Abstract
Incompletely resectable ependymomas are associated with a poor prognosis despite intensive radio- and chemotherapy. Novel treatments have been difficult to develop due to the lack of appropriate models. Here, we report on the generation of a high risk cytogenetic group 3 and molecular group C ependymoma model (DKFZ-EP1NS) which is based on primary ependymoma cells obtained from a patient with metastatic disease. This model displays stem cell features like self renewal capacity, differentiation capacity and specific marker expression. In vivo transplantation showed a high tumorigenic potential of these cells, and xenografts phenotypically recapitulated the original tumor in a niche dependent manner. DKFZ-EP1NS cells harbor transcriptome plasticity, enabling a shift from a neural stem cell-like program towards a profile of primary ependymoma tumor upon in vivo transplantation. Serial transplantation of DKFZ-EP1NS cells from orthotopic xenografts yielded secondary tumors in half the time compared to the initial transplantation. The cells were resistant to temozolomide, vincristine and cisplatin, but responded to histone deacetylase inhibitor (HDACi)-treatment in therapeutically achievable concentrations. In vitro treatment of DKFZ-EP1NS cells with the HDACi Vorinostat induced neuronal differentiation associated with loss of stem cell-specific properties. In summary, this is the first ependymoma model of a cytogenetic group 3 and molecular subgroup C ependymoma based on a human cell line with stem cell-like properties, which we used to demonstrate the differentiation inducing therapeutic potential of HDACi.
Keywords: ependymoma, cancer stem cells, differentiation, histone deacetylase inhibitor
Introduction
Ependymomas are common brain tumors and make for approximately 9–10% of all primary CNS tumors and 2% of all childhood malignancies [25]. In pediatric patients 5-year progression-free survival (PFS) is 43–55%, and 5-year overall survival (OS) is 63–71% [29], differing significantly from adult patients with a 5-year PFS of 62–63% and a 5-year OS of 85% [29]. However, patients with incompletely resected tumors fare worse in several studies [3, 13, 57], with 3-year PFS of only 39% [57]. Primary metastatic dissemination at the time of diagnosis ranges from 6–18% [2, 28, 46, 57], and secondary dissemination occurs in 16–17% of patients [29, 50]. Current treatments consist of surgery with the aim of total resection, and irradiation, while the benefit of chemotherapy is controversial.
Genetic aberrations are a frequent event in ependymoma, and aberration patterns are associated with anatomical localization, suggesting distinct pathogenetic pathways in ependymoma development [17, 24, 35, 38, 55]. The most common aberrations described are gains of chromosomes 1q, 7, and 9, and losses of chromosomes 6 and 22q [19, 29, 35, 42, 55]. An algorithm proposed by Korshunov et al. efficiently classifies patients into three groups with distinct survival probabilities using patterns of cytogenetic aberrations [29]. According to this algorithm, patients of group 1 (indicator aberrations including loss of chromosome 6 and gain of chromosomes 9, 15q, and 18) have the best prognosis, of group 2 (balanced profile) have an intermediate prognosis, and of group 3 (defined by 1q gain and/or homozygous deletion of CDKN2A) show a particularly poor prognosis [29]. CDKN2A has been shown to be inactivated in up to 25% of primary intracranial ependymomas [35], and tumors with homozygous deletions of the CDKN2A locus and/or 1q gain carry a high risk for secondary metastases [29]. Only recently the first genetic ependymoma mouse model was established on a CDKN2A deficient background [24].
Despite past efforts to improve PFS and OS of patients with ependymoma, studies have been hampered by the lack of preclinical models allowing for reproducible and standardized conditions under which new therapeutic options can be tested. Indeed, for the past two decades, there has been no significant improvement in the PFS and OS of patients with ependymomas [53]. Many groups studying new treatment options have used short term primary in vitro culture [42, 44] or in vivo transplant models [14, 20, 47]. Only recently Yu et al. [61] and Hussein et al. [22] have reported the establishment of continuously propagatable adherent cell lines, displaying stem cell-like features. However, none of the published human cell lines are reported to harbor the typical homozygous deletion of chromosome 9p21, locus of CDKN2A, reflecting a most aggressive ependymoma subtype.
Cancer stem cells (CSC) are thought to be a subpopulation of the tumor cells distinct from the bulk with the ability to self-renew and driving tumor growth [6]. CSC have been described in many solid tumors including pediatric medulloblastomas [54]. Since the CSC model predicts that CSC are responsible for treatment resistance and relapse, they are thought to be an attractive target for novel therapeutic approaches. Related to the CSC but not to be confounded with is the tumor cell of origin. Taylor et al. described radial glia cells as candidate stem cells for ependymoma [55], and Johnson et al. matched ependymoma subgroups with distinct neural stem cell types, which are thought to represent the origin of the distinct ependymoma tumors [24]. It therefore seems evident that stem cell properties are an inherent capacity of ependymoma.
We here report on the generation of an ependymoma cell line termed DKFZ-EP1NS isolated from a patient with metastasizing supratentorial ependymoma [37], which grows under neurosphere-promoting conditions, displays self-renewal capacity and is usable for long-term propagation. This ependymoma model displays stem cell characteristics, i.e. it generates serial tumors resembling the tumor of origin phenotypically. We finally present data demonstrating that treatment with histone deacetylase inhibitors (HDACi) induces differentiation and loss of stem cell properties in this chemotherapy-resistant model.
Materials and Methods
Cell culture
DKFZ-EP1NS cells were isolated from a patient with a supratentorial anaplastic ependymoma WHO grade III and cultured in neurosphere medium (NSM) as described [37]. Cells were tested and proved to be free of mycoplasma, viral as well as cell contamination using the in-house Multiplex cell Contamination Testing (McCT) Service [51], and services of the German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, DSMZ, Braunschweig, Germany). DKFZ-EP1NS cells were frozen and stored in liquid nitrogen using the cryopreservation medium described by Ladewig et al. [32]. DKFZ-EP1NS cells were kept in culture for up to 9 months, corresponding to over 30 passages. Long-term self-renewing rosette-type human embryonic stem cells (hESC)-derived neural stem cells (NSC) were cultured as described [27]. Brightfield images were taken using an Olympus CX41 microscope with a Color View camera, and CellB 2.3 software (Olympus, Shinjuku, Tokyo, Japan).
Patient samples
Fresh frozen samples were collected between 1993 and 2003. Diagnoses were confirmed by assessment by at least two neuropathologists including a central pathology review. Approval to link laboratory data to clinical and pathological data was obtained from the Institutional Review Board. For patients’ data see Suppl. Table 1.
Immunophenotypical analysis and flow cytometry
Surface markers and intracellular proteins were stained using the antibodies listed in Suppl. Table 3. Staining for ALDH activity was performed using the Aldefluor kit (Stemcell Technologies, Vancouver, BC, Canada), staining with Hoechst 33342 was performed as described [16]. For analysis, the flow cytometer FACS Canto II or FACS Diva for Hoechst staining (both Beckton Dickinson, Franklin Lakes, NJ, USA) and the software FACSDiva (Version 6.1.2., Beckton Dickinson) were used, plots were generated using FlowJo (Version 7.2.5., Tree Star, Inc., Ashland, OR, USA).
Animal model and magnetic resonance imaging (MRI)
Animal experiments were approved by the German Cancer Research Center institutional animal care and use committee and the Regierungspräsidium Karlsruhe. CB17-SCID mice were purchased from Charles River Laboratories (Sulzfeld, Germany) and held under standard animal care conditions at the animal facility of the DKFZ. A total of 2.5 × 105 DKFZ-EP1NS cells in 4 μl NSM were injected as described [26], coordinates were 0 mm rostral or caudal, 2.5 mm lateral (left) and 3 mm depth, allowing for injection into the striatum of the left hemisphere. All MRI examinations were performed using a custom-developed transmit/receive small animal coil in a conventional whole-body 1.5 T MRI scanner (Symphony, Siemens, Erlangen). Lesions including brain tumors were located on T2w turbospinsecho images (TE: 109 ms; TR: 4000; FoV: 40×30 mm; matrix: 128; voxel size: 0,3×0,3×1 mm3).
Immunohistochemistry (IHC), fluorescence in situ hybridization (FISH) and electron microscopy
All IHC stainings were performed on 5 μm thick sections of formalin-fixed, paraffin-embedded microdissected specimens. Antibodies used are listed in Suppl. Table 3. IHC was performed with an automated stainer (Benchmark XT, Ventana, Strasbourg, France) following the protocols of the manufacturer. Microscopic images were captured using a ScanScope scanner (Aperio Technologies, Vista, CA, USA), and the software ImageScope V. 10.2.2.2 352 (Aperio Technologies). FISH was performed as described [29]. For electron microscopy, untreated neurospheres were fixed, embedded and analyzed as described [12].
Gene expression microarray
RNA from DKFZ-EP1NS cells, neural stem cells and from mouse tumors was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany), RNA from primary ependymoma was isolated as described [36]. Hybridized microarrays were scanned at 5 μm resolution in a two-color Agilent Scanner G25505B (Agilent, Santa Clara, CA, USA) with automatically adjusted PMT voltages according to manufacturer’s specification. Array raw data were generated from scanned images using Feature Extraction software (version 9.1; Agilent). The data was preprocessed, quality controlled and analyzed with our in-house developed ChipYard framework for microarray data analysis (http://www.dkfz.de/genetics/ChipYard/) using R [1] and Bioconductor [15] software packages. Feature signals had to fulfill the following criteria to be considered for analysis: minimal signal to background ratio ≥ 1.2 in at least one channel; mean to median spot intensity ≤ the 75% quantile + 3 times the IQR of all features on the array; feature replicate standard deviation ≤ 0.25 per array. Normalization of raw signals was performed using vsn [21]. Probes with more than 40% missing values across all samples were removed. Based on BLASTing the probes sequence information against the genome, biological annotations were retrieved from EnsEMBL (version 54, NCBI Build 36 of the human genome reference sequence).
Metabolic activity assay, treatment with compounds, and neurosphere initiation capacity assay (NSIC assay)
For measurement of metabolic activity, 5 × 103 DKFZ-EP1NS cells were seeded in 100 μl NSM per well in a 96-well plate 24 h before adding compounds and solvent controls in 100 μl NSM. After 72 h of treatment metabolic activity was measured using the WST-1 assay (Roche, Mannheim, Germany) according to manufacturer’s instructions. EC50 (half maximal) concentrations were calculated using “GraphPad Prism” version 3.03 for Windows (GraphPad Software, La Jolla, CA, USA). Compounds used were cisplatin (CDDP; Axxora, Lörrach, Germany), vincristine (VCR; Axxora), temozolomide (TMZ; Axxora), valproic acid (VPA; Sigma-Aldrich), Vorinostat (suberoylanilide hydroxamic acid/SAHA; Chemos, Regenstauf, Germany), Panobinostat (Selleck Chemicals, Houston, TX, USA) and Entinostat (MS275; Axxora) at concentrations indicated. For the NSIC assay, DKFZ-EP1NS cells were seeded in 100 μl of NSM at indicated cell numbers per well in a 96-well plate. 100 μl medium containing compounds or solvent control was added to each well. 16 wells per condition were seeded. After 10 days, number of spheres per well were scored manually for each well.
Western Blot analysis and image processing
Protein concentrations of cell lysates were determined using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) and the Bradford assay (Bio-Rad, Hercules, CA, USA) according to manufacturer’s instructions. For antibodies see Suppl. Table 3. Detection was performed using Amersham ECL Advance Western Blotting Detection System (GE Healthcare, Buckinghamshire, UK). Luminescence was detected using the Chemi Capt detector system (Vilber Lourmat, Eberhardzell, Germany) and Chemi Smart software (version 15.01, Vilber Lourmat), or Amersham Hyperfilm ECL (GE Healthcare) and conventional film development. Uncropped images were cropped using Photoshop CS2 Version 9.0 (Adobe Systems, San Jose, CA, USA).
Cell counts, analysis of viability and cell cycle analysis
Cell counts and analysis of viability by trypan blue exclusion staining was performed using a ViCell XR counter (Beckmann Coulter, Brea, CA, USA). Cell cycle analysis was performed using propidium iodide staining as described [39].
Quantitative real-time RT-PCR
RNA extraction from cell culture experiments, cDNA synthesis, quantitative real-time polymerase chain reaction (qPCR) was performed as described [36]. The delta delta Ct (ddCt) method was used to obtain relative quantification. PGK1 and SDHA were used as control genes, RNA from solvent control treated cells or from normal total brain was used as reference sample as indicated. Primers were obtained through Thermo Electron (Ulm, Germany) and Qiagen (Hilden, Germany) (Suppl. Table 4). Genes defining molecular subgroups were identified from the data set published by Johnson et. al. [24]. Criteria were applied to group A, B, C or D as follows: log2 ratio of gene expression for group A, B, C or D >5, log2 ratio of gene expression for all other groups (A-I) <1, t-test for gene expression for group A, B, C or D <0.00001, allowing for selection of genes that showed a >16-fold expression in the group A, B, C or D over all other groups with a high statistical significance. After filtering for these criteria, the top 4 annotated genes were selected for identification of subgroups.
Statistical analysis
Statistical analysis was performed using “GraphPad Prism” version 3.03 for Windows (GraphPad Software, La Jolla, CA, USA) and “R” (R version 2.12.0, 2010, The R Foundation for Statistical Computing). “GraphPad Prism”: logrank-test for survival analysis, two-sided t-test for analysis of NSIC. The following analyses were carried out with “R”: Pearson’s correlation coefficient was used to calculate pairwise sample correlation. Its multivariate generalization, the RV coefficient [49], was used to assess correlation between sets of samples. Correspondence at the top (CAT) plots were produced to further determine comparability of gene expression profiles across models [23]. CAT plots were based on mean log2 ratios per model. For each of the two models, the proportion of most up- and downregulated genes both models have in common was plotted against the list size. Principal component analysis was used to illustrate separation of tumor models. qRT-PCR log2 relative expression data were tested for up- and downregulation using two-sided t-tests. P-values were adjusted for multiplicity using Benjamini-Hochberg correction in order to control the false discovery rate. For each cell cycle parameter, the ratio of treated and DMSO control measurements were tested with Student’s t-test for increase/decrease. P-values were adjusted for multiple testing. P-values below 0.05 were considered significant.
Results
Generation and characterization of DKFZ-EP1NS cells in vitro and in vivo
Cultivation of isolated primary tumor cells derived from the malignant ascites of a patient with highly aggressive supratentorial anaplastic ependymoma WHO III [37] in neurosphere medium (NSM) gave rise to neurospheres after 3 weeks in culture. Neurospheres were dissociated every 7–10 days and subsequent neurospheres formed (Fig. 1a). After more than 9 months of passaging for over 30 passages, dissociated neurospheres still gave rise to new neurospheres, hence displaying long-term self-renewal (Fig. 1a, last panel). DKFZ-EP1NS could be frozen in cryopreservation medium [32] and thawed without impairing their ability to self-renew.
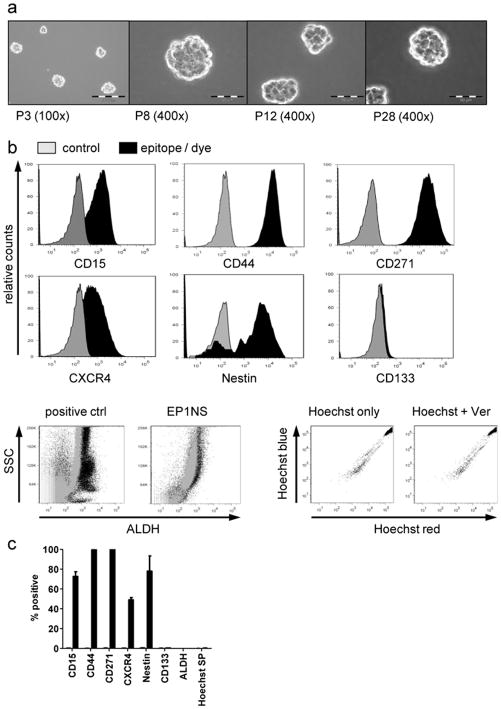
Fig. 1.
DKFZ-EP1NS cells grow in spheres and express markers associated with physiological or cancer stem cells (CSC)
a Brightfield images of DKFZ-EP1NS cells at different passages (P3-P28), original magnification as indicated. b Representative plots of cytometric analyses of markers associated with physiologic or CSC, showing expression of CD15, CD44, CD271, CXCR4 and Nestin on a high proportion of cells, while no consistent population expressing CD133, ALDH (positive control: bone marrow cells) or a side population as defined by Hoechst dye efflux were detected. c Quantification of cytometric analyses, bars represent means of at least three independent measurements performed at passages P10–25, error bars represent standard error of mean (SEM). ctrl: control; P: passage; SP: side population; SSC: sideward scatter; Ver: verapamil.
Because the serum-free neurosphere culture conditions enrich for cells with stem cell-like properties, DKFZ-EP1NS cells were analyzed for expression of markers associated with normal and cancer stem cells, namely CD15, CD44, CD271, CXCR4, Nestin, CD133, ALDH, and staining for Hoechst side population (SP). A majority of DKFZ-EP1NS cells stained strongly for CD15 (72.8% +/− 4.7%; mean +/− SEM), CD44 (99.5% +/− 0.4%), CD271 (99.9% +/− 0.1%), CXCR4 (49.1% +/− 2.3%) and Nestin (86.1% +/− 7.5%) (Fig. 1B and C). However, no distinct subpopulation was detected by staining for CD133, ALDH, or Hoechst SP (Fig. 1b and 1c).
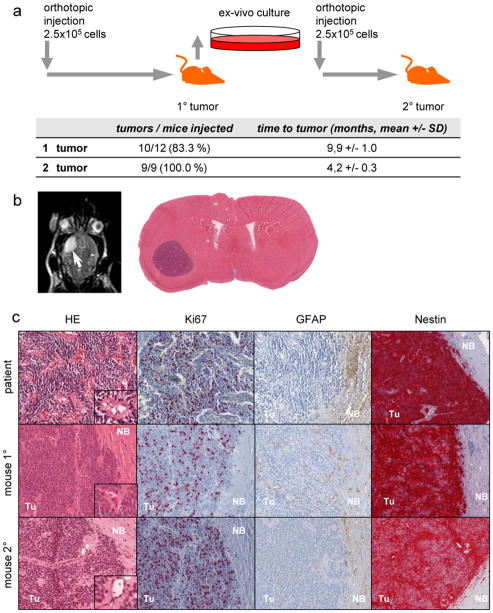
In order to assess the in vivo tumorigenicity of DKFZ-EP1NS cells, an orthotopic xenotransplant model employing CB17-SCID mice was used. DKFZ-EP1NS cells were transplanted into the striatum close to the subventricular zone, as this is thought to be the typical niche for supratentorial ependymoma. DKFZ-EP1NS cells gave rise to tumors after 9.9 +/− 1.0 months (mean +/− SD) in 83.3% (10/12) of the animals transplanted (Fig. 2a) as assessed by clinical signs, and subsequently MRI and HE staining (Fig. 2b and 2c). In order to assess the serial transplantability and hence the persistence of tumorigenicity after in vivo passaging, we explanted the primary tumors and cultured them ex vivo. After 10 days, primary neurospheres were generated and transplanted into mice (Fig. 2a). After 4.2 +/−0.3 months (mean +/− SD) tumors were now formed in all mice (9/9, 100%), as assessed by MRI. HE staining revealed a histology strongly reminiscent of human anaplastic ependymoma, including the formation of pseudorosettes [33], in both primary as well as secondary mouse tumors (Fig. 2c, insets). Ki67, GFAP and Nestin all stained similarly in the patient, the primary and the secondary mouse tumor (Fig. 2c), revealing a high similarity between the orthotopic xenograft tumors and the primary tumor of the patient.
Fig. 2.
DKFZ-EP1NS cells give rise to tumors in serial transplantation experiments and recapitulate the original tumor in vivo
a Schematic representation of serial transplantation experiments and table indicating engraftment rates and time to tumor detection. b Axial section of an MRI of a representative mouse with an orthotopic tumor (white arrow); and HE-stain of a transverse section of whole brain, showing a well-circumscribed tumor in the left hemisphere. c HE and immunohistochemical stainings of histological sections for markers in the primary tumor of the patient, 1° orthotopic in vivo tumor (mouse 1°) and 2° orthotopic in vivo tumor (mouse 2°). HE: mouse 1° and 2° tumors display typical features of ependymoma such as perivascular pseudorosettes (insets). Ki67: mouse 1° and 2° tumors show proliferation activity similar to the primary tumor in the patient, as detected by staining for Ki67. GFAP: tumor cells of both 1° and 2° tumors are virtually negative for GFAP, as was the primary tumor of the patient. Nestin: both 1° and 2° mouse tumors stained strongly for Nestin, similarly to the patients tumor. SD: standard deviation; HE : hematoxylin and eosin stain ; Tu: tumor; NB: normal brain.
When injected subcutaneously in matrigel, DKFZ-EP1NS cells equally gave rise to tumors. The subcutaneous tumors differed from the orthotopic tumors by displaying a clear cell phenotype, thus resembling the subcutaneous metastasis seen in the patient (Suppl. Fig. 1a). DKFZ-EP1NS cells in matrigel injected intraperitoneally revealed another distinct phenotype with compact small round cells, with no morphological correlate in the patient (Suppl. Fig. 1b).
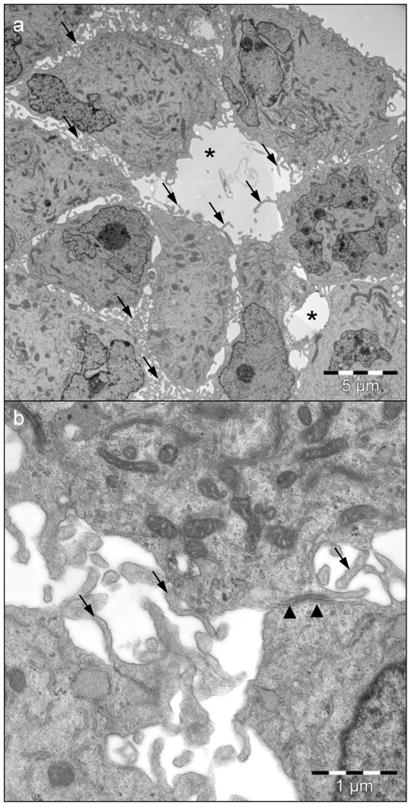
Staining for other markers used in the diagnosis of ependymoma, such as epithelial membrane antigen (EMA), smooth muscle antigen (SMA), vimentin, CD99, cytokeratin, S100 and synaptophysin, revealed strong staining for EMA, SMA and vimentin (Suppl. Fig. 2), and negativity for CD99, cytokeratin, S100 and synaptophysin (Suppl. Fig. 3), a staining pattern that can be found in anaplastic ependymoma. Furthermore, all markers stained similarly in the patient’s primary tumor, first and second recurrence, subcutaneous metastasis, 1° and 2° intracranial xenograft and subcutaneous xenograft (Suppl. Fig. 2), except for SMA, where an increase from medium positivity in the primary tumor to higher positivity in the recurrences was seen (Suppl. Fig. 2). Electron microscopy of DKFZ-EP1NS cells revealed intercellular lumina, junctions and microvilli, features typically found in ependymoma, but no ciliae (Fig. 3). In conclusion, extended IHC and ultrastructural examination confirms the diagnosis of anaplastic ependymoma WHO III.
Fig. 3.
Electron microscopy of DKFZ-EP1NS neurospheres reveals features typical of anaplastic ependymoma. a Overview showing intercellular lumina (*) and microvilli (arrows). b Detail depicting a junction (arrowheads) and microvilli (arrows).
Analysis of genomic aberrations by FISH showed stable genomic aberrations as described in the primary tumor [37], with loss of chromosome 1p36, monosomy of chromosome 9 including homozygous deletion of CDKN2A, in cultured DKFZ-EP1NS cells (late passage, P30, Suppl. Fig. 4a), primary and secondary mouse tumor (not shown). Hence, the cells and the arising tumors are classified into the high-risk cytogenetic group 3 according to Korshunov et al. [29]. Quantitative gene expression measurement of genes identifying subgroups A-D (encompassing supratentorial ependymomas) revealed that the ependymoma model described here consistently overexpressed genes classifying the DKFZ-EP1NS cells into subgroup C according to Johnson et al. [24] (Suppl. Fig. 4b).
In summary, the ependymoma model DKFZ-EP1NS expresses markers associated with normal adult and cancer stem cells, is tumorigenic in vivo, serially transplantable, retains the original genomic aberrations, and recapitulates the original tumor in a niche-dependent manner. Since these are characteristics of CSC [6], we conclude that DKFZ-EP1NS cells are CSC. Finally, genetic aberrations classify the cells into cytogenetic group 3, and the expression profile classifies them into molecular subgroup C. Hence, the tumor model represents a distinct molecular high risk supratentorial ependymoma with CSC properties.
Transcriptome analysis of DKFZ-EP1NS cells reveals model stability, a neural stem cell-like profile and differentiation-plasticity in a niche-dependent manner
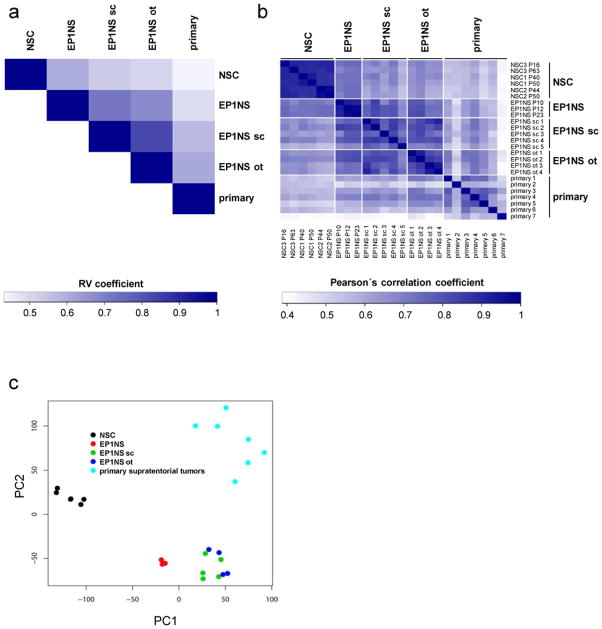
We next determined the transcriptional profile of the DKFZ-EP1NS model in relation to primary tumors and neural stem cells by performing microarray-based transcriptome analysis. In a first step, we compared the similarity of the mRNA expression profiles between different models (i.e. neural stem cells: NSC; in vitro cultured cells: EP1NS; subcutaneous tumors: EP1NS sc; orthotopic tumors: EP1NS ot; human primary supratentorial ependymomas WHO III° with recurrent clinical course: primary) rather than individual samples by computing the multivariate (RV) correlation coefficient. The RV coefficient allows for assessment of association of two sets of samples (models) [49], and here showed i) a higher correlation of NSC with EP1NS cells (RV coefficient: 0.59) than with all other models, and ii) the highest correlation of the orthotopic model with primary tumors (RV coefficient: 0.60) (Fig. 4a). Calculation of Pearson’s correlation coefficient showed i) a high degree of homogeneity in the NSC and the in vitro EP1NS models including cell culture passages 10, 12, 23 (median correlation coefficient/mcc: 0.90 and 0.92, respectively) and ii) a higher degree of heterogeneity in the in vivo models and the human primary tumors (orthotopic - mcc: 0.85; subcutaneous - mcc: 0.79; primary - mcc: 0.61) (Fig. 4b). Principal component analysis (PCA) revealed that transcriptomes of DKFZ-EP1NS cells located between NSC, subcutaneous and orthotopic tumors (Fig. 4c). Finally, “Correspondence-at-the-top” (CAT) plots were generated, showing a high degree of common proportion of up- and downregulated clones when comparing EP1NS in vitro, subcutaneous, orthotopic or primary tumors with NSC (Suppl. Fig. 5a) or EP1NS in vitro, subcutaneous, orthotopic or primary tumors with each other (Suppl. Fig. 5b).
Fig. 4.
Transcriptome analysis of DKFZ-EP1NS cells
a RV coefficient plot showing a higher correlation of neural stem cells (NSC) with in vitro DKFZ-EP1NS cells (EP1NS) than with all other models, and the highest correlation of the orthotopic xenograft model (EP1NS ot) with primary ependymoma tumors (primary). Shades of blue indicate different RV coefficients: the darker the shade of blue, the higher the RV coefficient, the higher the correlation between two models. b Plot of pairwise sample correlations shows a higher degree of homogeneity in the NSC and EP1NS, compared to the subcutaneous xenograft model (EP1NS sc), EP1NS ot, or primary. Shades of blue indicate different Pearson’s correlation coefficients: the darker the shade of blue, the higher the Pearson’s correlation coefficient, the higher the correlation between two individual samples. c Principal component analysis (PCA) of transcriptomes positions EP1NS between NSC and the derived in vivo models EP1NS sc and EP1NS ot.
In conclusion, these data show that i) the EP1NS in vitro model is stable over many passages, ii) the gene expression patterns of EP1NS cells show similarities to NSC and iii) compared to primary ependymoma, the orthotopic xenograft model most closely resembles primary ependymoma indicating that the transcriptional program of EP1NS cells shifts from a neural stem cell like program towards an ependymoma tumor program in a niche-dependent manner.
DKFZ-EP1NS cells show intrinsic resistance to chemotherapeutic agents but are susceptible to treatment with HDACi
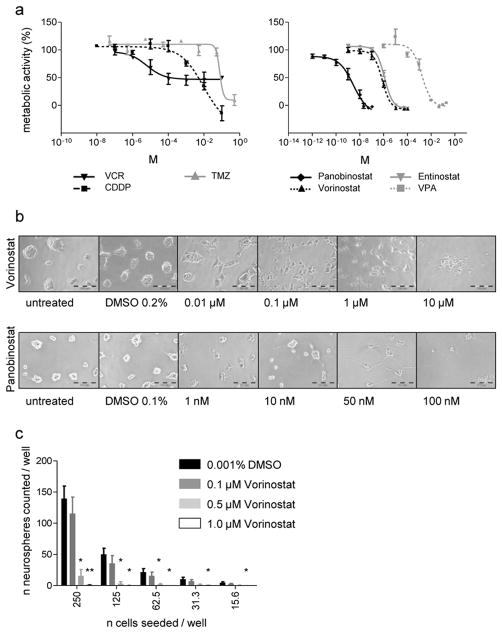
Because CSC have been hypothesized to be intrinsically resistant to chemotherapy treatment and may thus be responsible for treatment failure, we investigated the sensitivity of DKFZ-EP1NS cells to chemotherapeutic agents commonly applied in brain tumors including ependymoma. Exposure to vincristine (VCR), cisplatin (CDDP), and temozolomide (TMZ) indeed showed that these agents were not able to significantly reduce cell viability in therapeutically achievable concentrations (Fig. 5a, left panel). Calculation of half maximal effective concentration (EC50) values and comparison to published maximum peak plasma concentrations (max PPC) in patients [7, 10, 31, 41, 45, 58, 60] showed that the EC50/max PPC ratios ranged from 118-fold to 1064-fold (Suppl. Table 4).
Fig. 5.
DKFZ-EP1NS cells are relatively resistant to chemotherapeutic agents, but susceptible to treatment with HDAC inhibitors (HDACis)
a Measurement of metabolic activity by WST-1 assay after treatment with chemotherapeutics vincristine (VCR), cisplatin (CDDP) and temozolomide (TMZ) (left panel), and after treatment with HDACi Panobinostat, Vorinostat (syn.: SAHA), Entinostat (syn.: MS275) and valproic acid (VPA) (right panel). Note the relative resistance to VCR, CDDP and TMZ (find EC50 and peak plasma concentrations, PPC, in Suppl. Table 4). b Brightfield images of DKFZ-EP1NS treated with Vorinostat or Panobinostat at indicated concentrations: note the reduction in neurosphere-formation and induction of a differentiated phenotype at concentrations from 0.1–1 μM Vorinostat or 10–100 nM Panobinostat, and loss of morphology possibly due to cell death above 1 μM Vorinostat or 50 nM Panobinostat. c Neurosphere initiation capacity (NSIC) assay: NSIC is strongly reduced after 10 days of treatment with Vorinostat at indicated concentrations or DMSO control. Bars represent means of three independent measurements, error bars represent SEM; * = p-value < 0.05; ** = p-value < 0.01 (treated vs. control, two-sided t-test).
Because susceptibility of the ependymoma cell line EPN-2 to treatment with the HDACi TSA has been shown [43], and our own previous work demonstrated that HDACs play an important role in the biology of pediatric neural cancers and that HDACi are effective compounds in the treatment of tumors of neural origin [9, 36, 40], we investigated if DKFZ-EP1NS cells are susceptible to several, structurally divergent HDACi compounds. Indeed, DKFZ-EP1NS cells display a significant decrease in metabolic activity upon treatment with HDACi (Fig. 5a, right panel), and the EC50/max PPC ratios calculated ranged from 0.07 to 4.92 (Suppl. Table 4). The pan-HDAC inhibitors Vorinostat and Panobinostat (both hydroxamic acids) displayed EC50/max PPC ratios well below clinically achievable concentrations (0.17 and 0.07 respectively), while the benzamide Entinostat and the short chain fatty acid VPA, both preferentially inhibiting class I HDACs, showed a EC50/max PPC ratios of 3.08 and 4.92, respectively (Suppl. Table 4). We conclude that DKFZ-EP1NS cells show resistance to conventional chemotherapeutic drugs, but are sensitive to HDACi treatment at clinically achievable concentrations.
Treatment with the HDACi Vorinostat reduces neurosphere initiating capacity (NSIC) and induces neuronal differentiation
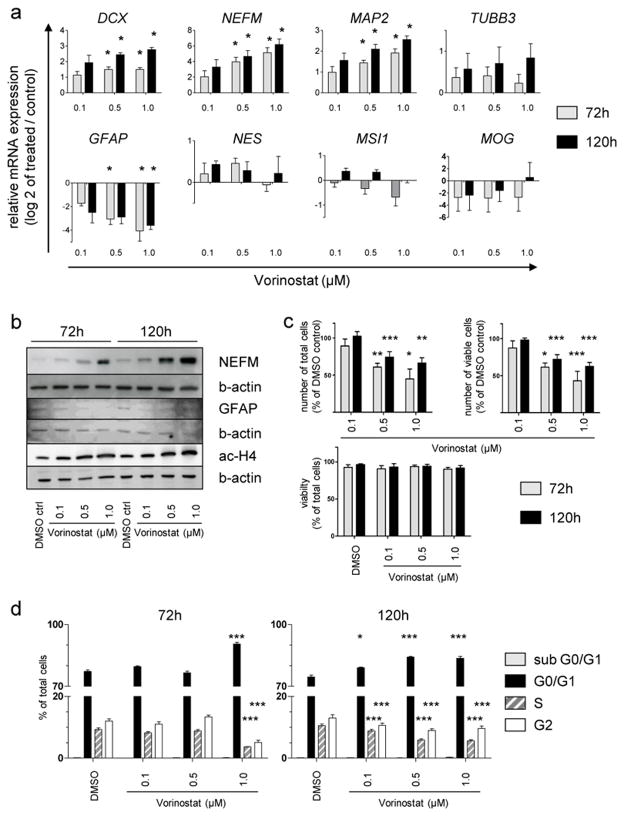
Because we observed morphological changes reminiscent of differentiation following treatment of DKFZ-EP1NS with HDACis in a concentration dependent manner (Fig. 5b), we assessed the potential of an HDACi to affect tumor stem cell properties, such as reduction of NSIC and induction of differentiation. We performed the NSIC assay as an in vitro surrogate assay for CSC tumorigenicity [11, 34]. Because Vorinostat is the HDACi that is most advanced in clinical trials, we chose this HDACi over Panobinostat. After 10 days of treatment with Vorinostat a marked reduction in NSIC in a dose dependent manner was demonstrated (Fig. 5c). Of note, NSIC was almost completely abolished at 0.5 μM Vorinostat, which is below the EC50 calculated in the metabolic activity assay (0.78 μM; Suppl. Table 4) and within a clinically achievable concentration range. We then assessed the expression of markers known to be associated with neuronal, glial, and oligodendroglial differentiation, respectively. As a control for effective targeting of HDACs by the compounds, we showed increase of acetylated histone H4 in a time- and dose-dependent manner (Fig. 6b). In accordance with the observed morphological change, we determined a significant induction of several markers of neuronal differentiation such as doublecortin (DCX), neurofilament M (NEFM), and microtubule-associated protein (MAP2) in a concentration- and time-dependent manner (Fig. 6a). Beta-3-tubulin (TUBB3) also tended to increase in a concentration- and time-dependent manner, however not significantly. In contrast, glial fibrillary acidic protein (GFAP), a marker associated with mature astrocytes but also with neural progenitors and specifically radial glia cells [5, 30], the proposed cells of origin of ependymoma, was strongly downregulated (Fig. 6a). Other markers investigated including stem cell markers Nestin (NES) and Musashi1 (MSI1) did not show significant alterations in expression. Myelin oligodendrocyte glycoprotein (MOG) showed no consistent and significant regulation, with very low to undetectable expression levels, indicating that no induction of oligodendrocyte lineage took place (Fig. 6a). Western blot confirmed the increase of NEFM and the decrease of GFAP on the protein level (Fig. 6b).
Fig. 6.
Treatment of DKFZ-EP1NS cells with Vorinostat induces a neuronal phenotype, and reduces cell numbers through G0/G1-arrest without increase in cell death
a Assessment of change in marker expression by quantitative real-time RT-PCR after treatment of DKFZ-EP1NS cells with Vorinostat. Note the upregulation of neuronal markers DCX, NEFM and MAP2, and downregulation of neural stem cell/astrocytic marker GFAP. Stem cell markers NES and MSI1 or oligodendrocytic marker MOG show no consistent up- or downregulation. Bars are calculated as relative expression of treated sample/DMSO-treated control (set at log2=0), and represent means of three independent measurements, error bars represent SEM; * = p-value < 0.05 (t-test, FDR-adjusted p-value). b Analysis of markers most strongly regulated on mRNA level and of histone4 acetylation by western blot confirms the upregulation of NEFM protein, the downregulation of GFAP protein and an increase in histone4 acetylation upon treatment with Vorinostat. c Analysis of cell counts shows a significant decrease of total as well as viable cell counts at 72 and 120h at 0.5 and 1 μM Vorinostat, while there is no decrease in cell viability at both timepoints or any of the concentrations tested. Bars represent means of four independent measurements, error bars represent SEM; * = p-value < 0.05, ** = p-value < 0.01, *** = p-value < 0.001 (t-test). d Cell cycle analysis by the Nicoletti method shows a G0/G1 cell cycle arrest at 72h for the highest concentration of Vorinostat, and at 120h for all concentration of Vorinostat tested. Bars represent means of four independent measurements, error bars represent SEM; * = p-value < 0.05, ** = p-value < 0.01, *** = p-value < 0.001 (t-test). ctrl.: control, b-actin: beta-actin, ac-H4: acetylated histone4.
Since differentiation is typically accompanied by a stop in proliferation and cell cycle arrest in G0/G1, we performed analyses of total as well as viable cell numbers, cell viability and cell cycle upon treatment with clinically achievable concentrations of Vorinostat (Fig. 6c and d). We found a significant decrease in cell number associated (Fig. 6c) with a significant cell cycle arrest in G0/G1 (Fig. 6d). No signs of cell death such as a decrease in viability as measured by trypan blue exclusion staining (Fig. 6c) or an increase of the subG1 fraction (Fig. 6d) were observed.
In conclusion, treatment of DKFZ-EP1NS cells with HDACis such as Vorinostat at clinically achievable concentrations reduces neurosphere initiation capacity and induces neuronal differentiation accompanied by cell cycle arrest in G0/G1 without induction of cell death.
Discussion
Progress in the treatment of ependymoma has been considerably hindered by the lack of preclinical models. The model reported here shows expression of stem cell markers, self-renewal, neurosphere-initiation capacity, in vivo tumorigenicity, serial transplantability, and similarity to gene expression profiles of neural stem cells. Furthermore, genetic alterations typical for high-risk ependymoma are found, classifying our model in the cytogenetic group 3 [29], making it a suitable preclinical model for high-risk ependymoma. Analysis of gene expression classifies our model in molecular subgroup C as described by Johnson et al. [24]. The cells are positive for many of the stem cell markers tested, but are negative for CD133, ALDH and Hoechst side population. Although CD133 has been reported to be expressed by candidate stem cells of ependymoma [55] as well as ependymoma neurospheres [22], there has been much controversy about the ability of CD133 to mark CSC populations [4, 52]. Considering especially the serial transplantability of our model, demonstrating the key feature of stem cell properties [6], we conclude that neither CD133, nor ALDH or the presence of a Hoechst side population is important for the model described here.
Although the tumor material from which the DKFZ-EP1NS cells were derived from is an extraneural metastasis, an uncommon but not undocumented event [48, 59], thorough immunohistochemical and ultrastructural evaluation of the cultured cells and xenografts reveals typical features of anaplastic ependymoma. In summary, the immunohistochemical and ultrastructural evaluation in combination with the genetic data leads us to the conclusion that the DKFZ-EP1NS cells are indeed a model for anaplastic ependymoma WHO III.
Because the DKFZ-EP1NS model can be used in cell culture, the model is amenable to standard cell culture assays facilitating preclinical drug testing. Resistance to chemotherapeutic agents was expected from the clinical course and the treatment of the patient the cells were derived from, and was indeed recapitulated in the cell culture model. Hence, our model mirrors challenges commonly encountered in ependymoma treatment. As chemotherapy resistance is a hallmark of CSC [8], our data points towards a role of ependymoma stem cells in therapeutic failure, as has been suggested previously by studies showing increased resistance to etoposide of ependymoma neurospheres [22]. The susceptibility of DKFZ-EP1NS cells to HDACi treatment at clinically achievable concentrations, and the effect on stem cell specific properties such as the neurosphere initiating capacity (NSIC) and the differentiation status, suggests that inhibition of HDACs may be a promising novel therapeutic strategy in the treatment of ependymoma. It is important to note in this context, that NSIC is indeed correlated with survival in pediatric brain tumors [56], and supratentorial ependymomas with a neuronal differentiation phenotype showed a favorable prognosis [3]. Therefore, it will be important to determine, if a pharmacological induction of decrease in NSIC and neuronal differentiation correlates with an improved survival. Furthermore, the varying EC50/PPC max ratios for the different HDACi, indicate that the pan-HDACi Vorinostat or Panobinostat may be more effective in a clinical setting than other HDACis. Whether the effects of HDACi treatment observed in our model are mediated epigenetically and can be recapitulated in vivo remains to be determined.
It has been reported that exposure of the ependymoma cell line nEPN2 to the HDACi TSA leads to increased apoptosis [43]. In contrast, we could not detect an increase of cell death upon treatment of DKFZ-EP1NS with Vorinostat. This may be due to differing concentration ranges used in the experiments, since we used low Vorinostat concentrations (0.1–1 μM, corresponding to 0.12–1.29-fold of the EC50). As the brightfield images (Fig. 5b) suggest, higher Vorinostat concentrations could possibly lead to increased cell death. However since the published peak plasma concentrations in patients are 4.5 μM max. [10] (see also Suppl. Table 4), we believe it is of high clinical relevance to study the effects of lower Vorinostat concentrations. It remains to be elucidated if other ependymoma models with stem cell-like properties will show similar effects upon HDACi treatment.
To date, only five other xenograft models (1425EPN, nEPN1, nEPN2, BT44, and BT57) with corresponding cell lines for human ependymoma have been described [18, 22, 61]. In contrast to the published models, our model recapitulates the original tumor in a niche dependent manner and is the first human model with a homozygous deletion for CDKN2A, typical for supratentorial ependymomas and cytogenetic group 3. It is furthermore the first ependymoma stem cell model that is directly derived from primary patient material and can be continuously passaged in neurosphere cultures in vitro.
In summary, we have established a novel ependymoma cell line with stem cell-like properties that can be propagated in vitro and is tumorigenic in vivo. The model recapitulates high risk ependymoma at the molecular, cellular and therapeutic level. Treatment with HDACis shows the applicability of this cell line in preclinical testing, as well as the differentiating potential of HDACi in ependymoma treatment.
Supplementary Material
Acknowledgments
The authors wish to thank the family of the patient for their strong support of this study. We thank Sandra Riedinger, Carina Konrad, Mathias Koch, Andreas Lacher, Diana Jäger, Sylvia Kaden, Tina Wiesner and Andrea Wittmann for excellent technical assistance. TM is supported by a grant from the B.Braun Foundation; TM, IO and SP by a grant from the Wilhelm Sander Foundation; HD and OW through the NGFNplus program by a grant of the Bundesministerium für Bildung und Forschung (BMBF), Germany; HD by the University of Heidelberg through both the FRONTIER and the OLYMPIA MORATA programs; OB by the EU (FP7-HEALTH-F5-2010-266753-SCR&Tox), BMBF grants 01GNO813 and 0315799 (BIODISC), BIO.NRW (project StemCellFactory) and the Hertie Foundation.
References
- 1.R: A language and environment for statistical computing. R: A language and environment for statistical computing. 2 2009. [Google Scholar]
- 2.Agaoglu FY, Ayan I, Dizdar Y, Kebudi R, Gorgun O, Darendeliler E. Ependymal tumors in childhood. Pediatr Blood Cancer. 2005;45:298–303. doi: 10.1002/pbc.20212. [DOI] [PubMed] [Google Scholar]
- 3.Andreiuolo F, Puget S, Peyre M, et al. Neuronal differentiation distinguishes supratentorial and infratentorial childhood ependymomas. Neuro Oncol. 2010;12:1126–34. doi: 10.1093/neuonc/noq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 5.Casper KB, McCarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cell Neurosci. 2006;31:676–84. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 7.Corona G, Casetta B, Sandron S, Vaccher E, Toffoli G. Rapid and sensitive analysis of vincristine in human plasma using on-line extraction combined with liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:519–25. doi: 10.1002/rcm.3390. [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 9.Deubzer HE, Ehemann V, Westermann F, et al. Histone deacetylase inhibitor Helminthosporium carbonum (HC)-toxin suppresses the malignant phenotype of neuroblastoma cells. Int J Cancer. 2008;122:1891–900. doi: 10.1002/ijc.23295. [DOI] [PubMed] [Google Scholar]
- 10.Fakih MG, Fetterly G, Egorin MJ, et al. A phase I, pharmacokinetic, and pharmacodynamic study of two schedules of vorinostat in combination with 5-fluorouracil and leucovorin in patients with refractory solid tumors. Clin Cancer Res. 2010;16:3786–94. doi: 10.1158/1078-0432.CCR-10-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X, Salford LG, Widegren B. Glioma stem cells: evidence and limitation. Semin Cancer Biol. 2007;17:214–8. doi: 10.1016/j.semcancer.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Feuerborn A, Srivastava PK, Kuffer S, et al. The Forkhead factor FoxQ1 influences epithelial differentiation. J Cell Physiol. 226:710–9. doi: 10.1002/jcp.22385. [DOI] [PubMed] [Google Scholar]
- 13.Figarella-Branger D, Civatte M, Bouvier-Labit C, et al. Prognostic factors in intracranial ependymomas in children. J Neurosurg. 2000;93:605–13. doi: 10.3171/jns.2000.93.4.0605. [DOI] [PubMed] [Google Scholar]
- 14.Gaspar N, Grill J, Geoerger B, Lellouch-Tubiana A, Michalowski MB, Vassal G. p53 Pathway dysfunction in primary childhood ependymomas. Pediatr Blood Cancer. 2006;46:604–13. doi: 10.1002/pbc.20532. [DOI] [PubMed] [Google Scholar]
- 15.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grill J, Avet-Loiseau H, Lellouch-Tubiana A, et al. Comparative genomic hybridization detects specific cytogenetic abnormalities in pediatric ependymomas and choroid plexus papillomas. Cancer Genet Cytogenet. 2002;136:121–5. doi: 10.1016/s0165-4608(02)00516-2. [DOI] [PubMed] [Google Scholar]
- 18.Guan S, Shen R, Lafortune T, et al. Establishment and characterization of clinically relevant models of ependymoma: a true challenge for targeted therapy. Neuro Oncol. 2011;13:748–58. doi: 10.1093/neuonc/nor037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirose Y, Aldape K, Bollen A, et al. Chromosomal abnormalities subdivide ependymal tumors into clinically relevant groups. Am J Pathol. 2001;158:1137–43. doi: 10.1016/S0002-9440(10)64061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houghton PJ, Morton CL, Tucker C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer. 2007;49:928–40. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 21.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 22.Hussein D, Punjaruk W, Storer LC, et al. Pediatric brain tumor cancer stem cells: cell cycle dynamics, DNA repair, and etoposide extrusion. Neuro Oncol. 2011;13:70–83. doi: 10.1093/neuonc/noq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irizarry RA, Warren D, Spencer F, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–50. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–6. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaatsch P, Spix C. German Childhood Cancer Registry - annual Report 2006/2007 (1980–2006) 2008. [Google Scholar]
- 26.Kleber S, Sancho-Martinez I, Wiestler B, et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell. 2008;13:235–48. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106:3225–30. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korshunov A, Golanov A, Sycheva R, Timirgaz V. The histologic grade is a main prognostic factor for patients with intracranial ependymomas treated in the microneurosurgical era: an analysis of 258 patients. Cancer. 2004;100:1230–7. doi: 10.1002/cncr.20075. [DOI] [PubMed] [Google Scholar]
- 29.Korshunov A, Witt H, Hielscher T, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28:3182–90. doi: 10.1200/JCO.2009.27.3359. [DOI] [PubMed] [Google Scholar]
- 30.Kriegstein AR, Gotz M. Radial glia diversity: a matter of cell fate. Glia. 2003;43:37–43. doi: 10.1002/glia.10250. [DOI] [PubMed] [Google Scholar]
- 31.Kummar S, Gutierrez M, Gardner ER, et al. Phase I trial of MS-275, a histone deacetylase inhibitor, administered weekly in refractory solid tumors and lymphoid malignancies. Clin Cancer Res. 2007;13:5411–7. doi: 10.1158/1078-0432.CCR-07-0791. [DOI] [PubMed] [Google Scholar]
- 32.Ladewig J, Koch P, Endl E, et al. Lineage selection of functional and cryopreservable human embryonic stem cell-derived neurons. Stem Cells. 2008;26:1705–12. doi: 10.1634/stemcells.2008-0007. [DOI] [PubMed] [Google Scholar]
- 33.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahller YY, Williams JP, Baird WH, et al. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One. 2009;4:e4235. doi: 10.1371/journal.pone.0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–9. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 36.Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–52. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 37.Milde T, Pfister S, Korshunov A, et al. Stepwise accumulation of distinct genomic aberrations in a patient with progressively metastasizing ependymoma. Genes Chromosomes Cancer. 2009;48:229–38. doi: 10.1002/gcc.20635. [DOI] [PubMed] [Google Scholar]
- 38.Modena P, Lualdi E, Facchinetti F, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–33. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 39.Oehme I, Bosser S, Zornig M. Agonists of an ecdysone-inducible mammalian expression system inhibit Fas Ligand- and TRAIL-induced apoptosis in the human colon carcinoma cell line RKO. Cell Death Differ. 2006;13:189–201. doi: 10.1038/sj.cdd.4401730. [DOI] [PubMed] [Google Scholar]
- 40.Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–9. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 41.Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10:3728–36. doi: 10.1158/1078-0432.CCR-03-0807. [DOI] [PubMed] [Google Scholar]
- 42.Puget S, Grill J, Valent A, et al. Candidate genes on chromosome 9q33–34 involved in the progression of childhood ependymomas. J Clin Oncol. 2009;27:1884–92. doi: 10.1200/JCO.2007.15.4195. [DOI] [PubMed] [Google Scholar]
- 43.Rahman R, Osteso-Ibanez T, Hirst RA, et al. Histone deacetylase inhibition attenuates cell growth with associated telomerase inhibition in high-grade childhood brain tumor cells. Mol Cancer Ther. 2010;9:2568–81. doi: 10.1158/1535-7163.MCT-10-0272. [DOI] [PubMed] [Google Scholar]
- 44.Ramachandran C, Khatib Z, Petkarou A, et al. Tamoxifen modulation of etoposide cytotoxicity involves inhibition of protein kinase C activity and insulin-like growth factor II expression in brain tumor cells. J Neurooncol. 2004;67:19–28. doi: 10.1023/b:neon.0000021738.77612.1b. [DOI] [PubMed] [Google Scholar]
- 45.Rathkopf D, Wong BY, Ross RW, et al. A phase I study of oral panobinostat alone and in combination with docetaxel in patients with castration-resistant prostate cancer. Cancer Chemother Pharmacol. 66:181–9. doi: 10.1007/s00280-010-1289-x. [DOI] [PubMed] [Google Scholar]
- 46.Rezai AR, Woo HH, Lee M, Cohen H, Zagzag D, Epstein FJ. Disseminated ependymomas of the central nervous system. J Neurosurg. 1996;85:618–24. doi: 10.3171/jns.1996.85.4.0618. [DOI] [PubMed] [Google Scholar]
- 47.Rich JN, Sathornsumetee S, Keir ST, et al. ZD6474, a novel tyrosine kinase inhibitor of vascular endothelial growth factor receptor and epidermal growth factor receptor, inhibits tumor growth of multiple nervous system tumors. Clin Cancer Res. 2005;11:8145–57. doi: 10.1158/1078-0432.CCR-05-0319. [DOI] [PubMed] [Google Scholar]
- 48.Rickert CH. Extraneural metastases of paediatric brain tumours. Acta Neuropathol. 2003;105:309–27. doi: 10.1007/s00401-002-0666-x. [DOI] [PubMed] [Google Scholar]
- 49.Robert P, Escouffier Y. A Unifying tool for linear multivariate statistical methods: the RV-coefficient. Applied Statistics. 1976;25:257–265. [Google Scholar]
- 50.Rousseau P, Habrand JL, Sarrazin D, et al. Treatment of intracranial ependymomas of children: review of a 15-year experience. Int J Radiat Oncol Biol Phys. 1994;28:381–6. doi: 10.1016/0360-3016(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt M, Pawlita M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009;37:e119. doi: 10.1093/nar/gkp581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu HK, Sall WF, Maity A, et al. Childhood intracranial ependymoma: twenty-year experience from a single institution. Cancer. 2007;110:432–41. doi: 10.1002/cncr.22782. [DOI] [PubMed] [Google Scholar]
- 54.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 55.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–35. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Thirant C, Bessette B, Varlet P, et al. Clinical relevance of tumor cells with stem-like properties in pediatric brain tumors. PLoS One. 2011;6:e16375. doi: 10.1371/journal.pone.0016375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmermann B, Kortmann RD, Kuhl J, et al. Combined postoperative irradiation and chemotherapy for anaplastic ependymomas in childhood: results of the German prospective trials HIT 88/89 and HIT 91. Int J Radiat Oncol Biol Phys. 2000;46:287–95. doi: 10.1016/s0360-3016(99)00414-9. [DOI] [PubMed] [Google Scholar]
- 58.van Hennik MB, van der Vijgh WJ, Klein I, et al. Comparative pharmacokinetics of cisplatin and three analogues in mice and humans. Cancer Res. 1987;47:6297–301. [PubMed] [Google Scholar]
- 59.Varan A, Sari N, Akalan N, et al. Extraneural metastasis in intracranial tumors in children: the experience of a single center. J Neurooncol. 2006;79:187–90. doi: 10.1007/s11060-006-9123-3. [DOI] [PubMed] [Google Scholar]
- 60.Voso MT, Santini V, Finelli C, et al. Valproic acid at therapeutic plasma levels may increase 5-azacytidine efficacy in higher risk myelodysplastic syndromes. Clin Cancer Res. 2009;15:5002–7. doi: 10.1158/1078-0432.CCR-09-0494. [DOI] [PubMed] [Google Scholar]
- 61.Yu L, Baxter PA, Voicu H, et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 2010;12:580–94. doi: 10.1093/neuonc/nop056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.