Abstract
Morphological form in multicellular aggregates emerges from the interplay of genetic constitution and environmental signals. Bacterial macrocolony biofilms, which form intricate three-dimensional structures, such as large and often radially oriented ridges, concentric rings, and elaborate wrinkles, provide a unique opportunity to understand this interplay of “nature and nurture” in morphogenesis at the molecular level. Macrocolony morphology depends on self-produced extracellular matrix components. In Escherichia coli, these are stationary phase-induced amyloid curli fibers and cellulose. While the widely used “domesticated” E. coli K-12 laboratory strains are unable to generate cellulose, we could restore cellulose production and macrocolony morphology of E. coli K-12 strain W3110 by “repairing” a single chromosomal SNP in the bcs operon. Using scanning electron and fluorescence microscopy, cellulose filaments, sheets and nanocomposites with curli fibers were localized in situ at cellular resolution within the physiologically two-layered macrocolony biofilms of this “de-domesticated” strain. As an architectural element, cellulose confers cohesion and elasticity, i.e., tissue-like properties that—together with the cell-encasing curli fiber network and geometrical constraints in a growing colony—explain the formation of long and high ridges and elaborate wrinkles of wild-type macrocolonies. In contrast, a biofilm matrix consisting of the curli fiber network only is brittle and breaks into a pattern of concentric dome-shaped rings separated by deep crevices. These studies now set the stage for clarifying how regulatory networks and in particular c-di-GMP signaling operate in the three-dimensional space of highly structured and “tissue-like” bacterial biofilms.
INTRODUCTION
Biofilms are a multicellular form of bacterial life which confers resistance against environmental stresses, antibiotics, and host immune systems (1). In addition, the complex macroscopic architecture of some bacterial biofilms provides an intriguing opportunity to study the emergence of morphological form in multicellular aggregates driven by the interplay of genetic information and environmental influences. Within biofilms, gene expression is heterogeneous, and physiologically different zones and strata containing different extracellular matrix components can be observed. These zones form as a consequence of gradients of nutrients, signaling compounds or waste products that build up during growth of the biofilm (2, 3). Biofilms grown for several days on agar surfaces, i.e., in “macrocolonies,” can adopt elaborate three-dimensional structures (Fig. 1) that have been termed “wrinkled,” “rugose,” or “rdar” (for red, dry, and rough) (4–7). It is clear that extracellular polymeric substances (EPS), which form a matrix that includes adhesins, amyloid-forming proteins, and exopolysaccharides, are required to generate these morphological structures (8–10), but how their actual three-dimensional patterns are controlled and why they are formed has remained elusive.
Fig 1.
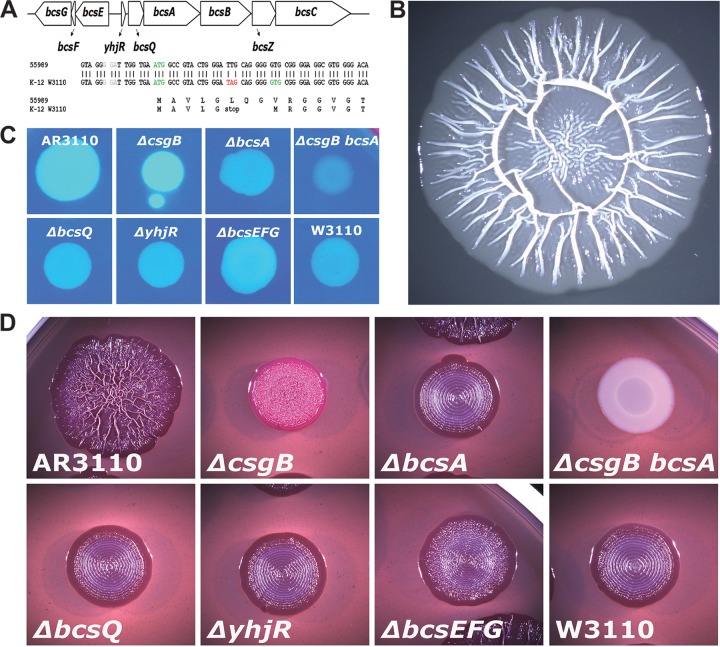
Cellulose biosynthesis and complex macrocolony morphology after chromosomal “repair” of the SNP that generates a stop codon early in bcsQ in E. coli K-12 strain W3110. (A) Operon structure of the bcs region and position of the TAG/TTG SNP in bcsQ. (B) Macrocolony structure of strain AR3110, a W3110 derivative with the bcsQ(TTG) allele after growth on salt-free LB plates for 5 days. (C) Fluorescence of 1-day-old colonies grown on Calcofluor plates of strain AR3110 and its indicated mutant derivates and strain W3110. (D) Colony morphology of AR3110 and the indicated mutant derivates after growth on CR plates for 5 days.
In Escherichia coli and Salmonella, macrocolony morphology depends on the presence of curli fibers and the exopolysaccharide cellulose in the biofilm matrix (7, 11, 12). During their secretion curli proteins are assembled into amyloid fibers (13, 14), which form basket-like structures around the small and starving cells in the upper layer of a macrocolony (2). Cellulose is a glucose polymer with β-1,4 glycosidic linkage, which in bacteria is synthesized and excreted by a membrane-inserted cellulose synthase complex consisting of two subunits, BcsA and BcsB (15, 16). This process is supported and regulated by several accessory proteins. The proteins involved are encoded in two divergently arranged and highly conserved operons, yhjR-bcsQABZC and bcsEFG (Fig. 1A) (12, 17). Both commensal and pathogenic E. coli strains are heterogeneous with respect to their ability to produce curli and/or cellulose (18, 19).
As first shown for Salmonella, biosynthesis of curli and cellulose occurs in early stationary-phase cells since it depends on the biofilm regulator CsgD, which in turn requires the stationary-phase sigma factor RpoS (σS) for expression (20, 21). In many E. coli strains, CsgD is temperature regulated and expressed only below 30°C (18, 22). Curli and cellulose production is also regulated by the ubiquitously biofilm-promoting second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), which is produced and degraded by multiple diguanylate cyclases (DGC; characterized by GGDEF domains) and phosphodiesterases (PDE; with EAL domains), respectively (23–25). c-di-GMP has multiple inputs into curli and cellulose production. First, the expression of the transcriptional regulator CsgD requires two c-di-GMP modules—each consisting of a DGC and a PDE—acting in a cascade (26–28). CsgD then drives the transcription of the curli genes (csgBAC), as well as of yaiC (adrA in Salmonella), which encodes yet another DGC that is essential to activate cellulose synthase (12, 21, 28, 29). Via the c-di-GMP-binding PilZ domain of BcsA, cellulose synthase is a direct target of c-di-GMP (30).
This regulatory network has been intensively studied at the molecular level. However, the question of how this complex control determines the spatial distribution of matrix components and thereby somehow translates into the intricate three-dimensional morphology of a macrocolony biofilm has been addressed only very recently. Macrocolonies of the E. coli K-12 laboratory strain W3110 form patterns of concentric rings surrounded by small wrinkles and a flat area of growing cells at the outer rim of the colony (Fig. 1D). Using fluorescence microscopy of cryosections, as well as scanning electron microscopy (SEM) at high resolution, small cells tightly surrounded by curli fibers were observed in the top layer, whereas rod-shaped and dividing cells entangled by a mesh of flagella were found in the bottom layer and in the outer growth zones of W3110 macrocolonies. In the transition zones in between, patches and small chains of curli-surrounded and “naked” flagellated cells occur side by side, which is also reflected in heterogeneous CsgD expression (2). That study was the first to demonstrate physiological differentiation in a macrocolony biofilm in situ at cellular resolution and thereby clarified that curli fibers and RpoS-dependent gene expression (of csgD and curli genes) occur in the top layer and at the surface.
However, cellulose could not be observed in that study, since E. coli K-12 strains do not produce it (12, 19). The reason for this deficiency has been unknown, since E. coli K-12 does have the bcs operons and expresses all the regulatory genes, including csgD and the DGC gene yaiC (27, 28). In the present study, we traced the cellulose-negative phenotype to a “domesticating” single nucleotide mutation. “Repairing” this SNP in the chromosome allowed us to address a number of questions in the otherwise well-characterized E. coli K-12 strain, such as where in the biofilm do we find cellulose? What does its microstructure look like at the cellular level, in particular in contact with other matrix components such as curli fibers and flagella? How does cellulose contribute to the formation of three-dimensional structure and the physical properties of macrocolony biofilms? Our data demonstrate that cellulose is an architectural element with a distinct spatial distribution within the biofilm. Together with the amyloid curli fibers, it forms a nano-composite “building material” which confers tissue-like properties, i.e., cohesion, elasticity, and stability, which in combination with spatial constraints in growing colonies generate the remarkable three-dimensional structure elements of macrocolony biofilms.
MATERIALS AND METHODS
Bacterial strains.
All strains used are derivatives of the E. coli K-12 strain W3110 (31). The rpoS::cat, csgD::cat, csgB::cat, and bcsA::cat mutations are full open reading frame deletion (Δ)/resistance cassette insertions previously described (2, 27, 28). For generating the bcsQ::cat, bcsEFG::kan, and yhjR::cat deletion/insertion mutants, the one-step-inactivation protocol was applied (32) using oligonucleotide primers listed in Table S1 in the supplemental material. When required, cassettes were flipped out (32). These mutations were transferred by P1 transduction (33).
For changing codon 6 of bcsQ from TAG(stop) to TTG(leucine) in the chromosomal copy of bcsQ in strain W3110, a two-step replacement procedure related to the one-step-inactivation protocol (32) was applied. The oligonucleotides for this procedure are listed in the Table S1 in the supplemental material. The procedure uses a fragment of plasmid pKD45 (32), encoding a kanamycin-resistance cassette and a ccdB toxin under the control of a rhamnose-inducible promoter, which is first introduced into the target locus in the chromosome, followed by its replacement by recombination with the desired allele (on a PCR fragment transformed into the recipient) which is selected for by growth in the presence of rhamnose (34). Allelic states of transformants were verified by PCR and DNA sequencing.
Construction of single copy lacZ reporter fusions.
Oligonucleotide primers used to construct single-copy lacZ reporter gene fusions of yhjR, bcsQ, and bcsA fusions are listed in Table S1 in the supplemental material. The procedure using lacZ fusion vector pJL28 was described previously (28). All fusions are translational fusions that include 9 (yhjR), 18 (bcsQ; counting from the ATG start codon; Fig. 1A), and 10 (bcsA) codons of the corresponding open reading frames. Upstream regions of all three lacZ reporter gene fusions contain the whole intergenic region between the yhjR-bcsQABZC and bcsEFG operons and start 406 bp upstream of the yhjR start codon (see Fig. 3; see also Fig. S1 in the supplemental material). All reporter fusions were transferred to the att(λ) location of the chromosome of a W3110 derivative containing a Δlac(I-A)::scar deletion via phage λRS45 or λRS74 (35). Single lysogeny was tested by a PCR approach (36).
Fig 3.
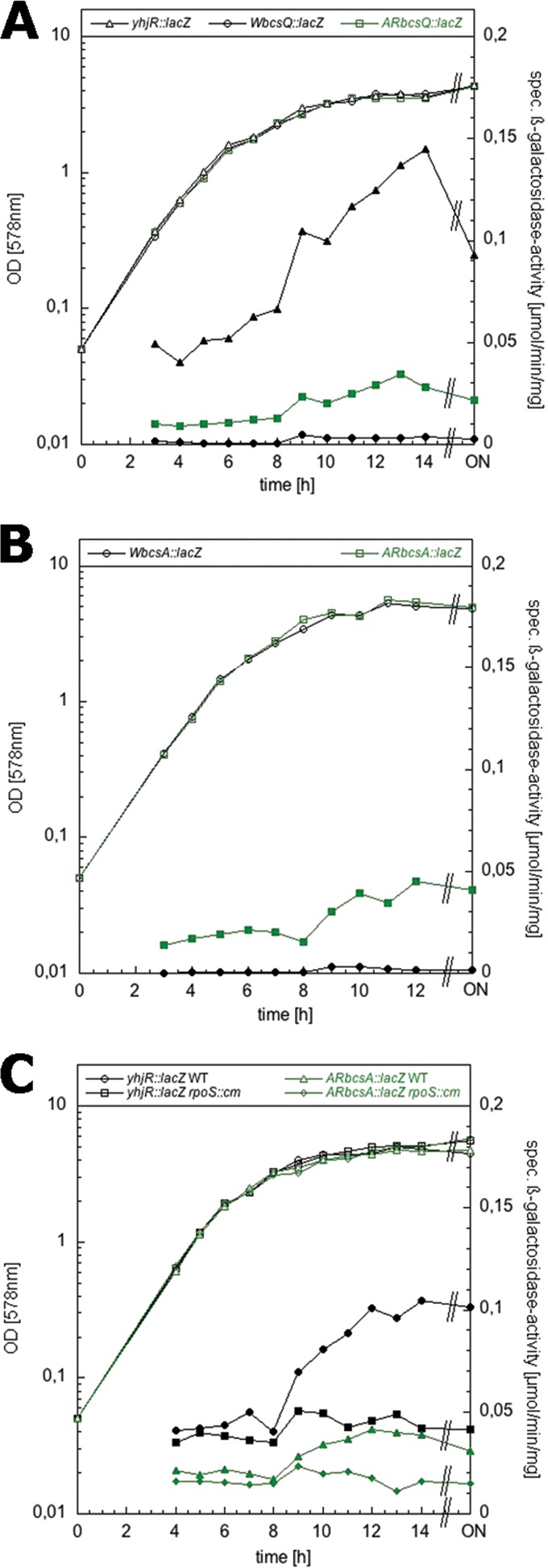
Expression of lacZ reporter fusions to yhjR, bcsQ, and bcsA in the presence or absence of the E. coli K-12-specific stop codon in bcsQ. Derivatives of strain W3110 carrying single copy lacZ fusions in yhjR or in bcsQ or bcsA with codon 6 in bcsQ being either TAG (W fusions) or TTG (AR fusions) were grown in LB medium at 28°C. The optical densities (open symbols) and specific β-galactosidase activities (closed symbols) were determined.
Generation of chromosomal alleles encoding 3×FLAG-labeled proteins.
C-terminally 3×FLAG-tagged chromosomally encoded constructs were generated using plasmid pSUB11 as a PCR template (37) and the oligonucleotide primers listed in Table S1 in the supplemental material according to a procedure based on λRed technology (32).
Growth of bacterial macrocolonies.
Cells were grown overnight in liquid Luria-Bertani (LB) medium (33) under aeration at 37°C. A total of 5 μl of the overnight cultures were spotted on salt-free LB agar plates. Where indicated, agar plates were supplemented with Congo red (CR; 40 μg ml−1) and Coomassie brilliant blue (20 μg ml−1; CR plates), Calcofluor (100 μg ml−1), or Thioflavin S (TS; 40 μg ml−1). In order to grow all strains to be compared in parallel on a single agar plate, 140-mm-diameter petri dishes (VWR) were used, which allows the spotting of up to 25 macrocolonies in a 5×5 array on a single plate. Plates with macrocolonies were incubated at 28°C for up to 7 days. Cellulose was detected as the fluorescence of colonies on a Calcofluor plate under 366-nm UV light.
Stereomicroscopy.
E. coli macrocolony biofilms were visualized at ×10 magnification with a Stemi 2000-C stereomicroscope (Zeiss, Oberkochen, Germany). Digital images were captured with an AxioCamICC3 digital camera coupled to the stereomicroscope, operated via the AxioVision 4.8 software (Zeiss).
Video recording.
E. coli macrocolonies on an agar plates were fully covered with culture medium and subject to slight shaking. Movies of macrocolony dissolution by shear forces were recorded at real time using a Cyber-Shot DSC camera (Sony, Tokyo, Japan).
Cryosectioning of macrocolony biofilms and fluorescence microscopy.
The procedure and materials used to cryosection macrocolony biofilms and to further examine and visualize thioflavine S fluorescence in cross-sections were exactly as described previously (2).
SEM of bacterial macrocolonies.
Preparation and subsequent SEM analysis of macrocolony biofilm samples were performed exactly as described previously (2).
Determination of β-galactosidase activity.
The β-galactosidase activity was assayed by use of o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate and is reported as μmol of o-nitrophenol per min per mg of cellular protein (33). Experiments showing the expression of lacZ fusions along the entire growth cycle were done at least twice, and the results of a representative experiment are shown.
RESULTS
Restoring cellulose biosynthesis in the “domesticated” E. coli K-12 drastically alters morphology and physical properties of macrocolony biofilms.
In a search for putative mutations that may explain the cellulose-negative phenotype of E. coli K-12, we compared the bcs operons in the genome sequences of the E. coli K-12 strains W3110 and MG1655 with the corresponding genomic regions of cellulose-synthesizing E. coli, in particular the enteroaggregative (EAEC) strain 55989, which not only produces cellulose but also develops into structurally complex macrocolonies (19). In the E. coli K-12 strains, a GTG codon has been annotated as the start codon of the bcsQ gene (Fig. 1A). However, we observed a putative stop codon (TAG) just three codons upstream with another putative start codon located five codons further upstream, which not only is an optimal ATG but is also preceded by a seemingly better Shine-Dalgarno sequence than the GTG start codon originally annotated. This suggested that bcsQ translation may actually start at the ATG codon, but could be terminated only five codons downstream, which may result in polarity on the expression of the genes downstream of bcsQ. Moreover, the cellulose-producing strain 55989 (as well as many other E. coli strains of different origins and properties) carry a TTG codon instead of this putative TAG stop codon in bcsQ. In addition, we observed a few more single nucleotide polymorphisms (SNPs) in the bcs operons of E. coli K-12 and EAEC 55989, but these SNPs result either in synonymous codons or in conservative amino acid exchanges.
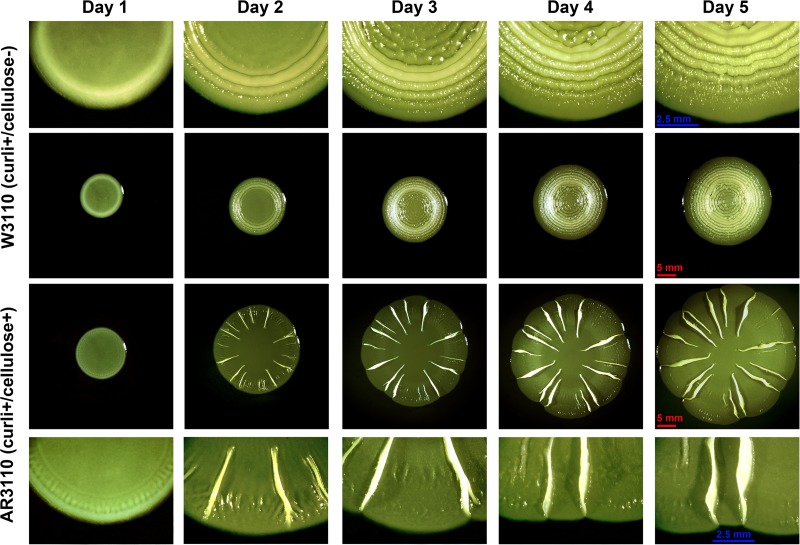
Thus, the TAG/TTG SNP early in bcsQ was clearly the one with the largest potential consequences for bcs gene expression. We therefore exchanged TAG in the chromosome of strain W3110 by TTG using a two-step replacement procedure based on λRed technology. The “repair” of this SNP produced a dramatically altered colony morphology. Macrocolonies of the resulting strain AR3110 were very large (with diameters of >20 mm after 5 to 7 days), were extremely flat, and featured radial ridges and elaborate wrinkles buckling up from the flat areas of the colonies (Fig. 1B). These macrocolonies stained dark red with the amyloid dye Congo red (CR), but morphological structures also formed on CR-free agar plates. The number of ridges and wrinkles increased on relatively moist agar plates, thereby producing patterns resembling the “rdar” morphotype (Fig. 1D) originally described for Salmonella (7) and also observed for certain pathogenic E. coli (18, 38), whereas macrocolonies grown on somewhat dryer agar plates generated fewer radial ridges up to 5 mm high (Fig. 2). These radial ridges started to form in the outer regions of macrocolonies between one and 2 days of growth, with a fine pattern of radial lines being already visible on day 1, some of which buckled up into the ridges, whereas those in between seemed to flatten out. The ridges then slowly increased in height and propagated toward the central region of the macrocolony (Fig. 2).
Fig 2.
Formation of macrocolony architecture of over time. Central panels show low-magnification images of W3110 and AR3110 macrocolonies developing over time on TS-containing salt-free LB plates. Upper and lower panels show enlarged view of macrocolonies presented in the central panels.
Calcofluor binding and fluorescence indicated that AR3110 macrocolonies generated cellulose (Fig. 1C). Moreover, a precise deletion of the cellulose synthase gene bcsA in AR3110 reconstituted the ring-structured morphology previously observed for strain W3110 (Fig. 1D). Similarly, precise and nonpolar deletions of bcsQ and yhjR, and—to a lesser degree—the deletion of the bcsEFG operon in AR3110 produced macrocolonies with ring patterns indicating no or reduced cellulose biosynthesis. A csgB-deleted derivative which synthesizes cellulose but no curli fibers produced pink staining on CR plates and a pattern of dense small wrinkles (Fig. 1D).
We conclude that the SNP that generates the stop codon early in bcsQ interferes with cellulose biosynthesis in the “domesticated” E. coli K-12 laboratory strains. In order to quantify this effect with respect to the expression of bcs genes downstream of this stop codon, we generated single-copy lacZ fusions in yhjR, bcsQ, and bcsA (see Fig. S1 in the supplemental material), with the fusions in the latter two genes containing either the TAG stop codon like in W3110 (′WbcsQ::lacZ′ and ′WbcsA::lacZ′) or the TTG codon as present in AR3110 (′ARbcsQ::lacZ′ and ′ARbcsA::lacZ′). Expression of the reporter fusions with the TTG codon was indeed >10-fold higher than that of the fusions carrying the stop codon at this position (Fig. 3), independently of whether expression of the fusions was assayed in the absence or presence of cellulose production (i.e., in the W3110 or AR3110 genetic backgrounds). The lacZ fusion data also show that the expression of yhjR, i.e., the first gene in the operon whose function in cellulose biosynthesis is unknown, was ∼5-fold higher than that of bcsQ and bcsA. Moreover, all three genes (and therefore probably the entire operon) showed some expression in growing cells already that further increased in a σS-dependent manner, when cells entered into stationary phase (Fig. 3C). Using Flag tagging of bcsA in the chromosome and immunoblotting, we could also detect cellulose synthase in macrocolonies of the repaired strain AR3110 but not in W3110 (see Fig. S2 in the supplemental material).
Localization of cellulose and amyloid curli fibers within E. coli macrocolonies.
Reconstitution of cellulose biosynthesis in strain AR3110 allowed us to study the function and precise localization of cellulose in the matrix of macrocolony biofilms of this otherwise well-characterized E. coli K-12 strain. Macrocolonies were grown on salt-free LB medium supplemented with thioflavine S (TS), a fluorescent dye that—like CR—binds amyloid curli fibers, as well as cellulose without altering macrocolony patterns (Fig. 2). Macrocolonies were analyzed by cryosectioning combined with bright-field and fluorescence microscopy.
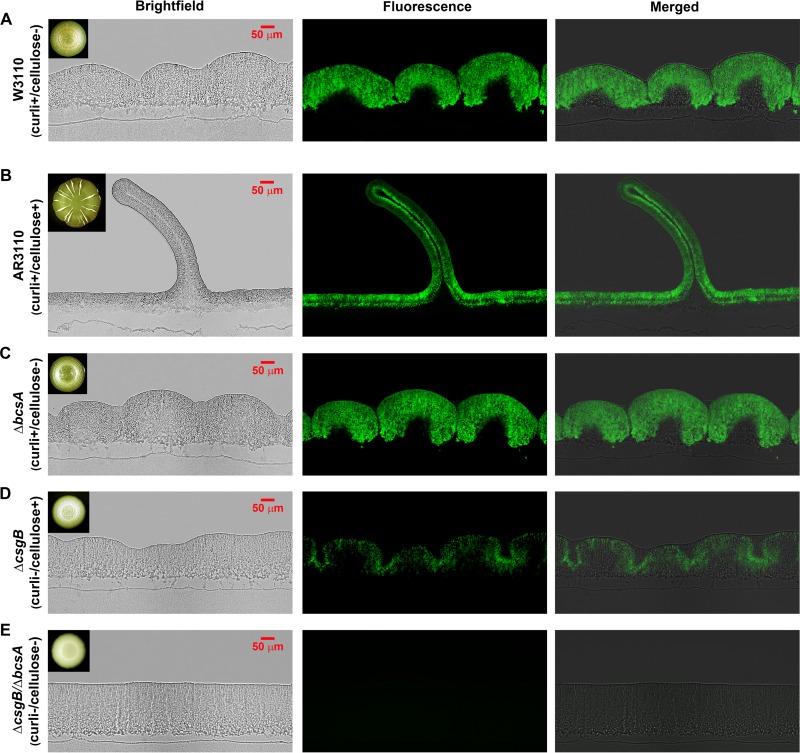
In line with macroscopic colony morphology, already low-magnification bright-field examination of cross-sections of mature AR3110 macrocolonies revealed dramatic architectural changes associated with restored bcs operon expression and cellulose production. Flat areas at the macrocolony center or between ridges exhibited a very regular height of ∼60 μm only, which nevertheless differentiated into two distinct layers (Fig. 4B). The upper bacterial layer (about 40 to 50 μm thick) showed intense TS fluorescence, whereas the bottom layer of cells (∼20 μm thick) was devoid of fluorescence. TS fluorescence is specific for curli and cellulose, as it was detected when either curli or cellulose alone were present (Fig. 4C and D), but was completely absent in the mutant that produced none of the two matrix components (Fig. 4E). In the very flat growth zone at the outer edge of AR3110 macrocolonies (see Fig. S3 in the supplemental material), no TS fluorescence could be detected, whereas at >200 μm further inside, the two-layer structure began to build up.
Fig 4.
Restricted localization of curli and cellulose defines a two-layer architecture of E. coli macrocolonies. Five-day-old E. coli macrocolony biofilms grown on salt-free LB medium supplemented with TS were cryoembedded and sectioned perpendicular to the plane of the macrocolony at a thickness of 5 μm. Thin sections were visualized at low magnification, with bright-field (left panels), fluorescence (middle panels), and merged (right panels) microscopic images representing macrocolony cross-sections of W3110 (A), AR3110 (B), and its ΔbcsA (C), ΔcsgB (D), and ΔcsgB/ΔbcsA (E) derivatives. Fluorescence images were false-colored green for TS. Images visualize the whole vertical section of the macrocolonies. The upper-left insets in bright-field images show top-view images of the respective macrocolonies.
Vertical ridge formation starts by buckling up in the outer zone of the flat colony, thereby generating double layers which then move upwards and maintain a constant width independent of the actual height of the ridge. Ridge formation depended on both curli and cellulose. Macrocolonies of strains that produce curli only exhibited concentric rings (Fig. 1D), which in cross-sections appear as a series of contiguous dome-like elevations >200 μm high (Fig. 4A and C). Instead, when cellulose was produced in the absence of curli, the macrocolony showed a pattern of small intertwined wrinkles in the central zone of the colony (Fig. 1D). In cross-sections this translated into a rippled surface (Fig. 4D). The absence of both curli and cellulose led to a thick (∼200 μm high) macrocolony without any morphological differentiation (Fig. 4E).
Vertical ridges featured a narrow inner layer of nonfluorescent cells (Fig. 4B). Remarkably, bacteria in this interior layer did not switch on curli/cellulose production, even when they were located close to the tip of the ridge, i.e., at a large distance from the nutrient-providing agar. It should also be noted that the space at the bottom of the ridges, which opens up during the buckling up of the macrocolony at this position, was entirely full of nonfluorescent cells, suggesting that this space was filled by growing cells.
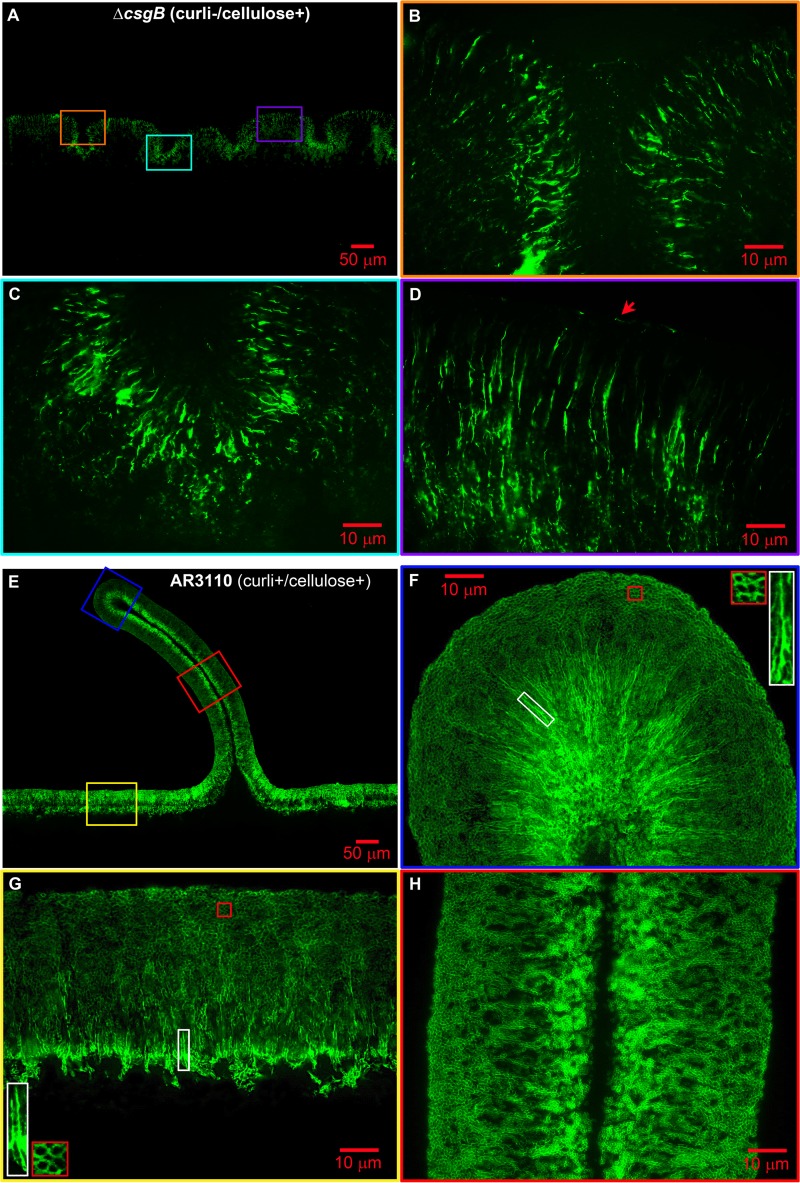
To learn about the detailed spatial arrangement of cellulose in macrocolonies, we first closely examined the strain that produces cellulose alone (AR3110 ΔcsgB). Here, cellulose was visualized by a TS fluorescence pattern that followed the shape of small wrinkles. The concave regions formed below and above the wavy cellulose-containing cell layer also appeared occupied by bacteria (Fig. 4D). Further zooming into the fluorescent layer at different locations (Fig. 5B to D) revealed a regular pattern of patches and especially bright strings, i.e., filaments and possibly sheet-like structures (between 2 and 15 μm long) that were orientated mostly parallel to each other, but perpendicular to the wrinkle line. At points where the fluorescent layer formed the colony surface, strings were also found laying on the surface (see the arrow in Fig. 5D). These results not only show cellulose as a filamentous matrix component, which is in agreement with its fibrillar nature, but demonstrate that its distribution and arrangement is not random, but localized and highly ordered. Unlike curli fibers, which form a “honeycomb”-like network tightly surrounding cells, cellulose filaments seem to extend in between and along cells.
Fig 5.
Cellulose and curli exhibit differential distribution and specific spatial arrangement in E. coli macrocolonies. (A) Low-magnification image showing TS fluorescence in a representative AR3110 csgB macrocolony cross-section, as presented in Fig. 4D. (B to D) Enlarged views of TS fluorescence pattern in respective color-coded macrocolony areas boxed in A. The arrow in D points to a cellulose string lying on the surface. (E) Low-magnification image showing TS fluorescence in a representative AR3110 macrocolony cross-section, as presented in Fig. 4B. (F to H) Enlarged views of TS fluorescence pattern in respective color-coded macrocolony areas boxed in panel E. The insets in panels F and G show enlarged views of the respective color boxed areas.
Having analyzed the distribution of curli fibers and cellulose each “in isolation,” we closely examined the pattern of TS fluorescence in AR3110 macrocolonies that produce both curli fibers and cellulose. Three representative macrocolony areas were examined: the flat area close to the agar surface and the middle part and the tip of the ridge (Fig. 5E). In the upper fluorescent layer of the flat area two patterns of TS fluorescence were clearly distinguished (Fig. 5G). The upper zone showed the “honeycomb”-like pattern around bacterial silhouettes. Further inside the upper layer, i.e., closer to the interface with the nonfluorescent bottom layer, these silhouettes of bacteria appeared more lined-up and vertically orientated, with intensely fluorescent filaments—that resemble the cellulose filaments in the absence of curli (Fig. 5B to D)—possibly forming sheath-like structures (see enlarged view of white boxed areas in Fig. 5G). These patterns suggest that the curli fiber network is relatively evenly distributed in the upper layer, whereas the bulk of cellulose would prevail in a somewhat lower zone of the upper layer, i.e., closer to the interface with the bottom layer.
This pattern was found to be stably maintained in the middle part of the ridge (Fig. 5H) and was even more pronounced, with even longer intensely fluorescent filaments (boxed area in Fig. 5F), at the tip, where the whole layer is massively curved (compare Fig. 5F and H). The vertically stretched appearance of filaments and putative sheaths, as well as the horizontally stretched appearance of the curli fiber network close to the surface, probably reflects the tension that extreme curvature at the tip imposes. The striking observation that the surface of the tip regions does not break under this tension and that ridges can actually become >30-fold higher than they are wide without losing their intricate spatial architecture demonstrates that cellulose confers strong cohesion and elasticity to the upper layer of the macrocolony. Overall, these findings reveal for the first time a highly differentially ordered spatial arrangement of cellulose filaments and curli fiber networks and suggest that the combination of these two matrix components confers tissue-like properties to E. coli macrocolony biofilms.
Fine structure of cellulose fibers and curli-cellulose composite structures at cellular resolution in different zones of E. coli macrocolonies.
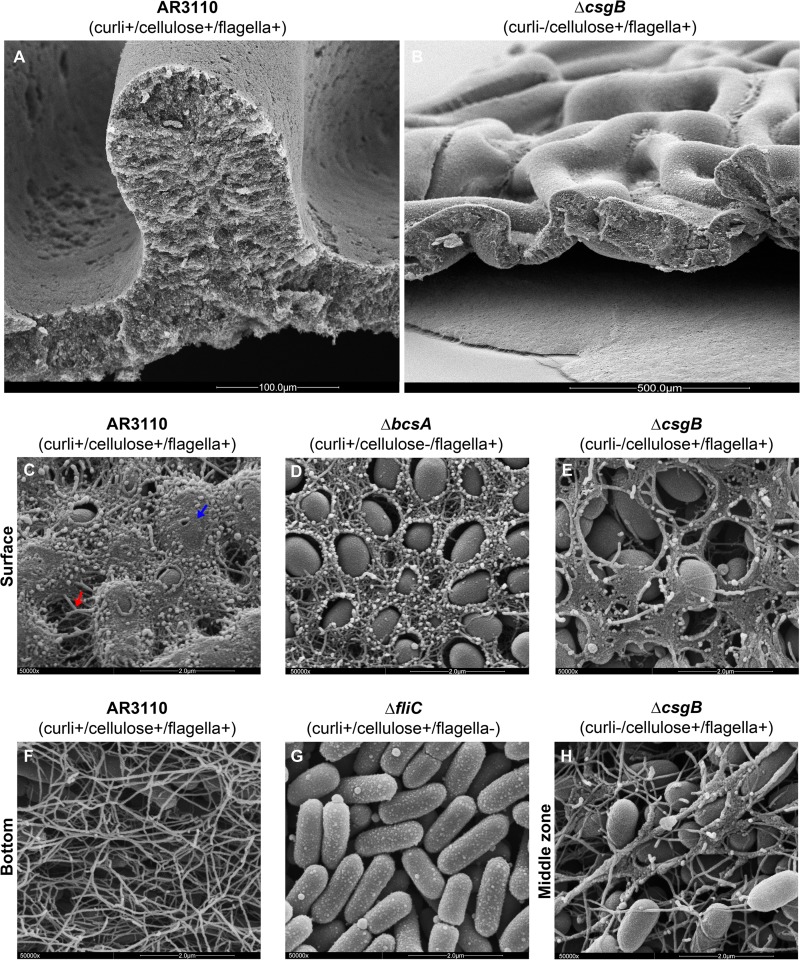
High-resolution SEM allowed us to study this spatial architecture and appearance of cellulose and curli in situ at the cellular level. SEM images of sections through a small AR3110 ridge (Fig. 6A) and a wrinkled area of the cellulose-only macrocolony (AR3110 ΔcsgB; Fig. 6B) show excellent correspondence to the morphological forms seen in the cryosections (Fig. 4 and 5). At higher resolutions, we focused not only on the presence and spatial arrangement of cellulose and curli but also on hallmarks of the bacterial physiological state, such as cell size and shape, cell division, and flagellum production.
Fig 6.
A composite of cellulose and curli fibers covers the AR3110 macrocolony surface, while flagella form a dense mesh at the bottom. (A) Low-magnification perspective-view SEM image of an AR3110 macrocolony area displaying a small ridge. (B) Low-magnification perspective-view SEM image of a macrocolony of the ΔcsgB derivative of AR3110 displaying small intertwined wrinkles. (C to E) High-resolution SEM images (50000x magnification) showing the surfaces of macrocolonies formed by strain AR3110 and its indicated derivatives. (F) High-resolution SEM image (×50,000 magnification) revealing details of the mesh of entangled filaments formed by bacteria at the bottom of the AR3110 macrocolony, which is not present in the AR3110 ΔfliC macrocolony showing flagellum-free elongated rod-shaped cells arranged in a plane (G). (H) High-resolution SEM image (50,000× magnification) showing long cellulose sheets interacting with flagella in the middle inner zone of a AR3110 ΔcsgB macrocolony.
High-magnification SEM imaging of the ΔbcsA macrocolony surface (Fig. 6D) showed the same curli-only pattern previously reported for W3110 macrocolonies (2), i.e., curli fibers forming a dense network around cells that left the cells partially visible (Fig. 6D). In contrast, bacterial cells at the surface of the cellulose-only ΔcsgB macrocolony were not encased but only partially covered by flat sheets of irregular shapes (Fig. 6E). This sheet-like matrix material was specifically cellulose, since knocking out bcsA in the already curli-negative ΔcsgB mutant background rendered bacteria at the surface fully devoid of matrix material (data not shown).
By contrast, the AR3110 macrocolony surface was nearly fully covered by a massive amount of matrix material such that bacteria were hardly visible anymore (Fig. 6C). Cells seemed to be smoothly coated as well as connected by filaments, but curli fibers and cellulose could not be distinguished anymore, suggesting that together they form a composite material. For all strains under investigation, very small and ovoid cells were found at the macrocolony surface (Fig. 6C to E), a finding indicative of cells being in stationary phase, i.e., a condition at which both matrix components are produced. This surface pattern was regularly observed in all surface areas of the macrocolony (e.g., flat sectors, ridges) that at the macroscale exhibited CR staining.
SEM examination of the agar-touching bottom zone of the AR3110 macrocolony showed the same pattern (Fig. 6F) as previously observed in W3110 macrocolonies (2), i.e., a dense mesh formed by long and entangled flagella that was not present in a ΔfliC mutant (Fig. 6G). These patterns were independent of curli and cellulose as they were also observed at the bottom of macrocolonies of ΔcsgB, ΔbcsA, and ΔcsgB ΔbcsA derivatives of AR3110 (data not shown). However, inside the macrocolonies, connections between cellulose and flagella could also be detected, with the network of entangled flagella apparently serving as a scaffold for the formation of cellulose filaments and elongated sheets (Fig. 6H).
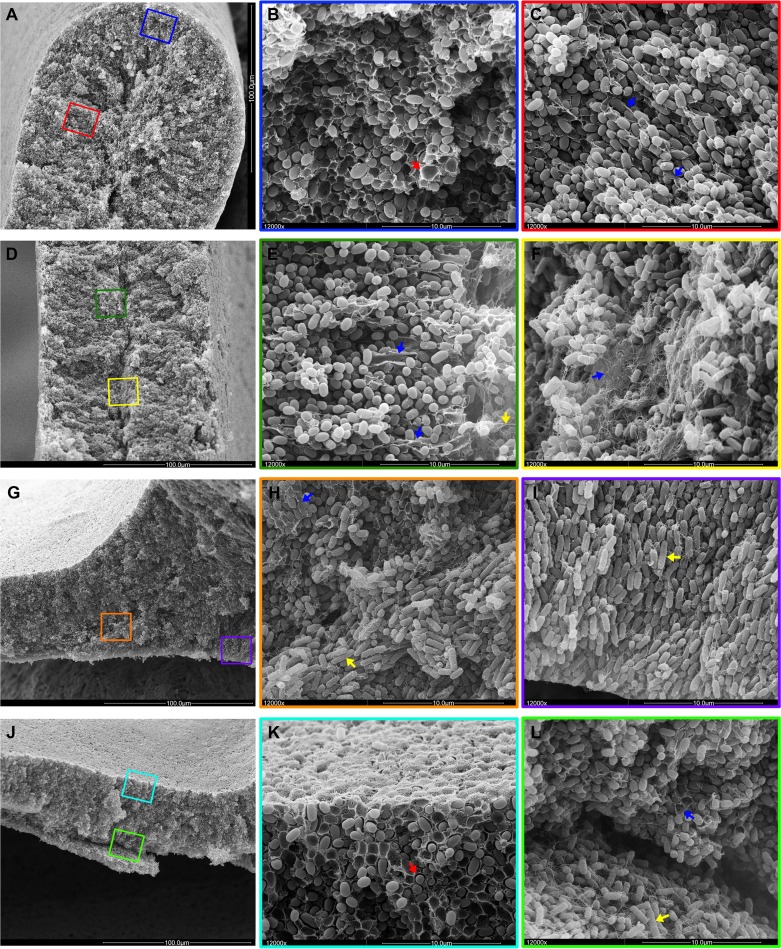
These inner biofilm zones were examined by SEM at four positions in the AR3110 macrocolonies: the tip, the middle part, and the base of the ridge and in flat areas (Fig. 7). At all of these positions the regions close to the colony surface were very compact with small ovoid cells encased by the basket-like structures typical for curli fiber networks (red arrows, Fig. 7B and K). Further inside at all four positions, this pattern changed with small bacteria only partially wrapped in long sheets and sheaths (blue arrows, Fig. 7C and E) that correlate to the elongated structures visualized by TS (Fig. 5F to H). Further inside, entire sheets of fibrillar material were observed (blue arrow, Fig. 7F), which correlates to strong TS fluorescence in this area (Fig. 5F to H). This apparently cellulose-rich zone delineated the boundary to the innermost cellular layer in the ridges and to the bottom layer in the flat areas. In the cross-sections of the flat area, these top and bottom layers also easily broke apart during preparation (Fig. 7J and L). The division into two physiologically distinct cell types was especially apparent at the base of the ridges, where small matrix-encased stationary-phase cells can be seen above the division line, whereas on the bottom side long rod-shaped cells grew in vertical rows and filled the space at the base of the ridge (Fig. 7G to I).
Fig 7.
Spatial arrangement of cellulose, curli and flagella and cell physiology in the context of the two-layer biofilm architecture. For an overview, low-magnification SEM images of AR3110 cross-sections at the tip (A), middle body (D), and base (G) of a ridge and a flat area (J) are shown in the left column of images. The middle and right columns show color-boxed images at ×12,000 magnification corresponding to the color-boxed zones in the overview images. A red arrow in panel B points to basket-like matrix structure typical for curli fiber networks. Blue arrows in panels C, E, F, and H label sheet-like material indicative of cellulose. A yellow arrow in panel E points to flagellum-like filaments in the inner region of the ridge characterized by elongated growing cells. Yellow arrows in panels H and I point to rod-shaped bacteria in the growth zone below the ridge. A red arrow in panel K points matrix-encased bacteria in the upper layer with starving cells. Blue and yellow arrows in panel L indicate cells in contact with sheet-like and filamentous matrix material at the base of the upper colony layer and elongated rod-shaped cells entangled by flagella in the lower layer, respectively.
Cellulose, together with curli fibers, generates cohesion and elasticity and thereby determines the morphological shapes of E. coli macrocolony biofilms.
Colonies of the cellulose-positive strain AR3110 exhibited stronger cohesion, e.g., during restreaking, than colonies formed by the parental W3110 strain, a phenotype also associated with the rdar colony morphotype in Salmonella (20, 21). This difference could be easily visualized by covering the fully grown macrocolonies with liquid and then exposing them to shear forces by slightly shaking the agar plates manually. A curli- and cellulose-deficient mutant just released a cellular haze dissolving into suspension during this procedure (see Movie S1 in the supplemental material). A W3110 macrocolony—with curli fibers as the only matrix component in its upper layer—broke into rigid pieces that swam away and broke down further (see Movie S2 in the supplemental material). In contrast, a AR3110 macrocolony with its composite matrix of curli fibers and cellulose, behaved more like a tissue (see Movie S3 in the supplemental material). Thus, its thick top layer detached from the agar support essentially as an entire macrocolony, with its edges “waving” back and forth with the liquid current, whereas the bottom layer—which reflects the pattern of wrinkles of the detached top layer—remained on the agar. This behavior indicates that cellulose in combination with curli fibers confers not only strong cohesion, but also high elasticity to the macrocolony biofilm—a property consistent with the almost complete absence of breaks in the high-resolution images of the surface of the macrocolonies AR3110 or its curli-deficient cellulose-only derivative (Fig. 5 and 6). This pronounced elasticity is likely to be a prerequisite for forming the highly curved surfaces associated with the ridges and wrinkles of macrocolonies.
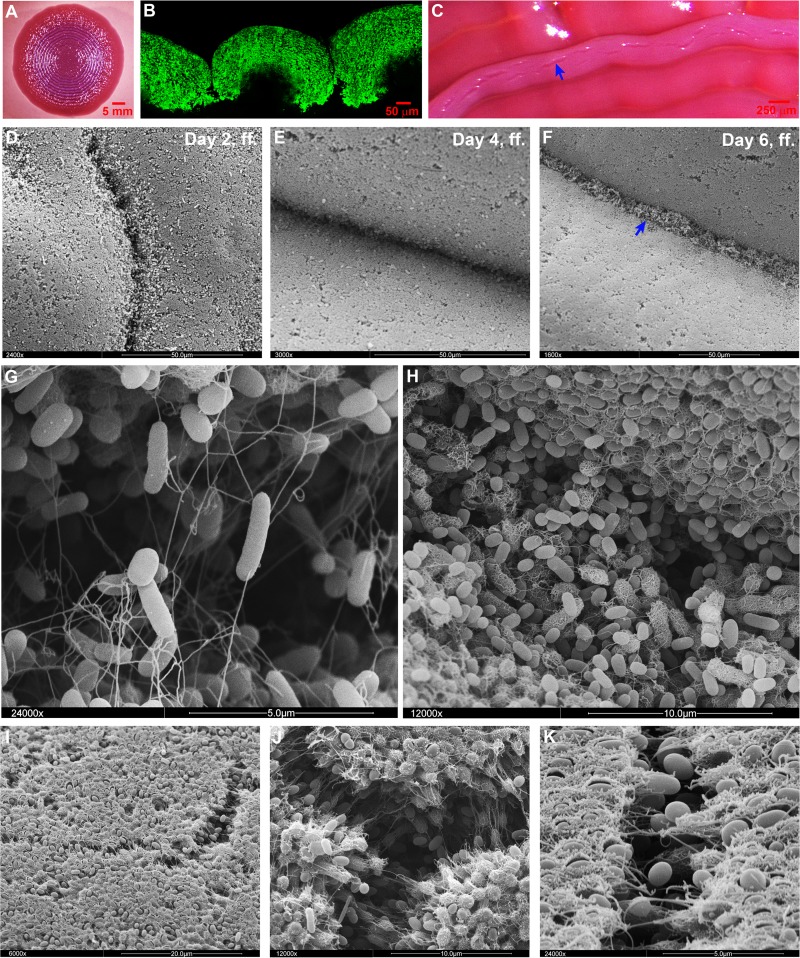
On the other hand, the rigidity and brittleness of the proteinaceous matrix consisting of the curli fiber network alone seems the basis for the formation of the macroscopic ring structures in the macrocolonies of W3110 and the other curli+ cellulose− strains generated in the present study (Fig. 8). The formation of these rings seemed to start with surface breaks in the circular zone of the growing macrocolony where the layer of curliated cells was still relatively thin and less confluent. These large breaks, which probably arose due to pressure generated by the still propagating cells below, widened into grooves (Fig. 8D), with the adjacent curliated cell layers rounding up and finally adopting the dome-like shapes in cross-sections through the ring pattern of the mature macrocolony (Fig. 8B). During this process, the grooves further deepened into crevices (Fig. 8E and G) which extended deep down into the colonies (Fig. 8B) and which were subsequently colonized by cells that seemed to be pushed up from the bottom layers (Fig. 8F). Initially, these cells emerged as rod-shaped flagellated cells (Fig. 8G) but probably due to starvation at their new position, they later on switched on curli production (Fig. 8H), which could eventually close the opening in the macrocolony structure by a “secondary” curli network layer. In addition, small microbreaks also formed in confluently curli-covered surface areas (Fig. 8C and I to K). These tiny fissures seemed to open slowly with occasional curli fibers stretching across them and did not affect the macroscopic overall structure of the colony.
Fig 8.
Deep breaks in the macrocolony surfaces of strains that produce curli but not cellulose generate concentric ring morphology. (A) Top-view image of a mature W3110 macrocolony grown at 28°C on a CR plate showing the curli-dependent ring pattern. (B) Fluorescence image at low magnification of a cross-section through a W3110 macrocolony grown in the presence of TS. (C) Enlarged top-view image of W3110 macrocolony rings. The arrow indicates naturally occurring breaks at the top of the rings. (D, E, and F) SEM images showing changes at the W3110 macrocolony surface over the course of ring formation. The arrow in panel F indicates bacteria colonizing the crevice at late time points. (G) SEM image of the colony surface (at ×24,000 magnification) showing the initial bacterial colonizers of a crevice (day 6, ff., in panel F), which are rod-shaped and carry flagella. (H) SEM image (at ×12,000 magnification) showing bacteria later during the colonization of a crevice (day 7, ff.), which have almost filled the crevice, have become shorter and in part have started to generate curli around their cell bodies. (I, J, and K) SEM images showing microbreaks at the top of a mature W3110 macrocolony (at ×6,000, ×12,000, and ×24,000 magnifications, respectively).
In conclusion, curli fibers and cellulose either alone or in combination determine which three-dimensional structural elements arise in a macrocolony biofilm. Concentric rings originate as breaks in the brittle macrocolonies that produce exclusively curli fibers in high amounts, whereas the formation of ridges and elaborate wrinkles starts by buckling up of the elastic tissue-like macrocolonies containing cellulose, with further elevation of stable ridges requiring a curli/cellulose composite matrix.
DISCUSSION
E. coli macrocolony biofilms show a two-layer architecture that reflects fundamentally different physiological states.
We have previously shown that cellulose-deficient E. coli K-12 (strain W3110) forms macrocolony biofilms consisting of two physiologically distinct layers of cells (2): growing flagellated cells at the outer colony edges, as well as in the bottom layer, as opposed to small stationary-phase cells surrounded by a thick network of curli fibers in the top layer and at the surface (except at the outer rims). Despite its drastically altered macroscopic morphology, the cellulose-“repaired” strain AR3110 equally shows a microscopic two-layer architecture (Fig. 4B). Due to the flatness of the colonies, the transition between these layers is even much sharper, with the thin bottom layer of growing cells—only ∼20 μm high—forming a tight mesh of flagella indistinguishable from that of the cellulose-free W3110 (Fig. 6F; compare to images shown in reference 2). The top layer, which is about 40 to 50 μm high, now produces curli fibers as well as cellulose (Fig. 4B, Fig. 5F and H; see also Fig. S3 in the supplemental material), with cellulose forming thick filaments and sheath-like structures that extend vertically into the top layer, as well as thick and flat sheets that delineate the border between the two layers, in particular within the ridges (Fig. 7).
It is important to note that this two-layer architecture is not a consequence of matrix formation. Rather, it is a function of the gradient of nutrients which are provided from the solid growth medium below and around the colony. In fact, it also forms in mutants free of cellulose and curli, as can be concluded from morphological hallmarks such as the different cell shape, the absence or presence of cell division, and flagellum formation in the two layers (2). Actually, the small and ovoid cells in the top layer grow slowly and finally cease to grow as they have switched to a RpoS-based stationary-phase physiology, which not only confers high stress resistance but is also a prerequisite for the production of curli fibers and cellulose (39). Once these matrix components are assembled the two layers can be physically disrupted (Fig. 7J and see also Movies S1 to S3 in the supplemental material). Since this stratification reflects the two fundamentally different physiological strategies of bacteria—optimizing growth versus optimizing maintenance and survival (39, 40)—and the formation of nutrient gradients is associated with any biofilm formation, we expect a two-layer architecture to be rather general in bacterial biofilms. Whether the starving cells make up the upper or the lower layer, actually depends on species-specific metabolic properties, in particular whether oxygen is essential for growth or not (as discussed in reference 2).
When our E. coli K-12 strains are grown on salt-free LB medium, expression of the regulator CsgD is high enough to generate the high levels of curli and cellulose required for buckling up into ridges and wrinkles. However, when bacteria are grown on yeast extract-Casamino Acids (YESCA) medium, the production of curli and cellulose is insufficient to generate complex morphology (S. Herbst and R. Hengge, unpublished data). Such conditions are therefore convenient for studying wrinkling-inducing conditions or mutations. In a recent study with the uropathogenic E. coli strain UTI89 grown on YESCA medium, the presence of ferric chloride could induce macrocolony wrinkling, which allowed to float off an upper colony layer. This was interpreted as “bimodal population development” into two distinct layers induced by oxidative stress, although the csgBAC and yaiC (adrA) genes were not induced further by this treatment (41). These floated layers were shown to contain cells that were H2O2 resistant and expressed curli (41), which are both RpoS-dependent functions (39), a finding consistent with physiological two-layer architecture being a function of the nutrient gradient building up in macrocolonies under all conditions rather than being induced de novo by oxidative stress. Taken together, this indicates that oxidative stress may specifically stimulate cellulose formation and therefore the tissue-like cohesion and elasticity required to generate the macroscopic colony structure observed by DePas et al. (41), as well as in our study.
Cellulose is a distinctly localized architectural element that forms a “nano-composite” with curli fibers and confers tissue-like properties.
At the cellular level, cellulose fibers assemble into irregular thick filaments and sheet-like structures that extend between cells (Fig. 6E) rather than tightly surrounding cells as the “baskets” formed by amyloid curli fibers (2). Cellulose is not randomly distributed within the upper macrocolony layer (summarized in Fig. 9A). Rather, a massive accumulation of vertically arranged cellulose filaments, sheets and possibly even long sheaths along strings of cells can be seen close to the interface with the lower layer, especially in the massively curved region of the tips of the ridges (Fig. 5F to H, Fig. 7C and E). Along the interface itself, large cellulose sheets are formed (Fig. 7F). In addition, a composite of cellulose and curli fibers can be visualized at the colony surface (Fig. 6C), in which the two types of fibers in fact cannot be distinguished. This matrix material even visually combines the properties of curli and cellulose, as it surrounds the cells, but also provides for filamentous and sheet-like connections between cells, which in toto generates a relatively smooth and confluent protective surface. This may explain the earlier observation that a Salmonella biofilm matrix consisting of curli and cellulose is not digested with either cellulase or proteinase K (12). In other words, E. coli and Salmonella produce naturally in their biofilm matrix what material scientists have been trying to optimize—a cellulose “nanocomposite” material (42, 43). Notably, cellulose can also assemble along flagella, as could be detected in the transition zone between the growing and starving cell layers (Fig. 6H), suggesting that cellulose in general easily assembles along or around filamentous scaffold structures.
Fig 9.
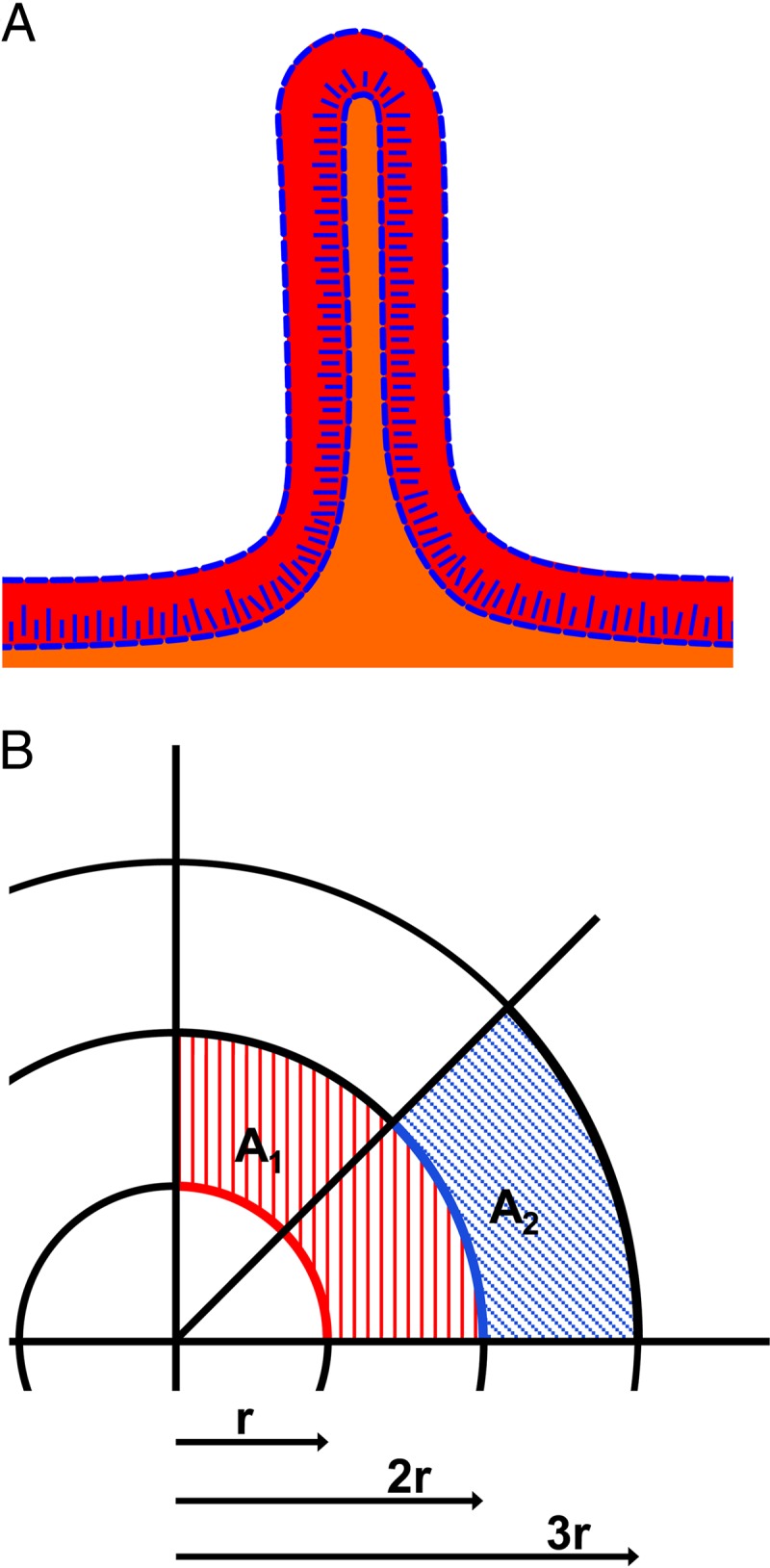
Cellulose localization and the generation of ridges in macrocolonies. (A) Schematic summary of cellulose location and two-layer architecture in AR3110 macrocolonies. Red and orange layers illustrate zones of stationary and post-exponential-phase-like physiology, respectively. Blue lines indicate zones rich in cellulose. (B) Schematic representation of radial expansion of a colony. Circle lines of a radius of r, 2r, and 3r represent the border of hypothetical colonies. Red and blue lines in colonies with a radio of r and 2r, respectively, have the same length but different curvature angles, i.e., bacteria positioned at these outer rim lines of an expanding colony can grow into successively smaller areas (illustrated by the hatched red and blue areas). Due to “tissue-like” cohesion and elasticity of a colony that generates both curli fibers and cellulose, this results in the colony buckling up into radially oriented ridges.
Only the combination of cellulose and curli fibers generates the pronounced cohesion, stability, and elasticity of the macrocolonies of strain AR3110 required to develop complex morphology. Elasticity in particular may be due to high water incorporation by cellulose (42). In contrast, an upper layer matrix with curli fibers only stabilizes cells at their place but is brittle and breaks into the ring structures typical strains producing high levels of curli only (Fig. 8). On the other hand, in a macrocolony that produces cellulose only, small wrinkles are initially generated, but then cellular crowding seems to push still growing cells into the initially forming cavities and seem to overgrow the wrinkles over time (Fig. 4D). In conclusion, curli fibers function as a cellular “mortar” that keeps cells in place, whereas cellulose confers elastic cohesion to this cellular “building material.” It should be noted that a recent report attributed ethanol-induced or dimethyl sulfoxide-induced increased cohesion and elasticity of pellicle biofilms of the uropathogenic E. coli UTI89 to increased curli expression (44, 45). Unfortunately, cellulose was not considered in those studies, and our observations of the distinct biofilm properties conferred by curli and cellulose, as well as the coregulation of both components by CsgD and c-di-GMP, suggest that cellulose biosynthesis may well be stimulated in parallel to curli expression under these conditions.
Tissue-like properties conferred by cellulose allow to generate the intricate three-dimensional morphology of E. coli macrocolonies.
It has long been known that curli fibers and cellulose are a prerequisite for complex macrocolony morphology (12). Based on our high-resolution localization studies, we can now address the question, how the combination of cellulose and curli fibers in specific zones of the E. coli matrix generates this particular three-dimensional architecture. In order to solve this question, it is helpful to conceptually deconstruct this complex morphology—summarily termed “rdar,” “wrinkled,” or “rugose”—into specific elements. These include the size and flatness of the macrocolony, as well as distinct morphological form elements such as “ridges,” “wrinkles,” and “rings,” which can occur in radial, concentric, and/or irregular patterns.
Mature AR3110 macrocolonies exhibit extreme size and flatness ≥20 mm in diameter but only about 60 μm high in the flat areas, as opposed to the >200-μm high macrocolonies of W3110 (Fig. 4). This is probably a function of cellulose filaments and sheets connecting to curli fibers and to flagella, which possibly tethers cells together such that progeny cells below cannot just pile up vertically but are forced to spread out laterally. Wrinkles and ridges arise when macrocolonies buckle up, which is only possible due to stability, cohesion, and elasticity of the cellulose/curli composite matrix in the upper macrocolony layer. Recently, it was proposed that buckling up of Bacillus subtilis macrocolonies into wrinkles happens in regions of local cell death, which “provides an outlet for lateral compressing forces” (46). Although that study focused on central and rather old regions of the macrocolonies, we observed that ridge formation started at the outer rims and then propagated toward the older center of the E. coli macrocolonies (Fig. 2). This points to an explanation, which relies on simple geometry combined to the tissue-like properties of the macrocolony. When a growing colony increases in size, sections of the same length along the outer colony edges (illustrated by the same length of the red and blue lines in colonies with a radius of r and 2r in Fig. 9B) exhibit a continuous reduction in curvature. Thus, although it seems counterintuitive, the larger the colony grows the less is the horizontal space that cells can grow into. In a colony with tissue-like cohesion and elasticity, the resulting tension cannot be released by just piling up single progeny vertically (note that the formation of the upper layer with curli and cellulose synthesis begins quite close behind the outer colony edge [see Fig. S3 in the supplemental material]) (2). Rather, an entire region of the colony has to locally bulge out upwardly. The resulting release of tension in fact allows the regions between two newly elevating ridges to flatten again (Fig. 2). At which positions the buckling actually happens, seems random and may be influenced by microheterogeneities at the agar surface.
Our data also show the amazing dimensions of the ridges. In mature macrocolonies these can be >4 mm high, which corresponds to >30-fold their width (of about 120 μm; Fig. 4B). In the “city of microbes” (47), these ridges are the equivalent of “skyscrapers” thus demonstrating the superb stability and elasticity conferred by the cellulose/curli nanocomposite material. Similar radial ridges are generally observed in macrocolony or pellicle biofilms of “undomesticated” bacteria. In the Gram-positive Bacillus subtilis, their formation depends on a combination of an amyloid-forming protein, TasA, and an exopolysaccharide (9, 10). Since amyloids are common in bacteria (48, 49), amyloid-polysaccharide nanocomposites in the matrix may stabilize complex biofilm architecture in many species.
Physiological roles of cellulose and the cellulose-generated morphology of macrocolony biofilms.
As we begin to understand how a particular macrocolony architecture is generated, another intriguing question is why bacteria produce this complex morphology. What is the benefit of growing in extremely large, flat, and wrinkled colonies; why invest all of the energy and carbon sources to produce a matrix that allows to build huge ridges? Oxygen concentration in macrocolonies decreases to ca. 10% of the oxygen content of air at a depth of ∼60 μm (41, 50). Since wrinkling of Pseudomonas aeruginosa or Bacillus subtilis is induced by impaired respiration, it has been suggested that surface enlargement by wrinkling facilitates access to oxygen as a terminal acceptor for respiration (50, 51). For E. coli, we suggest that the beneficial adaptation with respect to access to oxygen is the flatness of the macrocolonies generated by the curli/cellulose matrix. The buckling into wrinkles and ridges then seems a secondary and physical consequence of the tissue-like properties of these very flat biofilms.
In addition, the buckling up of certain regions of the colony has biological consequences as it relatively rapidly moves large groups of cells over large distances where they are exposed to novel microenvironmental conditions. For instance, during ridge formation, cells that used to be in the lower growth layer are rapidly elevated to a position where they are exposed to strict starvation. These cells form the thin layer inside the ridges that does not switch on curli and cellulose production, which is consistent with CsgD and curli genes being only weakly induced when cells are exposed to sudden starvation (e.g., in minimal medium; E. Klauck and R. Hengge, unpublished data). Also, at the bottom of a ridge, a new zone for growth opens up. Due to the largest distance from the colony surface, this zone can be expected to have the lowest oxygen content in the macrocolony. However, E. coli, which can easily switch to anaerobic growth, completely fills up these zones below the ridges (Fig. 4B and Fig. 7G to I). This is in contrast to the situation recently observed for the more oxygen-dependent B. subtilis, where the zones below ridges remain open and serve as channels that facilitate liquid transport (52).
From a more ecophysiological perspective, growing in very large, flat, and tissue-like macrocolonies in an outside environment—many E. coli strains produce curli and cellulose only below 30°C—may provide the advantage of rapidly covering nutrient-providing surfaces and therefore competing better with bacteria that are more dependent on oxygen for growth such as Bacillus or Pseudomonas species. Also, an outer colony layer covered by the curli/cellulose nanocomposite may provide superior protection against predators or exoenzymes produced by competing species in comparison to curli-only or cellulose-only surfaces. Furthermore, tissue-like cohesion of colonies may facilitate the transmission from the environment to a host and enhance survival during the passage through the stomach of a new host.
Cellulose also modifies properties of the fibers or filaments it assembles with and confers additional functions not associated with colony architecture. Thus, it reduces curli-mediated adhesion to surfaces (22). On the other hand, it allows Salmonella to adhere to the chitinous surface of Aspergillus hyphae in mixed biofilms (53), and it controls the adherence of Rhizobium to plant roots (54). Cellulose can also counteract the proinflammatory effect of curli fibers when expressed in the mammalian host (55, 56). Finally, cellulose can inhibit the rotation of flagella, which was observed in a mutant deficient for the c-di-GMP-binding protein YcgR, a protein that shuts down rotation by direct interaction with the flagellar basal body (57). In fact, we could visually demonstrate a tight entanglement of cellulose with flagella in the transition area between the growth and stationary phase layers of the macrocolony (Fig. 6H).
Common and differential regulation of amyloid curli fibers and cellulose and the environmental control of macrocolony architecture.
In the present study, we have focused on where in a macrocolony cellulose and curli fibers occur and how these matrix components together contribute to generating the specific morphological elements of macrocolony biofilms. In fact, macrocolony morphogenesis is regulated in response to many environmental cues such as the presence of salt (58), oxidative stress (41), ethanol (44), or humidity of the agar support. For instance, macrocolonies can be rather stiff with a few very high and radial ridges only, or they can present entire “landscapes” of elaborate wrinkles and a large number of high ridges (Fig. 1 and 2). It is obvious that not only location per se but also the amounts of and probably the ratio between cellulose and curli fibers in specific zones of a macrocolony determine stiffness and elasticity and therefore matter for generating a specific morphology. At the molecular level, curli and cellulose production are coregulated by CsgD (21), which in turn requires RpoS and several c-di-GMP-producing and -degrading enzymes acting in a complex regulatory network (26, 27, 59, 60). The ratio between the two matrix components can be affected by additional c-di-GMP signal input in the control of cellulose production (29).
Finally, zones of different conditions in a macrocolony are not restricted to the global physiological two-layer architecture. Actually, the formation of biofilm macrostructures such as ridges or wrinkles in turn generates novel microenvironments. In the future, it will be interesting to see, whether and how c-di-GMP signaling by multiple DGCs and PDEs spatially modulates the synthesis of curli, cellulose, and perhaps additional matrix components and thereby controls biofilm architecture in response to macro- and microenvironmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicole Sommerfeldt for generating the bcsA::Flag construct.
This study was funded by the European Research Council under the European Union's Seventh Framework Programme (ERC-AdG 249780 to R.H.). D.O.S. is the recipient of a postdoctoral fellowship from the Alexander von Humboldt Foundation.
Individual author contributions were as follows: concept and design of the study, R.H. and D.O.S.; design of the experiments, D.O.S., A.M.R., and R.H.; performance of the experiments, D.O.S. and A.M.R.; analysis of the data, D.O.S., A.M.R., and R.H.; and writing of the paper, R.H. and D.O.S.
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00946-13.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 2.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103–13. 10.1128/mBio.00103-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210 [DOI] [PubMed] [Google Scholar]
- 4.Aguilar C, Vlamakis H, Losick R, Kolter R. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 10:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331–348 [DOI] [PubMed] [Google Scholar]
- 6.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36:893–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Römling U. 2005. Characterization of the rdar morphotype, a multicellular behavior in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonas K, Tomenius H, Kader A, Normark S, Römling U, Belova LB, Melefors O. 2007. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7:70. 10.1186/1471-2180-7-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibres provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 107:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nature Rev. Microbiol. 11:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsén A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655 [DOI] [PubMed] [Google Scholar]
- 12.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
- 13.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan JLW, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitney JC, Howell PL. 2013. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 21:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solano C, García B, Valle J, Berasain C, Ghigo J-M, Gamazo C, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793–808 [DOI] [PubMed] [Google Scholar]
- 18.Bokranz W, Wang X, Tschape H, Römling U. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171–1182 [DOI] [PubMed] [Google Scholar]
- 19.Da Re S, Ghigo J-M. 2006. A CsgD-Independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Römling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behavior of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 21.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. 2000. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10–23 [DOI] [PubMed] [Google Scholar]
- 22.Gualdi L, Tagliabue L, Bertagnoli S, Ieranò T, De Castro C, Landini P. 2008. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 154:2017–2024 [DOI] [PubMed] [Google Scholar]
- 23.Hengge R. 2009. Principles of cyclic-di-GMP signaling. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 24.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 25.Römling U, Galperin MY, Gomelsky L. 2013. Cyclic-di-GMP: the first 25 years of a universal bacterial second messenger. Microb. Mol. Biol. Rev. 77:1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain phosphodiesterase YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in Escherichia coli biofilm control. EMBO J. 32:2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signaling within the σS network of Escherichia coli. Mol. Microbiol. 62:1014–1034 [DOI] [PubMed] [Google Scholar]
- 29.García B, Latasa C, Solano C, García-del Portillo F, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264–277 [DOI] [PubMed] [Google Scholar]
- 30.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 31.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34.Kolmsee T, Hengge R. 2011. Rare codons play a positive role in the expression of the stationary phase sigma factor RpoS (σS) in Escherichia coli. RNA Biol. 8:913–921 [DOI] [PubMed] [Google Scholar]
- 35.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 36.Powell BS, Court DL, Nakamura Y, Rivas MP, Turnbough CL., Jr 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss-Muszkat M, Shakh D, Zhou Y, Pinto R, Belausov E, Chapman MR, Sela S. 2010. Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl. Environ. Microbiol. 76:1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hengge R. 2011. The general stress response in Gram-negative bacteria, p 251–289 In Storz G, Hengge R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 40.Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1–8 [DOI] [PubMed] [Google Scholar]
- 41.DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. 2013. Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 110:2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klemm D, Kramer F, Moritz S, Lindström T, Ankerfors M, Gray D, Dorris A. 2011. Nanocelluloses: a new family of nature-based materials. Angew. Chem. Int. Ed. Engl. 50:5438–5466 [DOI] [PubMed] [Google Scholar]
- 43.Siró I, Plackett D. 2010. Microfibrillated cellulose and new nanocomposite materials: a review. Cellulose 17:459–494 [Google Scholar]
- 44.Lim JY, May JM, Cegelski L. 2012. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl. Environ. Microbiol. 78:3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C, Lim JY, Fuller GG, Cegelski L. 2012. Quantitative analysis of amyloid-integrated biofilms formed by uropathogenic Escherichia coli at the air-liquid interface. Biophys. J. 103:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asally M, Kittisopikul M, Rué P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Süel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. U. S. A. 109:18891–18896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watnick P, Kolter R. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. 2007. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9:3077–3090 [DOI] [PubMed] [Google Scholar]
- 49.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. 2012. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. 2013. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 195:1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolodkin-Gal I, Elsholz AK, Muth C, Girguis PR, Kolter R, Losick R. 2013. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 27:887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. 2013. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 110:848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandl MT, Carter MQ, Parker CT, Chapman MR, Huynh S, Zhou Y. 2011. Salmonella biofilm formation on Aspergillus niger involves cellulose-chitin interactions. PLoS One 6:e25553. 10.1371/journal.pone.0025553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laus MC, van Brussel AA, Kijne JW. 2005. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hais. Mol. Plant-Microbe Interact. 18:533–538 [DOI] [PubMed] [Google Scholar]
- 55.Tükel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Bäumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol. 12:1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Rochon M, Lamprokostopoulou A, Lünsdorf H, Nimtz M, Römling U. 2006. Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell. Mol. Life Sci. 63:2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zorraquino V, García B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C. 2013. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J. Bacteriol. 195:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kader A, Simm R, Gerstel U, Morr M, Römling U. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602–616 [DOI] [PubMed] [Google Scholar]
- 60.Simm R, Lusch A, Kader A, Andersson M, Römling U. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.