6-BzGua/DEB (B, D). O6-BzGua (80 mg/kg, ip) was administered 1 h prior to treatment with dibromoethane (30 mg/kg, ip) or DEB (25 mg/kg, ip) and BSO (8 mg/kg, ip) was administered 2 h prior to treatment with dibromoethane (30 mg/kg, ip) or DEB (25 mg/kg, ip). The statistical significance values were from compareisons with the cII mutant frequencies induced by dibromoethane or DEB and evaluated using Student’s t-test (shown in figure). In Part A, the relative mutations frequencies (compared to the highest value, with dibromethane treatment set at 100%) were 15% for vehicle, 42% for +BSO, and 88% for +O6-BzGua. In Part B, the relative mutations frequencies (compared to the highest value, with dibromethane treatment set at 100%) were 20% for vehicle, 43% for +BSO, and 88% for +O6-BzGua. In Part C, the relative mutations frequencies (compared to the highest value, with DEB treatment set at 100%) were 32% for vehicle, 57% for +BSO, and 67% for +O6-BzGua. In Part D, the relative mutations frequencies (compared to the highest value, with DEB treatment set at 100%) were 49% for vehicle, 68% for +BSO, and 80% for +O6-BzGua.
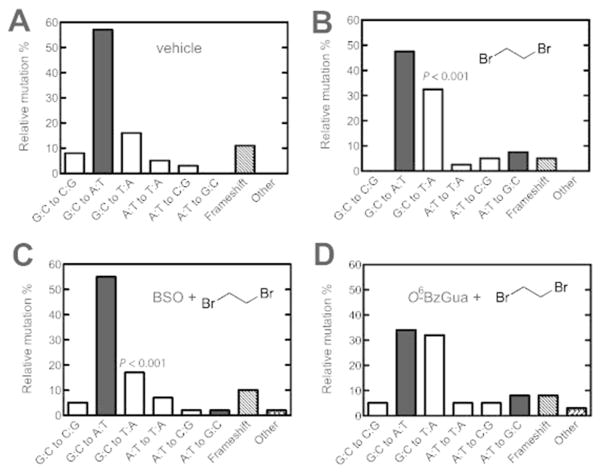
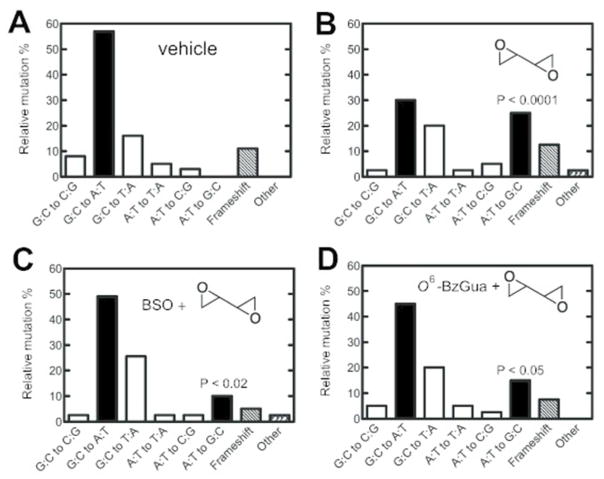
Mutation Spectra
Mutants (37 to 42 independent mutants in each case) were selected from each set obtained from livers of treated animals and subjected to nucleotide sequence analysis (Tables 1, 2 and Figures 3, 4). Details of the individual mutants are listed in the Supporting Information, Tables S3 and S4. The spontaneous (“vehicle”) mutants were characterized by the prominent GC to AT transitions (Figures 3A, 4A). Treatment with dibromoethane led to a statistically significant increase in GC to TA transversions (P < 0.001), which was suppressed by treatment with BSO (P < 0.001) but not O6-BzGua. Treatment with DEB yielded increased AT to GC transitions (P < 0.0001), which were suppressed by either BSO (P < 0.02)or O6-BzGua (P < 0.05) (Figure 4).
Table 1.
Summary of Independent Mutations in the cII Gene of Livers of Big Blue® Transgenic Mice Treated with Vehicle, Dibromoethane, BSO/Dibromoethane, or O6-BzGua/ Dibromoethane
| type of mutation | number of independent mutations (%)
|
|||
|---|---|---|---|---|
| vehicle | dibromoethane | BSO/dibromoethane | O6BzGua/ dibromoethane | |
| G:C to C:G | 3 (8) | 0 (0) | 2 (5) | 2 (5) |
| G:C to A:T | 21 (57) | 19 (47.5) | 23 (55) | 13 (34) |
| G:C to T:A | 6 (16) | 13 (32.5) | 7 (17) | 12 (32) |
| A:T to T:A | 2 (5) | 1 (2.5) | 3 (7) | 2 (5) |
| A:T to C:G | 1 (3) | 2 (5) | 1 (2) | 2 (5) |
| A:T to G:C | 0 (0) | 3 (7.5) | 1 (2) | 3 (8) |
| frameshift | 4 (11) | 2 (5) | 4 (10) | 3 (8) |
| other (tandem base substitution) | 0 (0) | 0 (0) | 1 (2) | 1 (3) |
| total mutants screened | 37 (100) | 40 (100) | 42 (100) | 38 (100) |
Table 2.
Summary of Independent Mutations in the cII Gene of Livers of Big Blue® Transgenic Mice Treated with Vehicle, DEB, BSO/DEB, or O6-BzGua/DEB
| type of mutation | number of independent mutations (%)
|
|||
|---|---|---|---|---|
| vehicle | DEB | BSO/DEB | O6-BzGua/DEB | |
| G:C to C:G | 3 (8) | 1 (2.5) | 1 (2.6) | 2 (5) |
| G:C to A:T | 21 (57) | 12 (30) | 19 (49) | 18 (45) |
| G:C to T:A | 6 (16) | 8 (20) | 10 (25.6) | 8 (20) |
| A:T to T:A | 2 (5) | 1 (2.5) | 1 (2.6) | 2 (5) |
| A:T to C:G | 1 (3) | 2 (5) | 1 (2.6) | 1 (2.5) |
| A:T to G:C | 0 (0) | 10 (25) | 4 (10) | 6 (15) |
| frameshift | 4 (11) | 5 (12.5) | 2 (5) | 3 (7.5) |
| other (tandem base substitution) | 0 (0) | 1 (2.5) | 1 (2.6) | 0 (0) |
| total mutants screened | 37 (100) | 40 (100) | 39 (100) | 40 (100) |
Figure 3.
Relative independent mutations induced by vehicle (A), dibromoethane (DBE) (B), BSO/dibromoethane (C), or O6-BzGua/dibromoethane (D) in liver of Big Blue® transgenic mice. O6-BzGua (80 mg/kg, ip) was administered 1 h prior to treatment with dibromoethane (30 mg/kg, ip) and BSO (8 mg/kg, ip) was administered 2 h prior to treatment with dibromoethane (30 mg/kg, ip). Transition mutations (■); transversion mutations (□); frameshifts and others (diagonal shading). The P values are for comparison of the GC to AT transversions between Parts A and B (P <0.001) and between Parts B and C (P <0.001).
Figure 4.
Relative independent mutations induced by vehicle (A), DEB (B), BSO/DEB (C), or O6-BzGua/DEB (D) in livers of Big Blue® transgenic mice. O6-BzGua (80 mg/kg, ip) was administered 1 h prior to treatment with DEB (25 mg/kg, ip) and BSO (8 mg/kg, ip) was administered 2 h prior to treatment with DEB (25 mg/kg, ip). Transition mutations (■); transversion mutations (□); frameshifts and others (diagonal shading). The P values are for comparison of the AT to GC transversions between Parts A and B (P <0.0001), between Parts B ad C ((P <0.02) and between Parts B and C (P <0.05).
DISCUSSION
Mouse studies were done to address the in vivo roles of GSH and AGT conjugation in the genotoxicity of dibromoethane and DEB. The results of the attenuation of the levels of each lead to the conclusion that GSH conjugation is a major factor in the in vivo genotoxicity of dibromoethane and that both GSH and AGT conjugation are major mechanisms of genotoxicity of DEB in vivo. These findings support in vitro findings with bacterial and mammalian cells and purified enzyme systems.14,51,54–58,72–74 The literature supports genotoxic mechanisms for the carcinogenicity of dihaloalkanes and DEB, and we propose that the conjugation pathways considered here are important in the carcinogenicity of both the industrial chemicals dibromoethane and 1,3-butadiene.
Several points are in order regarding the mutation analyses. First, the mutation spectra were dominated by single base pair mutations (~ 90% in all cases, Tables 1 and 2) as opposed to frameshifts and tandem mutations. This in vivo pattern differs from that reported in DEB-treated human lymphoblasts23 and splenic T cells isolated from mice treated with DEB.22, The discrepancy may be due to differences in concentrations, to the presence of conjugating enzymes (i.e., GST) in vivo (and in spleen T cells vs. liver), or to the nature of the phenotypic selection inherent in mutations in the cII vs. the hprt and tk loci.
Racemic DEB was used in this study, and the possibility can be considered that the individual stereoisomers of DEB may differ in their metabolism and genotoxicity. However, our own studies with the three isomers of DEB showed only ~ a 2-fold difference in the efficiency of GSH conjugation by individual GSTs or the genotoxicity of the GSH conjugates prepared from the three DEB isomers.12 Other work has shown only ~ 2-fold differences among the stereoisomers of DEB with regard to their crosslinking ability43 or genotoxicity in a yeast-based system.75 On the basis of these prior studies, we conclude that the use of racemic DEB is justified and that the conclusions are applicable to all three isomeric oxidation products (DEB) of 1,3-butadiene.
We cannot discount roles of some of the individual DNA adducts formed directly by reaction of dibromoethane, butadiene monoepoxide, or DEB (Scheme 2) with DNA.33–42 The literature shows that very little mutagenicity is seen with dibromoethane in the absence of GST.14,51,72,73 The mutagenicity of S-(2-hydroxy-3,4-epoxybutyl)GSH is ~ 20-fold greater than that of DEB.56,57 In the work presented here, in vivo depletion of 70% of the GSH (6 h time point, Supporting Information Figures S1, S2) was associated with a 70% decrease in mutant frequency (Figure 1A) for dibromoethane. Apparently AGT conjugation is not an important contributor to the in vivo mutagenicity of dibromoethane, in that 90% depletion of AGT (6 h, Supporting Information Figure S4) was not associated with a statistically significant change in the mutant frequency (Figure 2A). Conjugation reactions appear to play a major role in the in vivo genotoxicity of DEB, in that the 70% decrease in GSH and 90% decrease in AGT (Supporting Information Figures S2 and 3) were associated with 60 and 50% decreases in DEB genotoxicity (Figure 2B). Together these two pathways, if additive, can account for all of the in vivo genotoxicity of DEB, within the limits of statistical error.
One caveat about the role of GSH conjugation in the carcinogenicity of 1,3-butadiene is that monoepoxides (butadiene monoepoxide and 1,2-epoxy-3,4-dihydrobutane, the respective immediate oxidation product of 1,3-butadiene and the immediate hydrolysis product of DEB) can be conjugated with GSH and these pathways would be involved in detoxication. These two epoxides have low genotoxicity, however.56 Only the monoepoxide is an issue, because the diol epoxide would derive from DEB (which the mice were treated with). We did not measure the levels of adducts derived from the monoepoxide (Scheme 2) in our studies.
The mutation spectra arising from dibromoethane treatment showed an increase in GC to AT transitions (Figure 3). This finding is consistent with our previous work in bacteriophage M13mp1811 and yeast/human p5376 systems treated with S-(2-chloroethyl)GSH, the halogen analog of the dibromoethane half-mustard (Scheme 1). The results are probably not attributable to depurination, which would be expected to favor GC to TA transversions due to the “A Rule” for misinformational mispairing.77 The GC to AT transitions seen in the AGT-dibromoethane pathway in E. coli TGR8 cells65 were not apparently changed in mouse liver, but this result is consistent with the lack of an effect of O6-BzGua (i.e., lack of a significant role for the AGT pathway with dibromoethane in liver).
The increased AT to GC transitions seen with DEB (and attenuation by treatment with BSO or O6-BzGua) (Figure 4) are consistent with our finding that human DNA polymerase κ inserted dCTP opposite S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH.58 This finding (Figure 4) is also consistent with our previous report of enhanced AT to GC transitions in the rpoB gene of E. coli TRG8 cells treated with the DEB-GSH conjugate S-(2-hydroxy-3,4-epoxybutyl)GSH57 and the predominant A to G transitions in H-ras codon 61 in Harderian gland tumors of B6C3F1 mice treated with 1,3-butadiene by inhalation.78 However, increased AT to TA transversions were reported in the lacF gene in bone marrow of B6C3F1 mice treated with 1,3-butadiene.79 The incidence of this latter mutation was low in all of the DEB-derived mouse liver samples in our study (Figure 4). In all of the mutation studies, the contribution of phenotypic bias should not be discounted.
In summary, our results demonstrated that attenuation of GSH levels decreased hepatic levels of the major dibromoethane DNA adduct (N7-guanyl) and cII mutations in Big Blue® mice. We conclude that the GSH conjugation pathway has a major role in dibromoethane mutagenicity (and probably carcinogenicity) in vivo. AGT conjugation does not appear to play a major role. The results of similar attenuation studies lead to the conclusion that both the GSH and AGT conjugation pathways have major roles in the in vivo mutagenicity of DEB. The latter findings are considered to be of relevance to the carcinogenicity of 1,3-butadiene.
Supplementary Material
Acknowledgments
We thank K. Trisler for assistance in preparation of the manuscript and L. M. Folkmann for assistance with animal protocol submissions.
Funding
This work was supported in part by U.S. Public Health Service Grants R01 ES010546 and P30 ES000267 (F.P.G.).
ABBREVIATIONS
- AGT
O6-alkylguanine DNA-alkyltransferase
- O6-BzGua
O6-benzylguanine
- BSO
butathionine-S,R-sulfoximine
- DEB
(1,2,3,4-diepoxybutane
- DEB-GSH conjugate
S-(2-hydroxy-3,4-epoxybutyl)GSH
- dibromoethane
1,2-dibromoethane
- DTNB
5,5′-dithio-bis-(2-nitrobenzoic acid
- DTT
dithiothreitol
- ESI
electrospray ionization
- GSH
glutathione
- GST
GSH transferase
- N3A-(OH)2butyl-GSH
S-[4-(N3-adenyl)2,3-dihydroxybutyl)GSH
- IARC
International Agency for Cancer Research
- N6dA-(OH)2butyl-GSH
S-[4-(N6-deoxyadenosinyl)2,3-dihydroxybutyl)GSH
- N7G-(OH)2butyl-GSH
S-[4-(N7-guanyl)2,3-dihydroxybutyl)GSH
- N1dG-(OH)2butyl-GSH
S-[4-(N1-deoxyguanosinyl)2,3-dihydroxybutyl)GSH
- N4dC-(OH)2butyl-GSH
S-[4-(N4-deoxycytidinyl)2,3-dihydroxybutyl)GSH
- N3dT-(OH)2butyl-GSH
S-[4-(N3-thymidinyl)2,3-dihydroxybutyl)GSH
- N7G-DEB
N7-(2,3,4-trihydroxybutyl)guanine
- N3A-DEB
N3-(2,3,4-trihydroxybutyl)adenine
- N6A-DEB
N6-(2,3,4-trihydroxybutyl)adenine
- O6-MeGua
O6-methylguanine
- PBS
phosphate-buffered saline (10 mM potassium phosphate buffer, pH 7.4, containing 0.9% NaCl, w/v)
Footnotes
The authors declare no competing financial interest.
Measurements of GSH levels in livers of B6C3F1 and Big Blue® transgenic mice, gel electrophoresis imaging of AGT activity measurements, AGT activities in livers of B6C3F1 and Big Blue® transgenic mice at 6 and 24 h after treatment, quantitative analysis of DNA adducts in livers of Big Blue® transgenic mice, and independent mutations in the liver cII gene of Big Blue® transgenic mice. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wong LCK, Winston JM, Hong CB, Plotnick H. Carcinogenicity and toxicity of 1,2-dibromoethane in the rat. Toxicol Appl Pharmacol. 1982;63:155–165. doi: 10.1016/0041-008x(82)90036-9. [DOI] [PubMed] [Google Scholar]
- 2.Huff JE. 1,2-Dibromoethane (ethylene dibromide) Environ Health Perspect. 1983;47:359–363. doi: 10.1289/ehp.8347359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. Ethylene dibromide: worker exposure, use restricted. Chem Eng News. 1983;61:4. [Google Scholar]
- 4.Agency for Toxic Toxic Substances & Disease Registry. [accessed 26 Sept. 2013];Toxic Substances Portal - 1,2-Dibromoethane. http://www.atsdr.cdc.gov/toxprofiles/TP.asp?id=726&tid=131.
- 5. [accessed 24 Oct. 2013];Agents Classified by the IARC Monographs. 1–108 http://monographs.iarc.fr/ENG/Classification/ [Google Scholar]
- 6.Morrow NL. The industrial production and use of 1,3-butadiene. Environ Health Perspect. 1990;86:7–8. doi: 10.1289/ehp.90867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agency for Toxic Toxic Substances & Disease Registry. [accessed 24 Oct. 2013];Toxic Substances Portal-1,3-Butadiene. http://www.atsdr.cdc.gov/ToxProfiles/tp.asp?id=459&tid=81.
- 8.1,3-Butadiene. IARC Monographs, Evaluation of Carcinogenic Risk Chemicals to Humans. Int Agency Res Cancer. 1992 [PubMed] [Google Scholar]
- 9.Pelz N, Dempster NM, Shore PR. Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J Chromatogr Sci. 1990;28:230–235. doi: 10.1093/chromsci/28.5.230. [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum LS. A brief survey of butadiene health effects: a role for metabolic differences. Environ Health Perspect. 1993;101(Suppl 6):161–167. doi: 10.1289/ehp.93101s6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cmarik JL, Humphreys WG, Bruner KL, Lloyd RS, Tibbetts C, Guengerich FP. Mutation spectrum and sequence alkylation selectivity resulting from modification of bacteriophage M13mp18 with S-(2-chloroethyl)glutathione. Evidence for a role of S-[2-(N7-guanyl)ethyl]glutathione as a mutagenic lesion formed from ethylene dibromide. J Biol Chem. 1992;267:6672–6679. [PubMed] [Google Scholar]
- 12.Cho SH, Guengerich FP. Conjugation of butadiene diepoxide with glutathione yields DNA adducts in vitro and in vivo. Chem Res Toxicol. 2012;25:706–712. doi: 10.1021/tx200471x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury G, Cho S-H, Pegg AE, Guengerich FP. Detection and characterization of ethylene dibromide-derived DNA-crosslinks formed with O6-alkylguanine-DNA alkyltransferase. Angew Chem, Int Ed. 2013 doi: 10.1002/anie.201307580. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rannug U. Genotoxic effects of 1,2-dibromoethane and 1,2-dichloroethane. Mut Res. 1980;76:269–295. doi: 10.1016/0165-1110(80)90020-2. [DOI] [PubMed] [Google Scholar]
- 15.Ozawa N, Guengerich FP. Evidence for formation of an S-[2-(N7-guanyl)ethyl]glutathione adduct in glutathione-mediated binding of 1,2-dibromoethane to DNA. Proc Natl Acad Sci U S A. 1983;80:5266–5270. doi: 10.1073/pnas.80.17.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga N, Inskeep PB, Harris TM, Guengerich FP. S-[2-(N7-Guanyl)ethyl]glutathione, the major DNA adduct formed from 1,2-dibromoethane. Biochemistry. 1986;25:2192–2198. doi: 10.1021/bi00356a051. [DOI] [PubMed] [Google Scholar]
- 17.Peterson LA, Harris TM, Guengerich FP. Evidence for an episulfonium ion intermediate in the formation of S-[2-(N7-guanyl)ethyl]glutathione in DNA. J Am Chem Soc. 1988;110:3284–3291. [Google Scholar]
- 18.Kim DH, Guengerich FP. Formation of the DNA adduct S-[2-(N7-guanyl)ethyl]glutathione from ethylene dibromide: effects of modulation of glutathione and glutathione S-transferase levels and the lack of a role for sulfation. Carcinogenesis. 1990;11:419–424. doi: 10.1093/carcin/11.3.419. [DOI] [PubMed] [Google Scholar]
- 19.Csanády GA, Guengerich FP, Bond JA. Comparison of the biotransformation of 1,3-butadiene and its metabolite, butadiene monoepoxide, by hepatic and pulmonary tissues from humans, rats, and mice. Carcinogenesis. 1992;13:1143–1153. doi: 10.1093/carcin/13.7.1143. [DOI] [PubMed] [Google Scholar]
- 20.Duescher RJ, Elfarra AA. Human liver microsomes are efficient catalysts of 1,3-butadiene oxidation: evidence for major roles by cytochrome P450 2A6 and 2E1. Arch Biochem Biophys. 1994;311:342–349. doi: 10.1006/abbi.1994.1246. [DOI] [PubMed] [Google Scholar]
- 21.Bolt HM, Schmiedel G, Filser JG, Rolzhäuser HP, Lieser K, Wistuba D, Schurig V. Biological activation of 1,3-butadiene to vinyl oxirane by rat liver microsomes and expiration of the reactive metabolite by exposed rats. J Cancer Res Clin Oncol. 1983;106:112–116. doi: 10.1007/BF00395388. [DOI] [PubMed] [Google Scholar]
- 22.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: II. Mutational spectra of butadiene, 1,2-epoxybutene and diepoxybutane at the hprt locus in splenic T cells from exposed B6C3F1 mice. Carcinogenesis. 1994;15:719–723. doi: 10.1093/carcin/15.4.719. [DOI] [PubMed] [Google Scholar]
- 23.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2-butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- 24.Bolt HM. Butadiene and isoprene: future studies and implications. Toxicology. 1996;113:356–360. doi: 10.1016/0300-483x(96)03473-7. [DOI] [PubMed] [Google Scholar]
- 25.Jelitto B, Vangala RR, Laib RJ. Species differences in DNA dmage by butadiene: role of diepoxybutane. Arch Toxicol, Suppl. 1989;13:246–249. doi: 10.1007/978-3-642-74117-3_42. [DOI] [PubMed] [Google Scholar]
- 26.Kreiling R, Laib RJ, Filser JG, Bolt HM. Species differences in butadiene metabolism between mice and rats evaluated by inhalation pharmacokinetics. Arch Toxicol. 1986;58:235–238. doi: 10.1007/BF00297112. [DOI] [PubMed] [Google Scholar]
- 27.Bechtold WE, Strunk MR, Chang IY, Ward JB, Jr, Henderson RF. Species differences in urinary butadiene metabolites: comparisons of metabolite ratios between mice, rats, and humans. Toxicol Appl Pharmacol. 1994;127:44–49. doi: 10.1006/taap.1994.1137. [DOI] [PubMed] [Google Scholar]
- 28.Henderson RF, Thornton-Manning JR, Bechtold WE, Dahl AR. Metabolism of 1,3-butadiene: species differences. Toxicology. 1996;113:17–22. doi: 10.1016/0300-483x(96)03422-1. [DOI] [PubMed] [Google Scholar]
- 29.Humphreys WG, Kim DH, Cmarik JL, Shimada T, Guengerich FP. Comparison of the DNA alkylating properties and mutagenic responses caused by a series of S-(2-haloethyl)-substituted cysteine and glutathione derivatives. Biochemistry. 1990;29:10342–10350. doi: 10.1021/bi00497a008. [DOI] [PubMed] [Google Scholar]
- 30.Inskeep PB, Koga N, Cmarik JL, Guengerich FP. Covalent binding of 1,2-dihaloalkanes to DNA and stability of the major DNA adduct, S-[2-(N7-guanyl)ethyl]glutathione. Cancer Res. 1986;46:2839–2844. [PubMed] [Google Scholar]
- 31.Kim MS, Guengerich FP. Polymerase blockage and misincorporation of dNTPs opposite the ethylene dibromide-derived DNA adducts S-[2-(N7-guanyl)ethyl]glutathione, S-[2-(N2-guanyl)ethyl]glutathione, and S-[2-(O6-guanyl)ethyl]glutathione. Chem Res Toxicol. 1998;11:311–316. doi: 10.1021/tx970206m. [DOI] [PubMed] [Google Scholar]
- 32.Zhao C, Vodicka P, Sram RJ, Hemminki K. DNA adducts of 1,3-butadiene in humans: Relationships to exposure, GST genotypes, single-strand breaks, and cytogenetic end points. Environ Mol Mutag. 2001;37:226–230. doi: 10.1002/em.1031. [DOI] [PubMed] [Google Scholar]
- 33.Tretyakova NY, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem Res Toxciol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- 34.Citti L, Gervasi PG, Turchi G, Bellucci G, Bianchini R. The reaction of 3,4-epoxy-1-butene with deoxyguanosine and DNA in vitro: synthesis and characterization of the main adducts. Carcinogenesis. 1984;5:47–52. doi: 10.1093/carcin/5.1.47. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XY, Elfarra AA. Characterization of 1,2,3,4-diepoxybutane-2′-deoxyguanosine cross-linking products formed at physiological and nonphysiological conditions. Chem Res Toxicol. 2006;19:547–555. doi: 10.1021/tx0503395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tretyakova N, Sangaiah R, Yen TY, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: isolation and analysis in exposed calf thymus DNA. Chem Res Toxicol. 1997;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- 37.Selzer RR, Elfarra AA. Characterization of N1- and N6-adenosine adducts and N1-inosine adducts formed by the reaction of butadiene monoxide with adenosine: evidence for the N1-adenosine adducts as major initial products. Chem Res Toxicol. 1996;9:875–881. doi: 10.1021/tx960039a. [DOI] [PubMed] [Google Scholar]
- 38.Selzer RR, Elfarra AA. Chemical modification of deoxycytidine at different sites yields adducts of different stabilities: characterization of N3- and O2-deoxycytidine and N3-deoxyuridine adducts of butadiene monoxide. Arch Biochem Biophys. 1997;343:63–72. doi: 10.1006/abbi.1997.0164. [DOI] [PubMed] [Google Scholar]
- 39.Selzer RR, Elfarra AA. Synthesis and biochemical characterization of N1-, N2-, and N7-guanosine adducts of butadiene monoxide. Chem Res Toxicol. 1996;9:126–132. doi: 10.1021/tx950101o. [DOI] [PubMed] [Google Scholar]
- 40.Selzer RR, Elfarra AA. Characterization of four N-3-thymidine adducts formed in vitro by the reaction of thymidine and butadiene monoxide. Carcinogenesis. 1997;18:1993–1998. doi: 10.1093/carcin/18.10.1993. [DOI] [PubMed] [Google Scholar]
- 41.Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem Res Toxicol. 2010;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandes PH, Hackfeld LC, Kozekov ID, Hodge RP, Lloyd RS. Synthesis and mutagenesis of the butadiene-derived N3 2′-deoxyuridine adducts. Chem Res Toxicol. 2006;19:968–976. doi: 10.1021/tx060016o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: role of stereochemistry. J Am Chem Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- 44.Park S, Hodge J, Anderson C, Tretyakova N. Guanine-adenine DNA cross-linking by 1,2,3,4-diepoxybutane: potential basis for biological activity. Chem Res Toxicol. 2004;17:1638–1651. doi: 10.1021/tx0498206. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, Vodicka P, Sram RJ, Hemminki K. Human DNA adducts of 1,3-butadiene, an important environmental carcinogen. Carcinogenesis. 2000;21:107–111. doi: 10.1093/carcin/21.1.107. [DOI] [PubMed] [Google Scholar]
- 46.Tretyakova N, Chiang SY, Walker VE, Swenberg JA. Quantitative analysis of 1,3-butadiene-induced DNA adducts in vivo and in vitro using liquid chromatography electrospray ionization tandem mass spectrometry. J Mass Spectrom. 1998;33:363–376. doi: 10.1002/(SICI)1096-9888(199804)33:4<363::AID-JMS643>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Koc H, Tretyakova NY, Walker VE, Henderson RF, Swenberg JA. Molecular dosimetry of N-7 guanine adduct formation in mice and rats exposed to 1,3-butadiene. Chem Res Toxicol. 1999;12:566–574. doi: 10.1021/tx980265f. [DOI] [PubMed] [Google Scholar]
- 48.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanuri M, Nechev LV, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Mutagenic spectrum of butadiene-derived N1-deoxyinosine adducts and N6,N6-deoxyadenosine intrastrand cross-links in mammalian cells. Chem Res Toxicol. 2002;15:1572–1580. doi: 10.1021/tx025591g. [DOI] [PubMed] [Google Scholar]
- 50.Kotapati S, Maddukari L, Wickramaratne S, Seneviratne U, Groggia M, Pence MG, Villalta P, Guengerich FP, Marnett LJ, Tretyakova N. Translesion synthesis across 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2′-deoxyadenosine (1,N6-γ-HMHP-dA) adducts by human and archebacterial DNA polymerases. J Biol Chem. 2012;287:38800–38811. doi: 10.1074/jbc.M112.396788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thier R, Pemble SE, Taylor JB, Humphreys WG, Persmark M, Ketterer B, Guengerich FP. Expression of mammalian glutathione S-transferase 5–5 in Salmonella typhimurium TA1535 leads to base-pair mutations upon exposure to dihalomethanes. Proc Natl Acad Sci U S A. 1993;90:8576–8580. doi: 10.1073/pnas.90.18.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheever KL, Cholakis JM, El-Hawari AM, Kovatch RM, Weisburger EK. Ethylene dichloride: the influence of disulfiram or ethanol on oncogenicity, metabolism, and DNA covalent binding in rats. Fund Appl Toxicol. 1990;14:243–261. doi: 10.1016/0272-0590(90)90205-x. [DOI] [PubMed] [Google Scholar]
- 53.Guengerich FP, Kim DH, Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 54.Thier R, Müller M, Taylor JB, Pemble SE, Ketterer B, Guengerich FP. Enhancement of bacterial mutagenicity of bifunctional alkylating agents by expression of mammalian glutathione S-transferase. Chem Res Toxicol. 1995;8:465–472. doi: 10.1021/tx00045a019. [DOI] [PubMed] [Google Scholar]
- 55.Thier R, Pemble S, Kramer H, Taylor JB, Guengerich FP, Ketterer B. Human glutathione S-transferase T1–1 enhances mutagenicity of 1,2-dibromoethane, dibromomethane, and 1,2,3,4-diepoxybutane in Salmonella typhimurium. Carcinogenesis. 1996;17:163–166. doi: 10.1093/carcin/17.1.163. [DOI] [PubMed] [Google Scholar]
- 56.Cho SH, Loecken EM, Guengerich FP. Mutagenicity of a glutathime conjugate of butadiene diepoxide. Chem Res Toxicol. 2010;23:1544–1546. doi: 10.1021/tx100304f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho SH, Guengerich FP. Mutation spectra of S-(hydroxy-3,4-epoxybutyl)glutathione: Comparison with 1,3-butadiene and its metabolites in the Escherichia rpoB gene. Chem Res Toxciol. 2012;25:1522–1530. doi: 10.1021/tx3002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho SH, Guengerich FP. Replication past the butadiene diepoxide-derived DNA adduct S-[4-(N6-deoxyadenosinyl)2,3-dihydroxybutyl]glutathione by DNA polymerases. Chem Res Toxicol. 2013;26:1005–1013. doi: 10.1021/tx400145e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abril N, Luque-Romero FL, Prieto-Alamo MJ, Rafferty JA, Margison GP, Pueyo C. Bacterial and mammalian DNA alkyltransferases sensitize Escherichia coli to the lethal and mutagenic effects of dibromoalkanes. Carcinogenesis. 1997;18:1883–1888. doi: 10.1093/carcin/18.10.1883. [DOI] [PubMed] [Google Scholar]
- 60.Abril N, Ferrezuelo F, Prieto-Alamo MJ, Rafferty JA, Margison GP, Pueyo C. Contribution of ogt-encoded alkyltransferase to resistance to chloroethylnitrosoureas in nucleotide excision repair-deficient Escherichia coli. Carcinogenesis. 1996;17:1609–1614. doi: 10.1093/carcin/17.8.1609. [DOI] [PubMed] [Google Scholar]
- 61.Liu L, Pegg AE, Williams KM, Guengerich FP. Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2002;277:37920–37928. doi: 10.1074/jbc.M205548200. [DOI] [PubMed] [Google Scholar]
- 62.Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Human O6-alkylguanine-DNA alkyltransferase activation of a series of bis-electrophiles to produce mutagens. Chem Res Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 63.Kalapila AG, Pegg AE. Alkyltransferase-mediated toxicity of bis-electrophiles in mammalian cells. Mut Res. 2010;684:35–42. doi: 10.1016/j.mrfmmm.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guengerich FP, Fang Q, Liu L, Hachey DL, Pegg AE. O6-Alkylguanine-DNA alkyltransferase: low pKa and high reactivity of cysteine 145. Biochemistry. 2003;42:10965–10970. doi: 10.1021/bi034937z. [DOI] [PubMed] [Google Scholar]
- 65.Liu L, Hachey DL, Valadez JG, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- 66.Kim DH, Humphreys WG, Guengerich FP. Characterization of S-[2-(N1-adenyl)ethyl]glutathione formed in DNA and RNA from 1,2-dibromoethane. Chem Res Toxicol. 1990;3:587–594. doi: 10.1021/tx00018a015. [DOI] [PubMed] [Google Scholar]
- 67.Sedlak J, Lindsay RH. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman’s reagent. Anal Biochem. 1968;25:192–205. doi: 10.1016/0003-2697(68)90092-4. [DOI] [PubMed] [Google Scholar]
- 68.Zang H, Fung Q, Pegg AE, Guengerich FP. Kinetic analysis of steps in the repair of damaged DNA by O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 69.Cmarik JL, Inskeep PB, Meyer DJ, Meredith MJ, Ketterer B, Guengerich FP. Selectivity of rat and human glutathione S-transferases in activation of ethylene dibromide by glutathione conjugation and DNA binding and induction of unscheduled DNA synthesis in human hepatocytes. Cancer Res. 1990;50:2747–2752. [PubMed] [Google Scholar]
- 70.Chae MY, McDougall MG, Dolan ME, Swenn K, Pegg AE, Moschel RC. Substituted O6-benzylguanine derivatives and their inactivation of human O6-alkylguanine-DNA alkyltransferase. J Med Chem. 1994;37:342–347. doi: 10.1021/jm00029a005. [DOI] [PubMed] [Google Scholar]
- 71.Dolan ME, Chae MY, Pegg AE, Mullen JH, Friedman HS, Moschel RC. Metabolism of O6-benzylguanine, an inactivator of O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1994;54:5123–5130. [PubMed] [Google Scholar]
- 72.Rannug U, Beije B. The mutagenic effect of 1,2-dichloroethane on Salmonella typhimurium II Activation by the isolated perfused rat liver. Chem-Biol Interact. 1979;24:265–285. doi: 10.1016/0009-2797(79)90077-2. [DOI] [PubMed] [Google Scholar]
- 73.Rannug U, Sundvall A, Ramel C. The mutagenic effect of 1,2-dichloroethane on Salmonella typhimurium I Activation through conjugation with glutathione in vitro. Chem-Biol Interact. 1978;20:1–16. doi: 10.1016/0009-2797(78)90076-5. [DOI] [PubMed] [Google Scholar]
- 74.Wheeler JB, Stourman NV, Armstrong RN, Guengerich FP. Conjugation of haloalkanes by bacterial and mammalian glutathione transferases: Mono- and vicinal dihaloethanes. Chem Res Toxicol. 2001;14:1107–1117. doi: 10.1021/tx0100183. [DOI] [PubMed] [Google Scholar]
- 75.Kim MY, Tretyakova N, Wogan GN. Mutagenesis of the supF gene by stereoisomers of 1,2,3,4-diepoxybutane. Chem Res Toxicol. 2007;20:790–797. doi: 10.1021/tx700003b. [DOI] [PubMed] [Google Scholar]
- 76.Valadez JG, Guengerich FP. S-(2-Chloroethyl)glutathione-generated p53 mutation spectra are influenced by differential repair rates more than sites of initial DNA damage. J Biol Chem. 2004;279:13435–13446. doi: 10.1074/jbc.M312358200. [DOI] [PubMed] [Google Scholar]
- 77.Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 78.Goodrow TL, Nichols WW, Storer RD, Anderson MW, Maronpot RR. Activation of H-ras is prevalent in 1,3-butadiene-induced and spontaneously occurring murine Harderian gland tumors. Carcinogenesis. 1994;15:2665–2667. doi: 10.1093/carcin/15.11.2665. [DOI] [PubMed] [Google Scholar]
- 79.Sisk SC, Pluta LJ, Bond JA, Recio L. Molecular analysis of lacI mutants from bone marrow of B6C3F1 transgenic mice following inhalation exposure to 1,3-butadiene. Carcinogenesis. 1994;15:471–477. doi: 10.1093/carcin/15.3.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.