Abstract
Transforming growth factor-β1 (TGF-β1) is a multifunctional growth factor that has profound regulatory effects on many developmental and physiological processes. Disruption of the TGF-β1 gene by homologous recombination in murine embryonic stem cells enables mice to be generated that carry the disrupted allele. Animals homozygous for the mutated TGF-β1 allele show no gross developmental abnormalities, but about 20 days after birth they succumb to a wasting syndrome accompanied by a multifocal, mixed inflammatory cell response and tissue necrosis, leading to organ failure and death. TGF-β1-deficient mice may be valuable models for human immune and inflammatory disorders, including autoimmune diseases, transplant rejection and graft versus host reactions.
Transforming growth factor-β1 is the prototypic member of a family of structurally related polypeptides referred to as the transforming growth factor superfamily. Members of this family exhibit a variety of proliferative, inductive and regulatory properties, and include the TGF-βs, activins, inhibins, bone morphogenetic proteins, mullerian inhibiting substance, Drosophila decapentaplegic gene complex, and Xenopus Vg-l gene1-3 TGF-β1 is synthesized as a precursor polypeptide containing a hydrophobic signal sequence, pro-region and mature peptide. The biologically active growth factor is a disulphide-linked homodimer, with each monomer representing the carboxy-terminal 112 amino acids of the precursor which are cleaved from the amino-terminal glycopeptide at a tetrabasic cleavage site1-3.
Three TGF-β isoforms, termed TGF-β1, TGF-β2 and TGF-β3, have been identified in mammals. These isoforms exhibit both overlapping and distinct spatial and temporal patterns of expression throughout development. Pronounced embryonic expression of the TGF-βs in areas undergoing morphogenetic events, particularly those involving epithelial-mesenchymal interactions, suggests that these molecules playa critical role during embryonic development4,5. The large number of cells and tissues that express embryonic TGF-β1 messenger RNA or protein are described in refs 6-11. In the adult, TGF-β1 immunoreactivity is detected in many cell types12.
TGF-β1 elicits diverse cellular responses depending on cell type, state of differentiation and culture conditions1-3 Thus, TGF-β1 can have either stimulatory or inhibitory effects on the same cell, depending on the cellular environment1-3. Biological actions of TGF-β1 include regulation of cell proliferation, control of extracellular matrix protein production and degradation, and modulation of cellular differentiation1-3. In vitro studies with TGF-β1 have demonstrated inhibition of adipogenesis and myogenesis, inhibition of haematopoietic progenitor cell proliferation, promotion of angiogenesis, stimulation of chondrogenesis and osteogenesis, induction of bronchial epithelial cell differentiation, inhibition of intestinal epithelial cell growth, inhibition of keratinocyte proliferation, and modulation of numerous immune and inflammatory responses1-3. The relevance of these multiple activities to normal development and functioning of the whole animal are unknown.
To investigate the role of TGF-β1 in vivo, we disrupted one copy of the TGF-β1 gene in murine embryonic stem (ES) cells by homologous recombination and generated chimaeric mice carrying the disrupted TGF-β1 gene in the germ line. About 20 days after birth, animals homozygous for the mutated TGF-β1 allele exhibit an acute wasting syndrome followed by death. No gross developmental abnormalities are detected. The most prominent lesions observed are tissue necrosis in specific organs and multifocal, mixed inflammatory cell infiltration into numerous organs, particularly heart and stomach.
TGF-β1 gene targeting
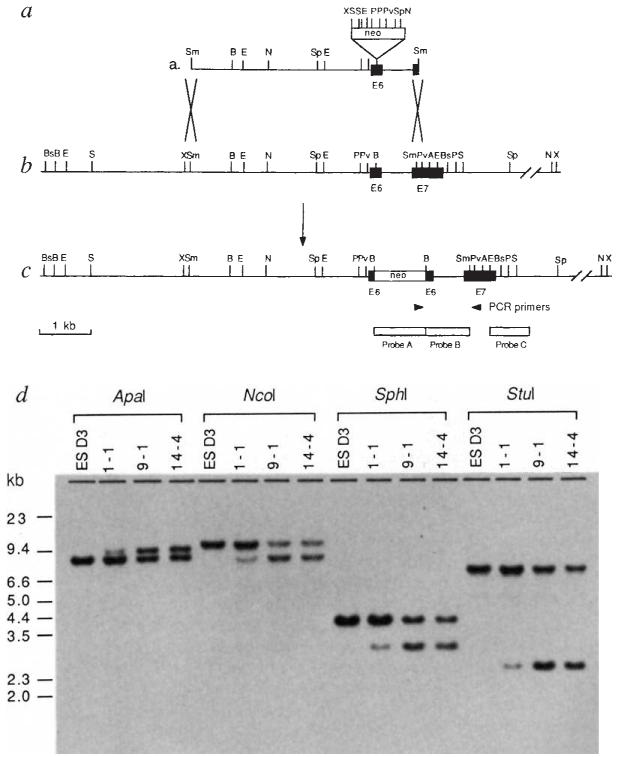
To ensure that strain differences between the ES cell and targeting DNA did not affect targeting efficiency, the murine TGF-β1 gene was isolated from a genomic library constructed with DNA isolated from a 129/SvJ mouse (Aw/Aw cch/c p/p), which is coisogenic to the 129/Sv strain (Aw/Aw+c/+c+P/+P) from which the D3 ES cell line13 used here was derived. The targeting strategy was designed to effect disruption of the mature peptide, while leaving the precursor pro-region intact. The targeting vector consisted of 4.0 kb of genomic TGF-β1 sequence containing a neor gene inserted into exon 6 (Fig. 1). The purified targeting fragment was introduced into D3 cells by electroporation, and the cells were grown in the presence of G418, which selects for neomycin resistance, for 12–14 days. ES cells containing a disrupted TGF-β1 gene were identified by polymerase chain reaction (PCR) and Southern blot hybridization of the PCR products using a TGF-β1 oligonucleotide probe spanning the 3′ insertion site. Four individual PCR-positive clones were obtained from a total of 700 G418-resistant colonies. Genomic Southern blot analysis using a TGF-β1 genomic fragment as probe (probe B; Fig. 1) confirmed that three of these clones contained the desired homologous recombination event, yielding an overall targeting efficiency of 1 in 2 × 106 electroporated cells. As shown in Fig. 1, cell lines 1-1, 9-1, and 14-4 were heterozygous, containing one copy of the disrupted TGF-β1 allele and one copy of the normal allele. Analysis of these cell lines using five additional enzymes and using both a neo probe (probe A; Fig. 1) and a probe external to the targeting fragment (probe C; Fig. 1) also confirmed that accurate targeting had occurred (data not shown).
FIG. 1.
Targeted disruption of the murine TGF-β1 gene in ES cells. a, Targeting construct consisting of a TGF-β1 4.0-kb Smal genomic fragment containing exon 6 and part of exon 7. A neor gene lacking the polyadenylation signal (XhoI-SaI1 fragment from pMC1 neo; ref. 52) was inserted into the BamHI site in exon 6 of TGF-β1, 102 nucleotides (34 amino acids) from the N terminus of the mature peptide, b, Restriction map of the wild-type TGF-β1 genomic locus surrounding the targeting site. e, Predicted structure of the disrupted TGF-β1 allele. Arrowheads represent positions of primers used for PCR analysis. The 5′ primer (5′-GCTTTACGGTATCGCCGCTC-3′) is located 68 nucieotides upstream of the stop codon in neo. The 3′ primer (5′-TGCGACCCACGT AGT AGACG-3′) is located 74 nucleotides down-stream of the Sma I site in exon 7 of TGF-β1 and is not contained in the targeting vector. The locations of probes used in Southern analysis are shown. Probe A is from the neor gene (XhoI-SaI1 fragment from pMC1 neo). Probe B is a 0.6-kb TGF-β1 genomic fragment (BamHI-SmaI). Probe C is a 0.7-kb TGF-β1 genomic fragment (EcoRI) not contained in the targeting vector. Restriction enzymes used in diagnostic digests are: A, Apal; Bs, BstI; E, EcoRI; N, NcoI; P, PstI; Pv, PvuII; Sp, SphI; S, StuI; and X, XbaI. d, Southern blot analysis of parental D3 embryonic stem (ESD3) cells and three PCR-positive clones. Genomic DNAs (15 μg) were digested with the enzymes shown, electrophoresed through 0.8% agarose, transferred to nylon membranes, and hybridized with probe B. The observed patterns correspond to those expected from homologous recombination of the targeting vector into the TGF-β1 locus. The difference in intensity of the bands in the digests of clone 1–1 is due to the fact that this clone contained a mixed population of targeted and untransfected cells.
METHODS. D3 embryonic stem (6×106 cells) were electroporated with 10 μg (6 nM) of purified targeting fragment using an IBI Gene Zapper at 800 V cm−1 and 200 μF. Cells were plated on mitomycin-treated neor mouse embryonic fibroblast feeder layers. After 24 h with no selection, 400 μg ml−1 G418 was added. After further 20 h, the medium was changed to 200 μg ml−1 G418 and cells were grown in this medium for 12–14 days. G418-resistant colonies (initially in pools and then individual colonies from PCR-positive pools) were analysed for homologous recombination by PCR. Targeting was confirmed by genomic Southern blot analysis.
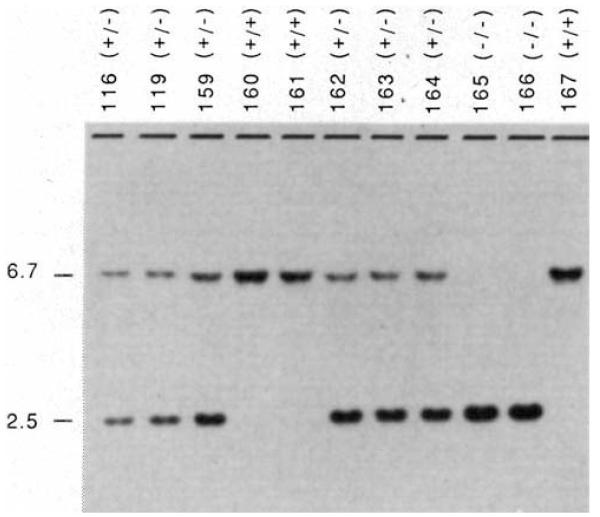
Chimaeric mice were generated by injecting ES cells from lines 9-1 and 14-4 into C57BL/6J blastocysts and implanting the blastocysts into the uteri of pseudopregnant F1 (C3H/HeN × C57BL/6J) recipients. Ten fertile coat colour chimaeric males were obtained, five of which exhibited >50% agouti coat colour. Mating of these five animals to strain CF1 females allowed identification of three animals that transmitted the ES cell-derived agouti coat colour to their offspring. Two of these males have produced a total of 18 litters and have transmitted the modified TGF-β1 allele to 50% of their agouti offspring, as expected. Heterozygous mice carrying one wild-type and one disrupted TGF-β1 allele are phenotypically normal and fertile. Heterozygous animals derived from both cell lines have been intercrossed to generate animals homozygous for the altered TGF-β1 allele. Genotypes of the progeny were determined by Southern blot analysis of tail DNA biopsies taken at roughly three weeks of age. A representative example of such a litter is shown in Fig. 2. Of 117 offspring from 14 heterozygote crosses, 34 were homozygous wild-type, 67 were heterozygous, and 16 were homozygous for the disrupted allele. A statistically significant (P < 0.005) deviation from the expected 1:2:1 ratio of homozygous wild-type, heterozygous, and homozygous mutant progeny was observed, because of an under-representation of the homozygous mutant class of animals. Here we report analyses of live homozygous mutant animals derived from targeted cell line 14-4. Homozygous mutant animals derived from cell line 9-1 exhibited the same phenotype as those derived from line 14-4.
FIG. 2.
Genotype of offspring from interbreeding mice heterozygous for the targeted TGF-β1 allele. Tail DNAs from parents (116 and 119) and offspring (159–167) were digested with StuI, fractionated by electrophoresis through 0.8% agarose, transferred to nylon membranes, and hybridized with a TGF-β1 probe (probe B; Fig. 1). The upper band (6.7 kb) represents the wild-type allele and the lower band (2.5 kb) represents the targeted allele. +/+, Homozygous wild-type; +/−, heterozygote; −/−, homozygous targeted.
Gross phenotype of homozygous mutants
Mice homozygous for the disrupted TGF-β1 allele were phenotypically indistinguishable from wild-type or heterozygous littermates until roughly three weeks after birth. At this time, all homozygotes exhibited severe wasting followed by death (or if they seemed so ill that survival until the next day was doubtful, they were killed). In some animals, wasting was accompanied by dishevelled appearance, hunched posture, conjunctivitis, and skin irritation. The age at which symptoms first appeared was variable, ranging from 17 to 34 days. Generally, progression of the illness was rapid, with initial onset of symptoms to death lasting only a few days. In one animal, number 181, the rapidity of the illness was dramatic, with no indication of illness before the day the animal died. Gross anatomical examination of animals homozygous for the mutant TGF-β1 allele revealed no apparent developmental abnormalities. But in most mutant animals, the spleens were smaller and Peyer’s patches less prevalent than those of normal littermates. This phenotype was not observed in any homozygous wild-type or heterozygous littermates, nor was it seen in any of 55 progeny resulting from interbreeding agouti animals which were homozygous wild-type at the TGF-β1 locus, that is, they had received the ES cell-derived unmodified TGF-β1 allele. These observations indicate that the mutant phenotype was due to homozygosity of the targeted TGF-β1 gene, and not to homozygosity of other D3 cell genes.
Histopathology
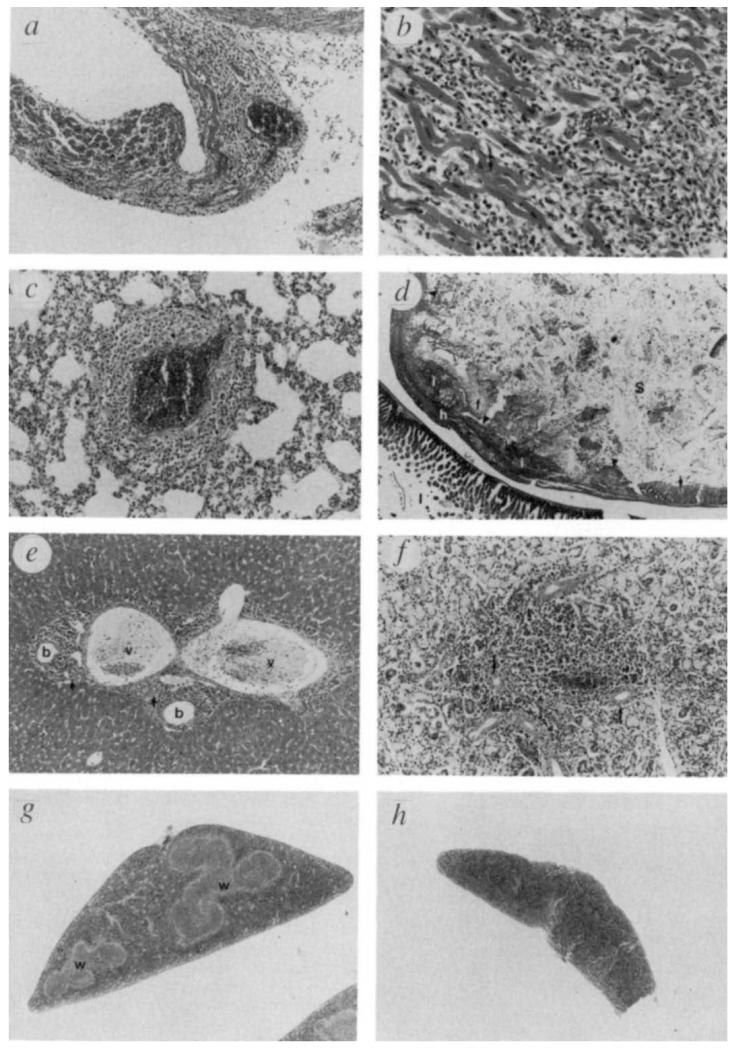
Histological analyses were performed on 15 homozygous mutant animals and six normal wild-type or heterozygous littermate controls (Table 1. Whereas the normal littermate controls exhibited no remarkable histopathology, all homozygous mutant animals exhibited a marked but variable degree of mixed inflammatory cell infiltration and tissue necrosis in several organs. The main organs involved were heart, stomach, liver, lung, pancreas, salivary gland and striated muscle. In liver, pancreas and salivary gland, the inflammatory cell infiltration was primarily periductal. The stomach inflammation appeared to be primarily granulocytic (−60% neutrophils and some eosinophils), whereas heart, lung, pancreas, striated muscle, salivary gland and brain infiltrates appeared primarily (−80%) lymphocytic and plasmacytic. In liver, granulocytes and lymphocytes appeared to be present in roughly equal numbers. Representative examples of affected tissues are shown in Fig. 3.
TABLE 1.
Histological analysis of TGF-β1-deficient mice
| Mouse | Control*† | 126 | 137 | 147 | 165 | 166 | 181 | 200 | 208 | 254 | 258 | 260 | 366 | 374 | 379 | 397 |
| Genotype | +/+,+/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| Sex | F,M | F | F | F | M | F | M | F | F | M | F | M | M | F | F | M |
| Age at death (days) | 21-37† | 29† | 31† | 21 | 25† | 22 | 27 | 19† | 20† | 21† | 34† | 21† | 23 | 23† | 23† | 21† |
| Weight at death (g) | 11-22 | ND | ND | ND | 8.0 | 7.0 | 12.0 | 6.0 | 70 | 8.0 | 9.8 | 11.8 | 9.1 | 7.5 | 7.9 | 8.5 |
| Heart (inflammation) | ||||||||||||||||
| Pericardium | − | +/− | − | − | − | + | ++ | − | − | ++ | ++++ | + | + | + | − | + |
| Myocardium | ||||||||||||||||
| Atria | − | +/− | + | + | + | + | +++ | +/− | + | ++ | ++++ | + | +++ | ++ | ++ | ++ |
| Ventricles | − | − | − | − | + | − | +++ | − | − | ++ | ++ | + | + | + | − | + |
| Endocardium | − | +/− | − | + | + | − | +++ | − | +/− | + | + | + | +/− | ++ | + | |
| Lung | ||||||||||||||||
| Perivasculitis | − | − | − | − | − | − | + | + | ++ | +/− | ++ | + | + | ++/− | +/− | |
| Perivascular oedema | − | − | − | − | − | − | +/− | − | − | − | − | − | − | − | − | − |
| Stomach | ||||||||||||||||
| Inflammation | − | +++ | ++++ | ++++ | ++++ | ++ | ++ | +++ | − | ++ | ++ | ++ | pmd | +/− | +++ | ++++ |
| Ulcer | − | +++ | ++++ | ++++ | ++++ | − | pmd | +++ | − | − | − | − | pmd | − | +++ | + |
| Alcanthosis | − | ++ | ++ | ++ | ++ | + | + | ++ | − | ++ | ++++ | +++ | pmd | +/− | +/− | ++++ |
| Hyperplasia | − | − | − | − | ++ | ++ | − | ++ | − | +++ | ++ | +++ | pmd | +/− | + | +++ |
| Liver | ||||||||||||||||
| Inflammation | − | +++ | ++ | + | +++ | + | ++ | +++ | + | ++/− | + | ++/− | + | + | ++/− | ++ |
| Microgranulomas | − | ++ | ++ | − | − | + | − | ++ | − | − | − | − | − | − | − | |
| Pancreas (inflammation) | − | − | + | +/− | +++ | − | − | − | ++ | ++ | ND | ++ | pmd | − | +++ | + |
| Salivary gland (inflammation) | − | − | − | − | +/− | − | ++ | − | − | − | + | ++ | + | − | +/− | ++/− |
| Spleen | ||||||||||||||||
| Size smaller | N | V | Y | Y | N | Y | Y | ND | Y | N | Y | Y | pmd | Y | Y | N |
| Indistinct white pulp | N | Y | Y | Y | N | Y | Y | ND | Y | N | Y | Y | pmd | N | N | Y/N |
| Lymph nodes | ||||||||||||||||
| Mediastinal | − | − | Ig | Ig | − | − | − | − | − | − | − | − | Ig | − | − | Ig |
| Mandibular | − | − | − | Ig | − | − | − | Ig | − | − | − | Ig | − | Ig | Ig | − |
| Mesenteric | − | Ig | − | Ig | − | − | − | Ig | − | Ig | − | − | − | − | Ig | − |
| Peyer’s patches‡ | 4-14 | 1 | 2 | pmd | 2 | 7 | pmd | 2 | 0 | 4 | 2 | 3 | pmd | 2 | 8 | 3 |
| Brain (inflammation) | − | ND | ND | − | − | − | − | +/− | ND | − | +/− | − | − | − | − | |
| Eye (inflammation) | ||||||||||||||||
| Conjunctiva | − | ND | ND | − | ND | + | ND | ND | ND | ++ | − | +/− | ND | − | ND | + |
| Lacrimal glands | − | ND | ND | − | ND | − | ND | ND | ND | − | − | − | ND | − | ND | ++ |
| Striated muscle | − | ND | ND | − | ND | − | ND | ND | ND | ++ | − | + | ND | − | ND | + |
| Striated muscle (inflammation) | − | − | ND | − | + | − | + | ND | − | + | + | ++ | ND | ND | +++ | +++ |
| Serosa (inflammation) | − | +/− | ND | ND | ND | +/− | ND | ND | ++ | +++ | + | ++ | pmd | ++ | − | +/− |
| Haematocrit§ | 24-50 | ND | ND | ND | ND | ND | ND | ND | ND | 27 | 52 | 24 | ND | 46 | 30 | 35.5 |
Degree of effect is indicated by – (no difference from control) to ++++ (large effect). N, no; Y, yes; Ig, large; ND, not done: pmd, post mortem degeneration.
Summary of results from three homozygous wild-type (3 females, aged 23, 29, and 31 days) and three heterozygous normal (1 female, 34 days; 2 males, 21 days) littermates.
Animals were killed.
Number of Peyer’s patches/total length of intestine
Leukocyte haematology is given in Table 2.
FIG. 3.
Histological analysis of TGF-β1 deficient mice. a, Heart, atrium, mouse 181, ×74. Myocarditis and pericarditis (arrow). The myocardium is infiltrated with inflammatory cells in all areas of the heart. Single cell to focal myocyte necrosis is evident. b, Heart, higher magnification (×148) ventricle, mouse 181. Inflammatory cells consist mostly of lymphocytes, but neutrophils, macrophages, and plasma cells are also present. Individual myocyte necrosis is evident (arrow). C, Lung, mouse 181, ×93, Perivasculitis. Inflammatory cells surround a vessel wall, with some oedema. The types and proportions of cells are similar to those seen in the heart. d, Stomach, gut roll, mouse 137, ×9. Severe, generalized necrosis (ulceration) of the squamous epithelium in the junction between the glandular (arrow) and nonglandular (arrowheads) portion. The submucosa is filled with mixed acute inflammatory cells (i), particularly neutrophils, and fibrin (f), with focal hemorrhage (h). Stomach lumen (S); intestine (I). e, Liver, mouse 126, ×46. Typical pattern of acute inflammatory cell infiltration in the portal triads of the liver. Infiltrating cells are most concentrated near bile ducts and consist mostly of lymphocytes and neutrophils. Bile ducts (b); portal veins (v); arteries (arrows). f, Salivary gland, mutant 181, ×74. Typical focal area of inflammatory cell infiltration seen particularly around ductal epithelium (arrows) of salivary glands and pancreas of some mutants. The inflammation is primarily lymphocytic. g, Spleen from normal littermate, 128, ×13. White pulp areas (w) are large and clearly delineated. h, Spleen from mutant 147, ×13. The white pulp areas are not detectable, and the entire spleen is smaller in cross-section. Tissues were prepared, sectioned, and stained with haematoxylin and eosin according to standard procedures.
The heart was affected in most homozygous mutant animals examined, although the degree of inflammation and necrosis was variable. In several cases, the presence of inflammatory cells was generalized and extensive, involving the pericardium, myocardium and endocardium of atria and ventricles (Fig. 3a and b). Striated muscle (masseter, leg and/or diaphragm) was examined in 11 mutant animals; eight of these exhibited some inflammation. In two animals (379 and 397), inflammation and necrosis of the diaphragm were severe enough to contribute to breathing difficulty and possibly death. A generalized mild to moderate perivasculitis was observed in the lungs of 10 of 15 mutant animals (Fig. 3c).
The non-glandular portion of the stomach exhibited some degree of acanthosis and inflammatory cell infiltration into the submucosa in 13 of 14 homozygous mutant animals in which this organ could be examined. Severe focal ulceration (seven animals) and/or hyperplasia of the basal epithelial layer (nine animals) accompanied the inflammation. As shown in Fig. 3d, severe, extensive ulceration was observed in the junction between the glandular and nonglandular portion of the stomach and the submucosa was filled with mixed inflammatory cells, fibrin and focal haemorrhage.
Inflammatory cell infiltration in the portal triad area of the liver, with cells most concentrated near the bile ducts, was observed in all 15 mutant animals (Fig. 3e). Some animals also exhibited multifocal hepatic necrosis and microgranulomas. Mild to moderate inflammation of the serosa of internal organs (stomach, intestine, kidney, ovary and testis) was seen in several animals. In 8 of 13 animals, some degree of multifocal inflammation was observed in the pancreas, and in 7 of 15 animals, a slight to moderate multifocal inflammatory cell infiltration in the salivary gland was detected (Fig. 3f). None of the mutant animals exhibited any kidney inflammation other than that of the renal serosa.
Histological analysis of lymphoid organs revealed that, compared with normal littermates, 10 of 15 homozygous mutant animals had slightly enlarged lymph nodes and 10 of 13 animals had spleens that were smaller and with less distinct white pulp (Fig. 3g and h). Peyer’s patches were usually fewer and possessed less distinct germinal centres than those of normal littermates. The thymus of mutant animals exhibited no significant lesions in the sections examined.
As one of the initial external symptoms in the homozygous mutant animals was conjunctivitis, the eyes of seven mutant animals were examined histologically, revealing conjunctivitis in four animals, inflammation of ocular striated muscle in three, and lacrimal gland inflammation in one. A slight focal inflammation in the brain choroid was observed in two animals.
Inflammation often accompanies infection and the inflammatory cell infiltration in some of the homozygous mutant animals resembled that seen in certain murine bacterial or viral diseases. However, the mutant phenotype was not observed in any wild-type or heterozygous littermates. Furthermore, mice were maintained in microisolator cages in a clean facility, with sentinel animals present for detection of antibodies against 14 murine bacterial and viral pathogens. Periodical comprehensive serology and histopathology revealed no indication of pathogens in the sentinels. Serum samples from two heterozygous animals producing litters containing homozygous mutant animals were assayed for the presence of antibodies against nine common murine bacterial and viral pathogens, including mouse hepatitis virus; no such antibodies were detected. Blood, liver, lung and fluid from intraperitoneal lavage from two mutants and one control animal were cultured in Columbia paediatric broth and analysed for the presence of gram-negative bacteria. No significant bacterial pathogens were detected. In addition, a C57BL/6 mouse doubly homozygous for the scid (severe combined immune deficiency) mutation14 and a non-responder allele at the Lps (lipopolysaccharide response) locus15 has been housed with the germ-line chimaera for over a month and has remained healthy. These results suggest that the phenotype observed in homozygous mutant animals is not due to primary infection by common mouse pathogens, although the possibility of secondary bacterial or viral involvement in the mutant phenotype cannot be eliminated completely.
Haematology
Peripheral blood samples from 9 homozygous mutants and an equal number of age-, sex- and parent-matched homozygous wild-type or heterozygous controls were analysed for total numbers and differential distributions of leukocytes. In mutant animals, the average number of white blood cells was usually elevated compared with controls, the increase being due to greater absolute numbers of neutrophils and monocytes. Even in this small group of mice, a statistically significant increase in monocytes and immature forms of neutrophils was observed (Table 2), indicating that a responsive left shift was present in the mutant mice. Haematocrits, determined for four control and six mutant animals, revealed no significant differences between the mutants and controls (Table 1).
TABLE 2.
Total numbers and differential distributions of blood leukocytes
| Control mice* | Mutant mice | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total WBC (per mm3) |
Neutrophils | Total WBC (per mm3) |
Neutrophils | ||||||||
| Mouse† | Lymphocytes | Total | Immature | Monocytes | Mouse | Lymphocytes | Total | Immature | Monocytes | ||
| 362F 25(+/+) | 2,400 | 1,896(79) | 432(18) | 0(0) | 24(1) | 166F 22(−/−) | 1,600 | 800(50) | 368(23) | 96(6) | 384(24) |
| 451F 20(ND) | 2,500 | 1,325(53) | 950(38) | 100(4) | 175(7) | 208F 20(−/−) | 1,300 | 715(55) | 429(33) | 39(3) | 156(12) |
| 447M 20(ND) | 2,800 | 1,568(56) | 952(34) | 84(3) | 196(7) | 254M 21(−/−) | 2,000 | 1,180(59) | 400(20) | 100(5) | 320(16) |
| 455F 19(ND) | 2,300 | 1,679(73) | 506(22) | 92(4) | 69(3) | 260F 21(−/−) | 2,500 | 1,650(66) | 600(24) | 250(10) | 225(9) |
| 377F 23(+/+) | 1,600 | 1,072(67) | 384(24) | 64(4) | 144(9) | 374F 23(−/−) | 4,200 | 2,730(65) | 1,050(25) | 168(4) | 378(9) |
| 532F 20(ND) | 2,100 | 1,428(68) | 630(30) | 63(3) | 42(2) | 379F 23(−/−) | 5,500 | 3,245(59) | 1.925(35) | 275(5) | 275(5) |
| 392M 22(+/−) | 2,500 | 1,825(73) | 500(20) | 175(7) | 150(6) | 397M 22(−/−) | 2,500 | 1,225(49) | 1,050(42) | 750(30) | 225(9) |
| 425M 37(ND) | 4,500 | 3.510(78) | 945(21) | 0(0) | 45(1) | 418M 37(−/−) | 4,000 | 2,080(52) | 1,640(41) | 600(15) | 200(5) |
| 471F 29(+/−) | 3,700 | 2,812(76) | 814(22) | 37(1) | 74(2) | 473F 29(−/−) | 5.000 | 2,350(47) | 2,150(43) | 500(10) | 400(8) |
| mean | 2,711 | 1,902(69) | 679(25) | 68(3) | 102(4) | 3,178 | 1,775(56) | 1,068(32) | 309(10) | 285(11) | |
| ±s.d. | ±877 | ±777(±9) | ±237(±7) | ±54(±2) | ±64(±3) | ±1,531 | ±883(±7) | ±689(±9) | 4251(±8) | ±89(±6) | |
| P value‡ | P> 0.05 | P> 0.05 | P>0.05 | P< 0.02 | P<0.01 | ||||||
Absolute number of cells per mm3 is given followed by percentage in parentheses. WBC, white blood cells.
Control mice were sex, parent and, as closely as possible, age matched.
The mouse number is followed by sex, age at death or sacrifice (days), and genotype (ND, not determined, phenotypically normal).
t-test of paired comparisons between control and mutant animals.
Cytokine analysis
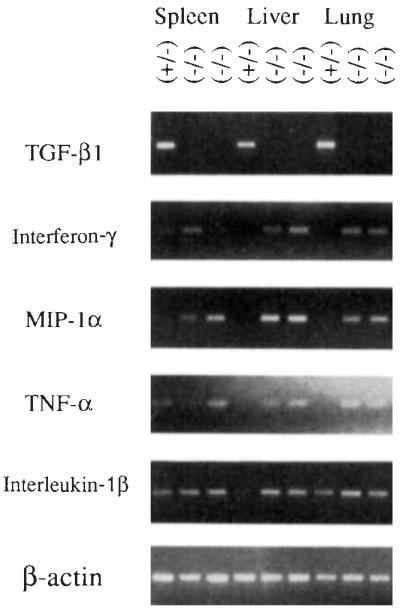
The expression of cytokines important in immune and inflammatory responses was investigated by PCR analysis of mRNAs from spleen, liver and lung from one heterozygous normal and two homozygous mutant animals (Fig. 4). As expected, no wild-type TGF-β1 mRNA was detected in the mutant animals, although it was detected in all tissues of the control. Interferon-γ tumour necrosis factor-α (TNF-α) and macrophage inflammatory protein-1α (MIP-1α), which are major mediators of inflammation, yielded strong amplification products in liver and lung of the mutant animals, but were not detected in these tissues in the control animal under the PCR conditions used. Interferon-γ, TNF-α and MIP-1α were detected in the spleens of both the control and mutant mice, although in one mutant, the amounts of product amplified were greater than those of the control, and in the other, less. Interleukin-1β, also an important mediator of inflammation, was present in all three tissues in both the control and mutant animals; in liver the amount of PCR product was slightly greater in the mutants than in the control. There were no significant differences between the control and mutant animals in the amounts of amplification products for TNF-β and interleukin-1α (data not shown).
FIG. 4.
PCR analysis of cytokine mRNAs in control and TGF-β1-deficient mice. Total RNAs from spleen, liver, and lung from control mouse 249 (+/−) and homozygous mutant animals 254 and 260 (−/−) were reverse-transcribed and examined by PCR analysis using primers specific for cytokines TGF-β1 (5′-GCGGACT ACT A TGCT AAAGAGG-3′ and 5′-GTIGTGTIGGTIGT AGAGGGCA-3′, 40 cycles), interferon-γ. (5′-TGGCTGTTICTGGCTGTIACTG-3′ and 5′-AATCAGCAGCGACTCCnnCC-3′, 35 cycles), MIP-1α: (5′-ACTGCCCTIGCTGTICTICTCT-3′ and 5′-AGGCA TICAGTICCAGGTCAGT-3′,40 cycles), TNF-α: (5′-CCAGACCCTCACACTCAGAT-3′ and 5′-AACACCCATICCCTICACAG-3, 31 cycles), and interleukin-1α (5′-TGACGGACCCCAAAAGATGAAG-3′ and 5′-CTGCTIGTGAGGTGCTGA TGT A-3′, 35 cycles). Amplification conditions using a Perkin–Elmer 9600 were 95 °C, 30 S; 58 °C, 1 min; 72 °c, 2 min (extended 1 s per cycle). The final extension was allowed to continue for 10 min. Amplified products were size-fractionated by electrophoresis through agarose and visualized by ultraviolet illumination of the ethidium bromide-stained gel. With each set of samples (except TNF-α: and β-actin), positive and negative PCR controls were run. The positive control consisted of total RNA from spleen cells cultured for 48 h with concanavalin A. The negative control contained all complementary DNA synthesis and PCR reagents but lacked RNA. Correct amplification products were observed in each positive control, whereas no products were amplified in the negative controls (data not shown). All samples gave comparable signals when amplified using primers specific for β-actin (5′-GTGGGCCGCTCTAGGCACCAA-3′ and 5′-CTCTTIGATGTCACGCACGA TTIC-3′).
Discussion
Histological analysis of animals homozygous for the disrupted TGF-β1 allele revealed a multifocal, mixed inflammatory cell infiltration, often with necrosis. The infiltrates, which seemed to contain primarily lymphocytes and neutrophils, affected many organs and often seemed so severe as to cause organ compromise or failure. Haematological analysis revealed a responsive increase in monocytes and immature neutrophils in the mutant animals. Important cytokine mediators of inflammation, including interferon-γ, TNF-α, and MIP-1α, were elevated in liver and lung of mutant animals relative to the control, consistent with the inflammation observed in these tissues. Thus, TGF-β1 deficiency resulted in severe pathology leading to death associated with dysfunction of the immune and inflammatory systems.
TGF-β1 is a potent regulator of the immune and inflammatory systems, generally functioning as an immunosuppressive agent. TGF-β1 suppresses B-cell proliferation and immunoglobulin (IgG and IgM) secretion16, 17; inhibits thymocyte, T lymphocyte, and large granular lymphocyte proliferation18-21; suppresses natural and lymphokine-activated killing by large granular lymphocytes22 ,23; and inhibits generation of cytotoxic T lymphocytes24. Both the initiation and resolution of general inflammatory responses seem to involve TGF-β1. TGF-β1 stimulates monocyte migration and growth factor production25, But after initiation of an inflammatory response, it also exhibits potent anti-inflammatory effects including inhibition of neutrophil and T-lymphocyte adhesion to endothelium26, down-regulation of macrophages25,27 and antagonism of TNF-α function24,28. Given these inhibitory effects, TGF-β1 deficiency might perturb normal immune and inflammatory system homeostasis, resulting in unregulated activity that is fatal to the organism. For example, dysregulated production of interferon-γ and TNF-α by activated T cells and macrophages, respectively, could contribute to the mutant phenotype as both cytokines mediate weight loss and inflammation29,30. The involvement of interferon-γ and TNF-α are consistent with the observed elevation of these cytokines in the mutant animals.
The inflammatory cell infiltration observed in some tissues of homozygous mutant animals resembles to a variable degree that seen in certain human autoimmune diseases such as myocarditis, polymyositis31, and Sjogren’s syndrome32, Pathological autoreactivity has been attributed to several processes in which TGF-β1 may playa regulatory role, including dysfunction of T cell suppression and aberrant expression of major histocompatibility complex (MHC) antigen by cells that normally do not present antigen33-35. TGF-β1 administration to mice protects against collagen-induced arthritis and relapsing experimental allergic encephalomyelitis36-39. T-cell suppression of experimental autoimmune encephalomyelitis after oral tolerization to myelin basic protein seems to be mediated by TGF-β1 (ref. 40), suggesting that the absence of TGF-β1 may reduce suppression by T cells and lead to autoreactivity in the affected animal. The autoimmune state may involve interferon-γ-induced expression of MHC class II molecules in a variety of cells34. TGF-β1 downregulates interferon-γ-induced MHC class II antigen expression on both lymphoid and non-lymphoid cell types41 and is a potent inhibitor of interferon-γ production by peripheral blood mononuclear cells28. Consequently, absence of TGF-β1 could lead to presentation of self antigens by inappropriate cells, thereby eliciting an autoimmune response.
TGF-β1 also seems to play an important role in regulating the mucosal immune system. Mucosal or secretory immunity involves induction of precursor IgA-secreting cells in bronchial-associated and gut-associated lymphatic tissue, including Peyer’s patches; migration of these cells to mucosal sites (including gut, respiratory tract, mammary gland, lacrimal gland and genitourinary system); and terminal differentiation to IgA-secreting cells in these tissues42. TGF-β1 specifically enhances IgA expression in stimulated Peyer’s patch and splenic B cells43,44 and seems to be involved in lymphocyte homing to mucosal sites, enhancing expression of human HML-l and murine β7αM290 integrins which are believed to mediate homing to the mucosa45,46. On the other hand, TGF-β1 inhibits adhesiveness of Peyer’s patch high endothelial venules for lymphocytes47. TGF-β1 deficiency could, therefore, result in dysfunction of the mucosal immune system, including abrogation of orally induced suppression, inhibition of IgA secretion and aberrant lymphocyte homing to mucosal sites, leading to adverse systemic consequences.
As the TGF-βs may play critical roles in embryogenesis1-5, it is intriguing that animals homozygous for the disrupted TGF-β1 gene exhibited no obvious developmental defects. It is proposed that functionally redundant genetic pathways may exist in critical developmental processes, involving structurally related genes or unrelated genes which have evolved overlapping functions48. The existence of multiple TGF-β isoforms, often exhibiting similar biological activities and overlapping patterns of expression during development, raises the possibility that expression of these other isoforms compensates for the lack of TGF-β1. But the under-representation of animals in the homozygous mutant class of progeny when heterozygous animals are interbred should be noted. Loss of half of the homozygous mutant animals seems to have occurred. The occurrence of this apparent embryonic lethality may depend on segregation of an additional gene(s) in the progeny and may reflect the genetic mix (129 and eF1) of the homozygous mutant animals.
An additional explanation for the failure to observe a developmental phenotype is that TGF-β1 is, in fact, required for embryonic and postnatal development and viability, but that maternal rescue of the TGF-β1-deficient animals occurs in utero by means of transplacental transfer of the growth factor and postnatally by transmission in the milk. TGF-β1 was first isolated from placenta49, but whether transplacental transfer occurs has not been determined. TGF-β-like activity has been detected in human and bovine colostrum50, and a TGF-β2-like protein has been isolated from bovine milk51. Thus, the onset of symptoms at the time of weaning may reflect maternal rescue before weaning. If this is the case, then this system provides a unique opportunity to study, in the maturing animal, the effects of a growth factor which is required during embryogenesis but which also has distinct functions in the adult.
Studies are in progress to determine the precise nature and mechanism of the immune and inflammatory dysfunction resulting from TGF-β1 deficiency; to rescue the mutant phenotype by TGF-β1 administration, anti-inflammatory drug therapy or appropriate mouse crosses; to investigate the potential prenatal lethality in homozygous mutant animals; and to investigate expression and function of the pro-region, which was not disrupted by the targeting strategy. TGF-β1-deficient mice may be valuable models for numerous inflammatory disorders, including autoimmune diseases, transplant rejection, graft versus host reactions, ulcer development, and malignancy-associated cachexia. TGF-β1-deficient cells are now available for examining the many developmental processes that TGF-β1 has been reported to affect, including cell proliferation and differentiation, extracellular matrix protein expression, cell adhesion and migration, and angiogenesis. Embryonic and fetal tissue explants from homozygous mutant animals can be used to study in vitro organ development in the absence of TGF-β1. Such studies should further elucidate the multifunctional roles of TGF-β1 as both a morphogenetic growth factor and immunoregulatory cytokine.
ACKNOWLEDGEMENTS
We thank G. Michael, E. Choi, P. Bonventre, J Roths and members of the Doetschman laboratory for discussion; J. Card for photographic assistance; L. Forrester and P. Holm for necropsy and histological techniques; and the Clinical Microbiology Division for analysing the organ cultures. This research was supported by grants from the NIH to T.D. and C.S.
References
- 1.Barnard JA, Lyons RM, Moses HL. Biochlm biophys.Acta. 1990;1032:79–87. doi: 10.1016/0304-419x(90)90013-q. [DOI] [PubMed] [Google Scholar]
- 2.Massague JA. Rev. Cell. Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AB, Sporn MB. In: Peptide Growth Factors and Their Receptors I. Sporn MB, Roberts AB, editors. Springer; New York: 1990. pp. 419–472. [Google Scholar]
- 4.Heine UI, et al. Cell. Biol. 1987;105:2861–2876. doi: 10.1083/jcb.105.6.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnert SA, Akhurst RJ. Development. 1988;104:263–273. doi: 10.1242/dev.104.2.263. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox JN, Derynck R. Molec. cell. Biol. 1988;8:3415–3422. doi: 10.1128/mcb.8.8.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gatherer D, Ten DiJke P, Baird DT, Akhurst RJ. Development. 1990;110:445–460. doi: 10.1242/dev.110.2.445. [DOI] [PubMed] [Google Scholar]
- 8.Pelton RW, Dickinson ME, Moses HL, Hogan BLM. Development. 1990;110:609–620. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 9.Schmid P, Cox D, Bilbe G, Maier R, McMaster GK. Development. 1991;111:117–130. doi: 10.1242/dev.111.1.117. [DOI] [PubMed] [Google Scholar]
- 10.Millan FA, Denhez F, Kondaiah P, Akhurst RJ. Development. 1991;111:131–144. doi: 10.1242/dev.111.1.131. [DOI] [PubMed] [Google Scholar]
- 11.Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Cell Biol. 1991;115:1091–1105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson NL, et al. J. Cell Biol. 1989;108:661–669. doi: 10.1083/jcb.108.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. J. Embryol. expo Morph. 1985;87:27–45. [PubMed] [Google Scholar]
- 14.Bosma GC, Custer RP, Bosma MJ. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 15.Glade LM, Rosenstreich DL. J Immun. 1976;117:2061–2066. [PubMed] [Google Scholar]
- 16.Kehrl JH, et al. J. Immun. 1986;137:3855–3860. [PubMed] [Google Scholar]
- 17.Kehrl JH, Thevenin C, Rieckmann P, Fauci AS. J. Immun. 1991;146:4016–4023. [PubMed] [Google Scholar]
- 18.Ristow HJ. Proc. natn. Acad. Sci. U.S.A. 1986;83:5531–5534. doi: 10.1073/pnas.83.15.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahl SM, et al. J. Immun. 1988;140:3026–3032. [PubMed] [Google Scholar]
- 20.Kehrl JH, et al. J. exp. Med. 1986;163:1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortaldo JR, et al. J. Immun. 1991;146:3791–3798. [PubMed] [Google Scholar]
- 22.Rook AH, et al. J. Immun. 1986;136:3916–3920. [PubMed] [Google Scholar]
- 23.Espevik T, Figari IS, Ranges GE, Palladino MA. J Immun. 1988;140:2312–2316. [PubMed] [Google Scholar]
- 24.Ranges GE, Figari IS, Espevik T, Palladino MA. J. expo Med. 1987;166:991–998. doi: 10.1084/jem.166.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahl SM. J. clin. Immun. 1992;12:61–74. doi: 10.1007/BF00918135. [DOI] [PubMed] [Google Scholar]
- 26.Gamble JR, Vadas MA. Science. 1988;242:97–99. doi: 10.1126/science.3175638. [DOI] [PubMed] [Google Scholar]
- 27.Tsunawaki S, Sporn M, Ding A, Nathan C. Nature. 1988;334:260–262. doi: 10.1038/334260a0. [DOI] [PubMed] [Google Scholar]
- 28.Espevik T, et al. J. exp. Med. 1987;166:571–576. doi: 10.1084/jem.166.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tracey KJ, et al. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 30.Matthys P, et al. Int J Cancer. 1991;49:77–82. doi: 10.1002/ijc.2910490115. [DOI] [PubMed] [Google Scholar]
- 31.Cronin ME, Miller FW, Plotz PH. In: Primer on the Rheumatic Diseases. Schumacher HR, editor. Arthritis Foundation; Atlanta: 1988. pp. 120–123. [Google Scholar]
- 32.Talal N. In: Primer on the Rheumatic Diseases. Schumacher HR, editor. Arthritis Foundation; Atlanta: 1988. pp. 136–138. [Google Scholar]
- 33.Schwartz RS, Datta SK. In: Fundamental Immunology. Paul WE, editor. Raven; New York: 1989. pp. 819–866. [Google Scholar]
- 34.Bottazzo GF, Pujol-Borrell R, Hanafusa T. Lancet. 1983;2:1115–1118. doi: 10.1016/s0140-6736(83)90629-3. [DOI] [PubMed] [Google Scholar]
- 35.Londei M, Lamb JR, Bottazzo GF, Feldman M. Nature. 1984;312:639–641. doi: 10.1038/312639a0. [DOI] [PubMed] [Google Scholar]
- 36.Brandes ME, Allen JB, Ogawa Y, Wahl SM. J. clin. Invest. 1991;87:1108–1113. doi: 10.1172/JCI115073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johns LD, Flanders KC, Ranges GE, Sriram S. J. Immun. 1991;147:1792–1796. [PubMed] [Google Scholar]
- 38.Kuruvilla AP, et al. Proc. natn. Acad. Sci. U.S.A. 1991;88:2918–2921. doi: 10.1073/pnas.88.7.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Racke MK, et al. J. Immun. 1991;146:3012–3017. [PubMed] [Google Scholar]
- 40.Miller A, Lider O, Roberts AB, Sporn MB, Weiner HL. Proc. natn. Acad. Sci. U.S.A. 1992;89:421–425. doi: 10.1073/pnas.89.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czarniecki CW, Chiu HH, Wong GHW, McCabe SM, Palladino MA. J. Immun. 1988;140:4217–4223. [PubMed] [Google Scholar]
- 42.Brandtzaeg P. Curr. Top. Microbiol. Immun. 1989;146:13–25. doi: 10.1007/978-3-642-74529-4_2. [DOI] [PubMed] [Google Scholar]
- 43.Coffman RL, Lebman DA, Shrader B. J.exp. Med. 1989;170:1039–1044. doi: 10.1084/jem.170.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lebman DA, Lee FD, Coffman RL. J. Immun. 1990;144:952–959. [PubMed] [Google Scholar]
- 45.Kilshaw PJ, Murant SJ. Eur. J. Immun. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- 46.Parker CM, et al. Proc. natn. Acad. Sci. U.S.A. 1992;89:1924–1928. doi: 10.1073/pnas.89.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin YH, Cai J-P, Xiu XM. J. Immun. 1992;146:1106–1112. [PubMed] [Google Scholar]
- 48.Tautz D. BioEssays. 1992;14:263–266. doi: 10.1002/bies.950140410. [DOI] [PubMed] [Google Scholar]
- 49.Frolik CA, Dart LL, Meyers CA, Smith DM, Sporn MB. Proc. natn. Acad. Sci. U.S.A. 1983;80:3676–3680. doi: 10.1073/pnas.80.12.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokuyama H, Tokuyama Y. Cell Biol. int. Rep. 1989;13:251–258. doi: 10.1016/0309-1651(89)90147-1. [DOI] [PubMed] [Google Scholar]
- 51.Cox DA, Burk RR. Eur. J. Biochem. 1991;197:353–358. doi: 10.1111/j.1432-1033.1991.tb15918.x. [DOI] [PubMed] [Google Scholar]
- 52.Thomas KR, Capecchi MR. Cell. 1987;51 doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]