Abstract
The present study was carried out for detection and molecular characterization of fowl adenoviruses (FAdVs) associated with hydropericardium syndrome or inclusion body hepatitis in commercial broiler chickens. The FAdVs were detected in liver samples from 33 commercial broiler chicken flocks by polymerase chain reaction (PCR) using hexon gene specific primers. The restriction enzyme analysis using StyI, BsiWI, MluI, AspI, BglI and ScaI enzymes of all the 33 FAdV-positive samples revealed FAdV-4 in 10 samples, FAdV-8 in five samples, FAdV-2 and FAdV-12 in two samples each, and FAdV-5 and FAdV-6 in one sample each. Twelve samples revealed the digestion pattern for more than one serotypes with FAdV-8 and FAdV-5, FAdV-8 and FAdV-7, FAdV-8 and FAdV-6, FAdV-8 and FAdV-12 being the predominant combinations indicating mixed infection. The serotypes FAdV-2 and FAdV-5 have not been detected previously in the country. The purified PCR products of FAdVs of four samples (HR 1, HR 2, HR 3 and HR 4) were cloned and sequenced. Phylogenetic analysis revealed that FAdVs of all four samples clustered in separate groups consistent with the REA pattern. In conclusion, this study reveals the presence of FAdV-2, FAdV-4, FAdV-5, FAdV-6, FAdV-7, FAdV-8 and FAdV-12 in broiler chickens affected with hydropericardium syndrome or inclusion body hepatitis in India.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-013-0183-7) contains supplementary material, which is available to authorized users.
Keywords: Molecular characterization, Broiler chickens, Fowl adenovirus, Hydropericardium syndrome, Inclusion body hepatitis
Introduction
Fowl adenoviruses (FAdVs) belong to the genus Aviadenovirus under the family Adenoviridae. The members of Aviadenovirus have been associated with a number of disease conditions including inclusion body hepatitis (IBH), hydropericardium syndrome (HPS), gizzard erosions, proventriculitis and tenosynovitis. The Aviadenoviruses have been subgrouped into five species based on their molecular structure and have been further divided into 12 serotypes based on cross-neutralization tests. The five species of FAdV including their serotypes are: fowl adenovirus A (FAdV-1), fowl adenovirus B (FAdV-5), fowl adenovirus C (FAdV-4 and -10), fowl adenovirus D (FAdV-2, -3, -9, and -11), and fowl adenovirus E (FAdV-6, -7, -8a, and -8b) [6].
Of the various disease conditions caused by FAdVs in poultry, IBH and HPS are the most important disease conditions and have been recorded from different parts of the world including India. All 12 serotypes have been incriminated in IBH [2], whereas serotype 4 has been implicated in HPS [9, 11].
Both conventional and molecular techniques are used for the diagnosis of IBH/HPS. Conventionally, IBH/HPS outbreaks are diagnosed on the basis of clinical signs, gross pathological lesions, histopathology and/or agar gel precipitation test [1, 14, 15]. Molecular techniques like polymerase chain reaction followed by restriction enzyme analysis (REA) and sequencing have been used for the rapid detection and differentiation of FAdVs [4, 5, 9, 10, 12, 16]. The present study was undertaken to detect and characterize the FAdVs from field outbreaks of IBH/HPS in broiler chickens in Haryana, a north-western state of India, to generate epidemiological information that could be helpful in the formulation of an effective vaccination strategy.
Materials and methods
Collection of samples
Liver tissue from 40 different flocks of commercial broiler chicken suspected to be suffering from IBH/HPS were collected from different parts of Haryana. The tissue samples from 3 to 4 affected birds in a flock were collected in 50 % buffered glycerin and were pooled to make a single pooled sample (hereinafter referred to as ‘sample’). The samples were stored at −20 °C until used. The initial diagnosis for IBH or HPS in these 40 flocks was based on clinical and necropsy findings. Two commonly used vaccines (VI and VII) were used in this study as positive virus controls. Liver tissue from an apparently healthy bird was taken as a negative virus control.
DNA extraction from samples
Total DNA was extracted directly from all 40 samples. For this, 50 mg sample was homogenized in 0.5 ml TNE buffer (50 mM Tris, 150 mM NaCl and 10 mM EDTA; pH 8.0), incubated with 1 mg/ml Proteinase K at 37 °C for 30 min followed by treatment with 0.5 % SDS at 55 °C with gentle agitation at least for 2 h. Total DNA was precipitated with ethanol after two rounds of phenol–chloroform extractions. The extracted DNA was stored at −20 °C till further use in PCR.
Polymerase chain reaction
The PCR was optimized to amplify FAdV hexon gene sequence of the viral DNA generating PCR product of size ≈900 bp using the primer pair as reported earlier [12]. The primer sequence was: Forward 5′CAARTTCAGRCAGACGGT3′ and Reverse 5′TAGTGATGMCGSGACATCAT3′. DNA amplification was carried out in a total volume of 50 μl containing 10 ng total DNA, 20 pmol of each forward and reverse primer, 200 μM dNTPs mix, 1.5 mM MgCl2 and 2.5U Taq DNA polymerase. The reaction was carried out in a thermal cycler (Biometra, UK) with initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 1 min with a step of final extension at 72 °C for 10 min. The PCR product was analysed in 1.0 % agarose gel.
Restriction enzyme analysis and cloning
The PCR products from positive samples and both vaccines were purified using QIAquick Gel Extraction Kit (QIAGEN). All FAdV positive purified PCR products were subjected to restriction digestion with three enzymes viz. BsiWI, StyI and MluI. In addition, AspI restriction enzyme was used to differentiate between FAdV-2 and FAdV-12, BglI between FAdV-4 and FAdV-9 and ScaI between FAdV-7 and FAdV-11. The number of restriction sites and distinctive restriction patterns in different FAdV serotypes were taken as criteria for selection of specific restriction enzymes [12].
Purified PCR products from four samples (HR 1, HR 2, HR 3 and HR 4) were cloned in pGEM®-T Easy vector (Promega, USA). These four samples belonged to flocks that showed high mortality. The recombinant plasmid clones were confirmed by PCR using hexon gene primers [12] and by restriction enzyme digestion using EcoRI.
Nucleotide sequencing and phylogenetic analysis
The recombinant plasmids containing 900 bp hexon gene insert of four field samples (HR 1, HR 2, HR 3 and HR 4) were sequenced in both directions using ABI automated sequencer. These nucleotide sequences are available in GenBank data base with accession numbers: HR 1 (HM748590), HR 2 (HM748589), HR 3 (HM748588) and HR 4 (HM748587).
Using Mega 4.0 programme, the forward and reverse sequences were aligned to make one sequence by ClustalW method. The aligned nucleotide sequences were truncated to 144–1041 nucleotide positions [3] and were compared to previously published sequences of different FAdV serotypes retrieved from the GenBank/EMBL databases (Supplementary Table 1). The nucleotide sequences were translated into amino acids (49–346 positions) which correspond to the L1 loop in the CELO strain. Percent divergence and similarities based on nucleotide and deduced amino acid sequences were also drawn. Aligned nucleotide sequences were subjected to phylogenetic analysis.
Results
FAdVs in clinical samples
As expected a band of ≈900 bp was observed in agarose gel in both VI and VII vaccines and no band at this position was observed in the negative control. Similarly, a band of ≈900 bp was also observed in 33 of the 40 field samples tested indicating them to be positive for FAdVs. No band was observed in seven field samples indicating them to be negative for FAdV.
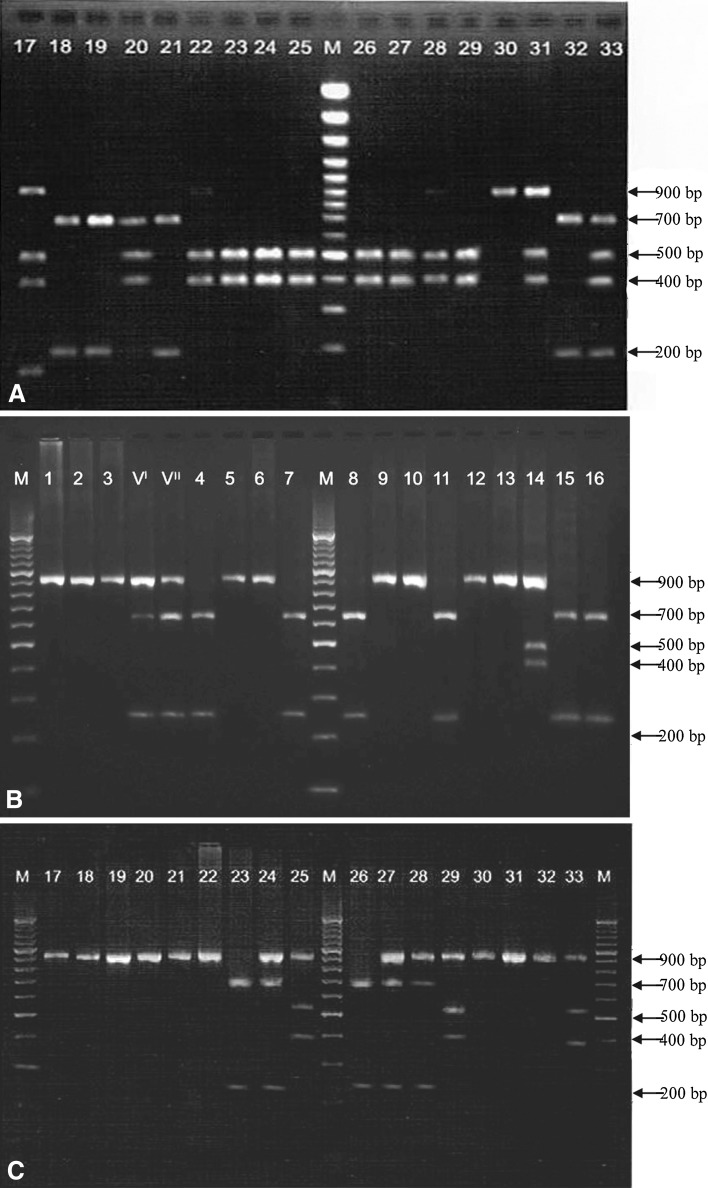
None of the FAdV-positive PCR products could be digested with enzymes BglI indicating the absence of FAdV-9 serotype in this study (Table 1). The REA pattern generated by different enzymes revealed the presence of only one serotype in 21 samples. The digestion patterns in the remaining 12 samples revealed the presence of more than one serotypes (Fig. 1). Both vaccines revealed the presence of FAdV-4 and FAdV-8 serotypes (Table 1; Fig. 1).
Table 1.
Restriction enzyme analysis pattern of the fowl adenovirus positive samples
| Number of samples in each categorya | Restriction enzyme analysisb | Serotype(s) | ||
|---|---|---|---|---|
| StyI | BsiWI | MluI | ||
| 2 | – | + | + | FAdV-2 |
| 10 | + | – | + | FAdV-4 |
| 1 | + | – | – | FAdV-5 |
| 1 | – | + | – | FAdV-6 |
| 5 | – | – | + | FAdV-8 |
| 2 | – | + | + | FAdV-12 |
| 1 | +/+ | –/+ | +/+ | FAdV-4 + FAdV-7 |
| 1 | –/+ | – | +/+ | FAdV-8 + FAdV-4 |
| 4 | –/+ | – | +/– | FAdV-8 + FAdV-5 |
| 2 | – | –/+ | +/– | FAdV-8 + FAdV-6 |
| 2 | – | –/+ | +/+ | FAdV-8 + FAdV-12 |
| 2 | –/+ | –/+ | +/+ | FAdV-8 + FAdV-7 |
| VI | +/– | – | +/+ | FAdV-4 + FAdV-8 |
| VII | +/– | – | +/+ | FAdV-4 + FAdV-8 |
a33 samples were positive for FAdVs by PCR
bAdditional REs were used for differentiation between FAdV-2 and FAdV-12 (AspI), FAdV-4 and FAdV-9 (BglI) or FAdV-7 and FAdV-11 (ScaI)
Fig. 1.
Restriction enzyme digestion patterns of fowl adenovirus positive-PCR products of field samples and vaccine strains [A=Digestion pattern with restriction enzyme MluI, (Lane M=100 bp DNA marker and lanes 17–33=FAdV-positive field samples); B=Digestion pattern with restriction enzyme StyI (Lane M=100 bp DNA marker, lanes VI and VII= Vaccine strains and lanes 1–16=FAdV-positive field samples); C=Digestion pattern with restriction enzyme BsiWI (Lane M=100 bp DNA marker and lanes 17–33=FAdV-positive field samples)
Four samples (HR 1, HR 2, HR 3 and HR 4) were cloned and confirmed by PCR and restriction enzyme digestion using EcoRI. Amplification of single band of 900 bp in each of the recombinant plasmid and release of 900 bp fragment after EcoRI digestion confirmed that the recombinant plasmid had the correct size and orientation of insert.
Sequence analysis
Comparison of partial sequences of FAdVs from these four samples with previously published hexon gene sequences of FAdVs revealed changes at the amino acid levels (Supplementary Fig. 1). These included: P128A, N138I, G151S, L234N, G246T and D266E in sample HR 1 and I188R, A193R, L263I and L264V in sample HR 2. Similarly, substitutions at S212I, S244R, G282R, M284K, L301M, V308A and N314D were observed in sample HR 3. The changes at T76A and E141D were observed in samples HR 1 and HR 3, and HR 1, HR 3 and HR 4, respectively. Nucleotide sequence analysis revealed that the FAdVs of this study had 67.6–95.2 % similarity among themselves. The FAdV of sample HR 1 had maximum similarity with FAdV-2 (96.4 %), HR 2 with FAdV-4 (98.2 %), HR 3 with FAdV-8 (96.7 %) and HR 4 with FAdV-12 (97.7 %).
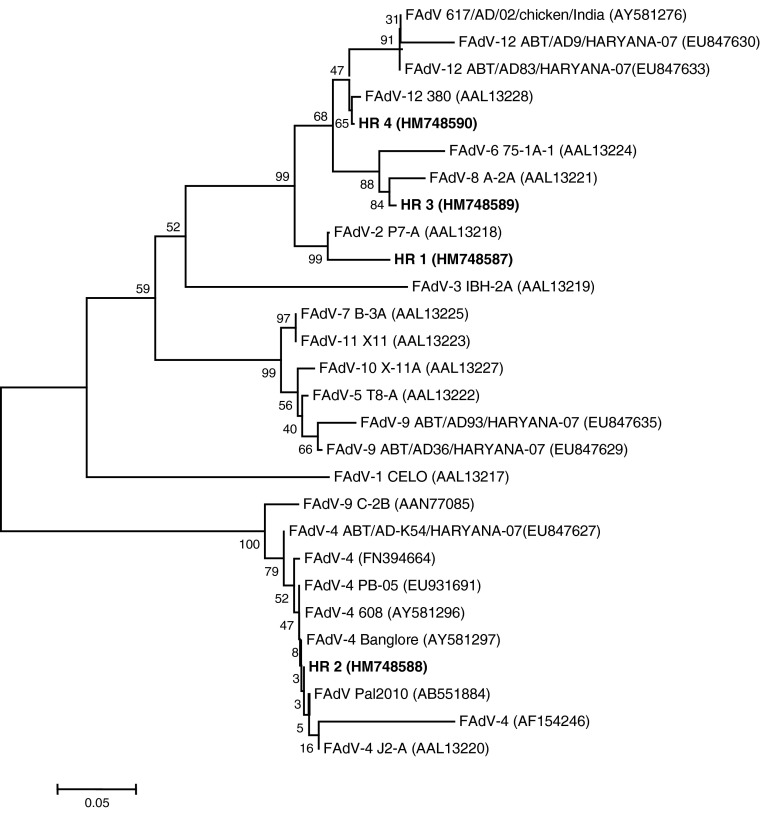
Phylogenetic analysis based on nucleotide sequence revealed that all four FAdVs were placed in separate clusters (Fig. 2). The HR 1 clustered with reference FAdV-2 P7-A (AAL13218). The HR 2 was closer to FAdV-4 previously reported from other parts [Bangalore (AY581297), FAdV C (AB551884), 608 (AY581296), PB-05 (EU931691) and FAdV-4 (FN394664)] than FAdV-4 strain from Haryana (ABT/AD-K54/Haryana-07; EU847627). The HR 3 clustered with FAdV-8 (VR 833 A-2A; AAL13221). The HR 4 clustered with FAdV-12 (380; AAL13228), a reference strain and was in a separate cluster from previously reported FAdV-12 strains from India [617/AD/02 (AY581276), ABT/AD9/HARYANA-07 (EU847630), ABT/AD83/HARYANA-07 (EU847633)] (Fig. 2). Clustering pattern indicated that the samples HR 1, HR 2, HR 3, and HR 4 had FAdV-2, FAdV-4, FAdV-8, and FAdV-12, respectively. The results of phylogenetic analysis of these four samples were consistent with the REA pattern.
Fig. 2.
Phylogenetic tree based on nucleotide sequences of the hexon gene of fowl adenoviruses. Strain names with accession number (in bold) are of the present study, and strain names with GenBank/EMBL accession numbers were retrieved from database
Discussion
Fowl adenoviruses have been incriminated as etiological agents for a number of clinical conditions in broiler chickens, breeder flocks and layers. Among the diseases caused by FAdVs, IBH and HPS are economically important. Almost all serotypes of FAdVs have been reported to cause IBH in broiler chickens [2], while only FAdV-4 causes HPS. The REA and sequencing have helped in the identification of FAdV serotypes involved with these two conditions [10, 12, 13]. The primers designed from hexon gene have been used in PCR for detection of fowl adenovirus [5, 13, 16, 22] and amplify all reference strains of FAdV [12].
Seven field samples that were negative for FAdVs by PCR were also negative for inclusion bodies by histopathology and for FAdV by AGPT. Since the primers used in this study can amplify DNA from all the 12 FAdV serotypes, it is unlikely that the negative results were due to PCR performance. Another possibility is that the viral load in the samples was below the detection limit [18]. In the field, certain disease conditions are associated with occurrence of IBH. The IBH outbreaks in broiler chicks and Japanese quail have been reported to be associated with mycotoxicosis especially aflatoxicosis [21].
The combination of PCR followed by REA allowed differentiation of FAdVs to species and presumptive serotypes. Based on REA pattern, 36.4 and 48.5 % of the positive samples had FAdV-4 and FAdV-8, respectively, indicating them to be the predominant serotypes. Predominance of FAdV-8 in Canada and the increase in isolations of this serotype over the years have been reported [16]. The REA pattern revealed the presence of FAdV-2, FAdV-5, FAdV-6, FAdV-7 and FAdV-12 serotypes in addition to FAdV-4 and FAdV-8 in this study. Overall, FAdV-2, FAdV-4, FAdV-5, FAdV-6, FAdV-7, FAdV-8 and FAdV-12 serotypes were identified in 2, 12, 5, 3, 3, 16 and 4 samples, respectively. The FAdV serotypes -1, -3, -9, -10 and -11 were not recorded in the present study. Various workers from India had identified serotypes FAdV-1, FAdV-4, FAdV-6, FAdV-7, FAdV-8 and FAdV-12 from IBH/HPS in poultry from different parts of the country [17, 19, 20]. However, the presence of FAdV-2 and FAdV-5 serotypes has not been reported previously. Earlier, only serotypes FAdV-4 and -8 were considered to be involved in cases of HPS and IBH, respectively [11]. Identification of serotypes other than FAdV-4 and FAdV-8 in this study suggests their possible role in causation of the disease.
Mixed infections with two or more adenovirus serotypes have been described before [12, 19]. In the present study, 12 of the 33 FAdV positive samples contained two serotypes indicating mixed infections with different serotypes during same outbreak. Mixed infections of FAdV-1 and FAdV-8, and that of FAdV-7 and FAdV-1 have been reported earlier in IBH affected flocks [8, 16]. It is not clearly understood whether the presence of two serotypes has any effect on the severity of clinical disease. Detection of multiple serotypes in a flock may be important; one serotype may act as reinforcing factor for the other serotype or both serotypes may act simultaneously to produce additive or synergistic effects. The occurrence of two or more serotypes from a single outbreak/flock indicates that there may be little cross protection among serotypes [8]. Further studies are needed to determine the serologic relationships among these serotypes and to identify levels of cross-protection that may or may not exist among these serotypes.
The vaccines used as positive control in this study contained only FAdV-4 and FAdV-8. It is not known whether these vaccines will accord full protection against challenges with serotypes other than FAdV-4 and FAdV-8. Further studies are needed to determine the nature of serotypes other than FAdV-4 and FAdV-8 as far as their pathogenicity and virulence are concerned. Such studies will be helpful in determining if modification of current vaccines is necessary for better protection.
The phylogenetic analysis has shown consistent results when compared with the REA. Based on REA pattern and phylogenetic analysis, the HR 1, HR 2, HR 3 and HR 4 were found to contain FAdV-2, FAdV-8, FAdV-4 and FAdV-12, respectively. The results of nucleotide sequence similarity of this study are consistent with earlier report of Ganesh et al. [7]. High degree of variation was reported to be due to shift in reading frame caused by deletions in nucleotide sequences [7].
In conclusion, this study reveals the presence of FAdV serotypes -2, -5, -6, -7, and -12 in addition to FAdV-4 and FAdV-8 in broiler chickens suffering from IBH or HPS. The role of these serotypes (except FAdV-4 and FAdV-8) in the severity of IBH or HPS needs to be elucidated further. Further studies are also required to determine whether the commercial vaccines would be able to accord protection against FAdV serotypes other than FAdV-4 and FAdV-8. Such studies can help in minimizing economic losses to poultry farmers as both diseases can cause considerable mortality.
Electronic supplementary material
Supplemental Fig. 1a Alignment of deduced amino acid sequences of hexon genevariable region of fowl adenovirus serotypes. Amino acid residuesdiffering from majority consensus are boxed. Amino acid positionsare as per Chiocca et al. [3] (JPG 4266 kb)
Supplemental Fig. 1b Alignment of deduced amino acid sequences of hexon genevariable region of fowl adenovirus serotypes. Amino acid residuesdiffering from majority consensus are boxed. Amino acid positionsare as per Chiocca et al. [3] (JPG 2469 kb)
References
- 1.Asthana M, Chandra R, Kumar R. Hydropericardium syndrome: current state and future developments. Arch Virol. 2013;158:921–931. doi: 10.1007/s00705-012-1570-x. [DOI] [PubMed] [Google Scholar]
- 2.Chandra R, Dixit VP, Kumar M. Inclusion body hepatitis in domesticated and wild birds: a review. Indian J Virol. 1998;14:1–12. [Google Scholar]
- 3.Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotton M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi KS, Kye SJ, Kim JY, Jeon WJ, Lee EK, Park KY, Sung HW. Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poult Sci. 2012;91:2502–2506. doi: 10.3382/ps.2012-02296. [DOI] [PubMed] [Google Scholar]
- 5.Dar A, Gomis S, Shirley I, Mutwiri G, Brownlie R, Potter A, Gerdts V, Tikoo SK. Pathotypic and molecular characterization of a fowl adenovirus associated with inclusion body hepatitis in Saskatchewan chickens. Avian Dis. 2012;56:73–81. doi: 10.1637/9764-041911-Reg.1. [DOI] [PubMed] [Google Scholar]
- 6.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus taxonomy-eighth report of the international committee on the taxonomy of viruses. San Diego: Elsevier Acad Press; 2005. [Google Scholar]
- 7.Ganesh K, Suryanarayana VVS, Raghavan R, Gowda S. Nucleotide sequence of L1 and part of P1 of hexon gene of fowl adenovirus associated with hydropericardium hepatitis syndrome differs with the corresponding region of other fowl adenoviruses. Vet Microbiol. 2001;78:1–11. doi: 10.1016/S0378-1135(00)00288-1. [DOI] [PubMed] [Google Scholar]
- 8.Gomis S, Goodhope AR, Ojkic AD, Willson P. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 2006;50:550–555. doi: 10.1637/7577-040106R.1. [DOI] [PubMed] [Google Scholar]
- 9.Kim JN, Byun SH, Kim MJ, Kim J, Sung HW, Mo IP. Outbreaks of hydropericardium syndrome and molecular characterization of Korean fowl adenoviral isolates. Avian Dis. 2008;52:526–530. doi: 10.1637/8178-112207-Case. [DOI] [PubMed] [Google Scholar]
- 10.Mase M, Nakamura K, Imada T. Characterization of fowl adenovirus serotype 4 isolated from chickens with hydropericardium syndrome based on analysis of the short fiber protein gene. J Vet Diagn Invest. 2010;22:218–223. doi: 10.1177/104063871002200207. [DOI] [PubMed] [Google Scholar]
- 11.Mazaheri A, Prusas C, Vob M, Hess M. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol. 1998;27:269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- 12.Meulemans G, Boschmans M, Van den Berg TP, Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenovirus. Avian Pathol. 2001;24:693–706. doi: 10.1080/03079450120092143. [DOI] [PubMed] [Google Scholar]
- 13.Meulemans G, Couvreur B, Decaesstecker M, Boschmans M, Van den Berg TP. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. 2004;33:164–170. doi: 10.1080/03079450310001652086. [DOI] [PubMed] [Google Scholar]
- 14.Mittal D, Khokhar RS, Jindal N. Diagnosis of the inclusion body hepatitis–hydropericardium syndrome using conventional techniques. Haryana Vet. 2011;50:53–56. [Google Scholar]
- 15.Nakamura K, Mase M, Yamamoto Y, Takizawa K, Kabeya M, Wakuda T, Matsuda M, Chikuba T, Yamamoto Y, Ohyama T, Takahashi K, Sato N, Akiyama N, Honma H, Imai K. Inclusion body hepatitis caused by fowl adenovirus in broiler chickens in Japan, 2009–2010. Avian Dis. 2011;55:719–723. doi: 10.1637/9813-052511-Case.1. [DOI] [PubMed] [Google Scholar]
- 16.Ojkic D, Martin E, Swinton J, Vaillancourt JP, Boulianne M, Gomis S. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 2008;37:95–100. doi: 10.1080/03079450701805324. [DOI] [PubMed] [Google Scholar]
- 17.Parthiban M, Manoharan S, Roy P, Chandran NDJ, Aruni AW, Koteeswaran A. Nucleotide sequence analysis of the L1 loop variable region of hexon gene of fowl adenovirus 4 isolates from India. Acta Virol. 2005;49:65–68. [PubMed] [Google Scholar]
- 18.Philippe C, Grgic H, Ojkic D, Nagy E. Serologic monitoring of a broiler breeder flock previously affected by inclusion body hepatitis and testing of the progeny for vertical transmission of fowl adenoviruses. Can J Vet Res. 2007;71:98–102. [PMC free article] [PubMed] [Google Scholar]
- 19.Rahul S, Kataria JM, Senthilkumar N, Dhama K, Sylvester SA, Uma R. Association of fowl adenovirus serotype 12 with hydropericardium syndrome of poultry in India. Acta Virol. 2005;49:139–143. [PubMed] [Google Scholar]
- 20.Singh A, Oberoi MS, Grewal GS, Hafez HM, Hess M. The use of PCR combined with restriction enzyme analysis to characterize fowl adenovirus field isolates from northern India. Vet Res Commun. 2002;26:577–585. doi: 10.1023/A:1020299700907. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Oberoi MS, Jand SK, Singh B. Epidemiology of inclusion body hepatitis in poultry in northern India from 1990 to 1994. Rev sci tech Off int Epiz. 1996;15(3):1053–1060. doi: 10.20506/rst.15.3.976. [DOI] [PubMed] [Google Scholar]
- 22.Steer PA, O’Rourke D, Ghorashi SA, Noormohammadi AH. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust Vet J. 2011;89:184–192. doi: 10.1111/j.1751-0813.2011.00695.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1a Alignment of deduced amino acid sequences of hexon genevariable region of fowl adenovirus serotypes. Amino acid residuesdiffering from majority consensus are boxed. Amino acid positionsare as per Chiocca et al. [3] (JPG 4266 kb)
Supplemental Fig. 1b Alignment of deduced amino acid sequences of hexon genevariable region of fowl adenovirus serotypes. Amino acid residuesdiffering from majority consensus are boxed. Amino acid positionsare as per Chiocca et al. [3] (JPG 2469 kb)