Abstract
Hepatitis C virus (HCV) reinfects liver allografts in transplant recipients by replicating immediately after transplantation, causing a rapid increase in blood serum HCV RNA levels. We evaluated dynamic changes in the viral genetic complexity after HCV reinfection of the graft liver; we also identified the characteristics of replicating HCV clones using a massively parallel ultradeep sequencing technique to determine the full-genome HCV sequences in the liver and serum specimens of five transplant recipients with genotype 1b HCV infection before and after liver transplantation. The recipients showed extremely high genetic heterogeneity before transplantation, and the HCV population makeup was not significantly different between the liver and blood serum specimens of the individuals. Viral quasispecies complexity in serum was significantly lower after liver transplantation than before it, suggesting that certain HCV clones selectively proliferated after transplantation. Defective HCV clones lacking the structural region of the HCV genome did not increase in number, and full-genome HCV clones selectively increased in number immediately after liver transplantation. A re-increase in the same defective clone existing before transplantation was detected 22 months after transplantation in one patient. Ultradeep sequencing technology revealed that the genetic heterogeneity of HCV was reduced after liver transplantation. Dynamic changes in defective HCV clones after liver transplantation indicate that these clones have important roles in the HCV life cycle.
INTRODUCTION
The hepatitis C virus (HCV) has an approximately 9.6-kb plus-strand RNA genome that encodes the viral core, envelope glycoprotein 1 (E1), E2, and p7 structural proteins and the NS2, NS3, NS4A, NS4B, NS5A, and NS5B nonstructural proteins (1). A characteristic of HCV infection is its remarkable genetic diversity with a high degree of genetic heterogeneity in each patient, which is referred to as a quasispecies. In heterogeneous HCV clones, a dominant viral population might evolve as a result of its viral replicative fitness and concurrent immune selection pressures that drive clonal selection.
In HCV-positive liver transplant recipients, HCV reinfection of the liver allograft occurs at the time of transplantation, and replication of HCV begins immediately after transplantation. Blood serum HCV RNA levels then rapidly increase to levels that are 10- to 20-fold higher than pretransplant levels. It is thus hypothesized that specific HCV clones that have growth advantages increase after liver transplantation. Although several studies have attempted to clarify the change in genetic heterogeneity following liver transplantation, the abundant diversity and complexity of HCV have been obstacles to a detailed evaluation of viral genetic heterogeneity. The recent introduction of ultradeep sequencing technology, which is capable of producing millions of DNA sequence reads in a single run, however, is rapidly changing the landscape of genome research (2, 3).
In this study, we performed ultradeep sequencing analyses to unveil the levels of viral quasispecies of genotype 1b HCV in the liver and the serum specimens from 5 patients who underwent living donor liver transplantation (LDLT) and clarified the changes in viral genetic complexity after reinfection of HCV in the graft liver. In the analyses, we found that the population of defective HCV clones that lack structural regions of the HCV genome changed after liver transplantation. We then clarified the dynamics and characteristics of the defective HCV clones.
MATERIALS AND METHODS
Patients.
The participants comprised 5 Japanese adult patients with end-stage liver disease with genotype 1b HCV infection who underwent LDLT at Kyoto University Hospital between May 2006 and September 2008. Serum samples were obtained before and 1 month after liver transplantation. In addition, a blood serum sample from a patient in the chronic hepatitis phase 22 months after liver transplantation was obtained and analyzed. Liver tissue samples were obtained from 4 patients (patients 1–4) at the time of transplantation, frozen immediately, and stored at −80°C until use.
Tacrolimus with a steroid or mycophenolate mofetil was administered to induce immunosuppression in the patients. A patient who received an ABO blood type-incompatible transplant was treated with rituximab, plasma exchange, and hepatic artery or portal vein infusion with prostaglandin E1 and methylprednisolone (4).
The ethics committee at Kyoto University approved the studies (protocol no. E1211), and written informed consent for participation in this study was obtained from all patients.
Virologic assays.
The HCV genotype was determined using a PCR-based genotyping system developed by Ohno et al. (5) to amplify the core region using genotype-specific PCR primers for the determination of the HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. The blood serum HCV RNA load was evaluated before LDLT 1 month post-LDLT and then every 3 months after LDLT using PCR and an Amplicor HCV assay (Cobas Amplicor HCV monitor; Roche Molecular Systems, Pleasanton, CA) until April 2008 or a real-time PCR-based quantitation method for HCV (Cobas AmpliPrep/Cobas TaqMan HCV test; Roche Molecular Systems) starting May 2008.
Direct population Sanger sequencing.
To define the representative reference sequences of full-length HCV in each clinical specimen, serum samples collected before liver transplantation were first subjected to direct population Sanger sequencing using the Applied Biosystems 3500 genetic analyzer (Applied Biosystems, Foster City, CA) (6). Total RNA was extracted from 140 μl of serum using a QIAamp viral RNA minikit (Qiagen, Valencia, CA) and reverse transcribed in a volume of 20 μl with the OneStep RNA PCR kit AMV (TaKaRa Bio, Ohtsu, Japan). The HCV genomes were amplified using Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland). Oligonucleotide primers were designed to amplify the first half (∼5,000 bp) and latter half (∼4,500 bp) of the genotype 1b HCV genome sequence. PCR products purified by the QIAquick gel extraction kit (Qiagen) were assayed for direct sequencing. The nucleotide sequences of the PCR products were determined using an ABI Prism BigDye Terminator ready reaction kit (Applied Biosystems). A blood serum sample from a healthy volunteer was used as a negative control.
Massively parallel ultradeep sequencing.
Paired-end sequencing with multiplexed tags was carried out using the Illumina Genome Analyzer II. End repair of DNA fragments, the addition of adenine to the 3′-ends of the DNA fragments, adaptor ligation, and PCR amplification by Illumina-paired end PCR primers were performed as described previously (6, 7). Briefly, the viral genome sequences were amplified with high-fidelity PCR and sheared by nebulization using 32 lb/in2 N2 for 8 min, and the sheared fragments were purified and concentrated using a QIAquick PCR purification kit (Qiagen). The overhangs resulting from the fragmentation were then converted into blunt ends using T4 DNA polymerase and Klenow enzymes, followed by the addition of terminal 3′-adenine residues. One of the adaptors containing six unique base pair (bp) tags, such as ATCACG and CGATGT (multiplexing sample preparation oligonucleotide kit; Illumina), was then ligated to each fragment using DNA ligase. Adaptor-ligated DNAs in the range of 200 to 350 bp were then size selected by agarose gel electrophoresis. These libraries were amplified independently using a minimal PCR amplification step of 18 cycles with Phusion high-fidelity DNA polymerase and then purified using a QIAquick PCR purification kit for a downstream assay. Cluster generation and sequencing were performed for 64 cycles on the Illumina Genome Analyzer II according to the manufacturer's instructions. The obtained images were analyzed and base called using the GA pipeline software version 1.4 with default settings provided by Illumina. Validation of the multiplex ultradeep sequencing of the HCV genome was performed using a plasmid encoding full-length HCV as a template, as reported previously (6). The overall error rates were determined to be, on average, 0.0010 per base pair. We also confirmed that high-fidelity PCR amplification with HCV-specific primer sets followed by multiplex ultradeep sequencing resulted in no significant increase in the error rates of viral sequencing data (ranging from 0.0012 to 0.0013 per bp; per-nucleotide error rate, 0.12% to 0.13%) (6).
Genome Analyzer sequence data analysis.
Using the high-performance alignment software NextGENe (SoftGenetics, State College, PA), the 64-base tags obtained from the Genome Analyzer II reads were aligned to the reference HCV RNA sequences of ∼9,200 bp that were determined by direct population Sanger sequencing in each clinical specimen. Entire reads were removed from the analysis when the median quality value score was <20 and when they contained >3 uncalled nucleotides. Low-quality bases were trimmed from the reads when >3 consecutive bases fell below a quality score of 16. Based on the above criteria, reads were aligned if ≥90% of their bases matched a particular position of the reference sequence. Each position of the viral genome was assigned a coverage depth representing the number of times that nucleotide position was sequenced.
Detection of defective HCV clones.
The methods for detecting defective HCV clones were reported previously (8). Briefly, reverse transcription-PCR (RT-PCR) was performed using the OneStep RNA PCR kit (TaKaRa) with the extracted RNA from liver and blood serum as a template and two pairs of primers, 5′-CGCCGACCTCATGGGGTACA-3′ and 5′-TGGTGTACATTTGGGTGATT-3′ for the first RT-PCR (HCV-P1) and 5′-TGCTCTTTCTCTATCTTCCT-3′ and 5′-GTGATGATGCAACCAAGTAG-3′ for the second PCR (HCV-P2). The PCR products were analyzed by electrophoresis in 0.8% agarose gels stained with ethidium bromide. Each purified DNA sample was sequenced at least three times using an ABI Prism BigDye Terminator ready reaction kit (Applied Biosystems). To determine defects in the HCV genome, the sequence of each sample was compared with the registered HCV genome sequence.
Statistical analysis.
The viral quasispecies nature was evaluated by analyzing the genetic complexity based on the number of different sequences present in the HCV population. The genetic complexity was determined by Shannon entropy index, calculated as follows:
where n is the number of different species identified, fi is the observed frequency of the particular variant in the quasispecies, and N is the total number of clones analyzed (9, 10). Statistical comparisons of the complexity between two groups were made using the Wilcoxon rank sum test or the Mann-Whitney U test. P values of <0.05 were considered to be statistically significant.
RESULTS
Patient characteristics.
The clinical and virological characteristics of the 5 patients are summarized in Table 1. Four of the 5 recipients were male, and the median age of the patients at the time of LDLT was 52 years (range, 47 to 65 years). All patients had decompensated cirrhosis caused by chronic hepatitis C, and 3 patients had hepatocellular carcinoma before liver transplantation. Right-lobe grafts were used for all patients. All patients had an HCV genotype 1b infection. The median blood serum HCV RNA load before transplantation was 5.5 log IU/ml (range, 4.6 to 6.6 log IU/ml) and was 5.9 log IU/ml (range, 5.8 to 6.4 log IU/ml) 1 month after liver transplantation; however, this difference was not significant (P = 0.18).
Table 1.
Baseline characteristics of 5 patients with chronic HCVa genotype 1b infection
| Patient characteristicb | Data for patient noc: |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Age (yr) | 65 | 52 | 47 | 58 | 48 |
| Sex | Female | Male | Male | Male | Male |
| Existence of HCC | + | + | − | + | − |
| Child-Pugh score | 10 | 10 | 9 | 10 | 10 |
| MELD score | 14 | 15 | 14 | 15 | 15 |
| HCV viral load (log IU/ml) | |||||
| Pre-LDLT | 4.6 | 6.6 | 4.9 | 5.5 | 5.9 |
| Post-LDLT | |||||
| 1 mo | 5.9 | 6.1 | 5.8 | 5.8 | 6.4 |
| 22 mo | 6.5 | ||||
| HCV infection | |||||
| Duration of hospital visit (yr) | 37 | 18 | 3 | 24 | 13 |
| Route of infection | Blood transfusion | Unknown | Unknown | Unknown | Unknown |
| Blood type | AB identical | A identical | A identical | A identical | A incompatible |
| Immunosuppressants | Tacrolimus, MMF | Tacrolimus, MMF | Tacrolimus, PSL | Tacrolimus, MMF | Tacrolimus, PSL |
HCV, hepatitis C virus.
HCC, hepatocellular carcinoma; MELD, model for end-stage liver disease; LDLT, living donor liver transplantation.
MMF, mycophenolate mofetil; PSL, prednisolone.
HCV population did not significantly differ between liver and serum samples.
To clarify the landscape of HCV heterogeneity as a quasispecies, we determined the viral full-genome sequences in liver and serum samples collected from the 5 recipients before transplantation using multiplex ultradeep sequencing and compared the results with those obtained by the direct population Sanger sequencing method. The HCV nucleotide sequence reads obtained by ultradeep sequencing were aligned to the consensus viral sequences in the serum specimen of each individual that were determined by direct population Sanger sequencing. A mean of 1,548-fold coverage was achieved at each nucleotide site of the HCV sequences in each specimen. First, the nucleotide sequence complexities expressed as the Shannon entropy index of HCV in the liver were compared with those in the serum. The overall viral complexity determined by the Shannon entropy index did not significantly differ between the liver and serum samples of each individual (see Fig. S1 in the supplemental material). Moreover, the patterns and distributions of genetic heterogeneity of the viral nucleotide sequences in the liver tissue sample were similar to those observed in the serum sample of the same patient (see Fig. S2 in the supplemental material). Next, we compared the viral genome sequences in the liver tissue with those in the serum in the same patient at the sites of the reported mutations that are related to the efficacy of interferon treatment and drug resistance against HCV protease and polymerase inhibitors (see Table S1 in the supplemental material). The prevalences of these mutations of the HCV genome in the liver were similar to those in the serum of the same patients. These findings suggested that a similar pattern of viral heterogeneity was maintained in the liver and serum of patients with chronic HCV infection.
Early dynamic decrease of viral complexity after liver transplantation.
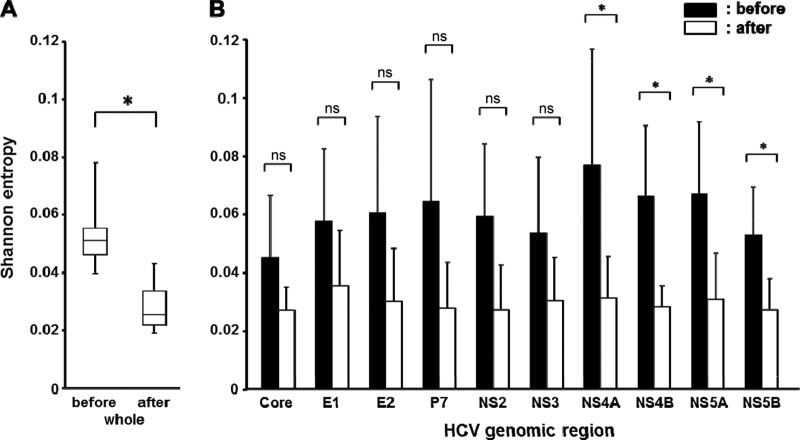
To clarify the changes in the viral quasispecies after liver transplantation, we investigated the change in the viral complexities of the serum specimens before and 1 month after liver transplantation in these 5 patients. The mean coverages of 1,284-fold and 1,141-fold were mapped to each reference sequence before and after liver transplantation, respectively. We then estimated the genomic complexity by calculating the Shannon entropy index for each nucleotide position before and after liver transplantation (Fig. 1A). The level of viral complexity of the blood serum HCV significantly differed between pretransplantation and posttransplantation (mean Shannon entropy index, 0.056 versus 0.029; P = 0.043), demonstrating that the viral quasispecies nature after reinfection and replication in the graft liver became more homogeneous than that before transplantation. To identify the specific regions in the HCV genome that were responsible for the selective increase in HCV after liver transplantation, we analyzed the changes in complexity of each region of the HCV genome (Fig. 1B). A decrease in the genetic complexity after liver transplantation was observed throughout the individual viral genetic regions. In particular, the complexity during pre- and posttransplantation was significantly different in the NS4A, NS4B, NS5A, and NS5B regions, suggesting that these regions are important for active proliferation of HCV at the early phase of reinfection in the graft liver. We then examined whether a specific nucleotide position was associated with a decrease in complexity after liver transplantation, but none of the specific nucleotide positions that changed by >50% after liver transplantation compared to before transplantation were commonly identified in the 5 patients (data not shown); this indicates that no association exists between a specific nucleotide position and the decrease in complexity after liver transplantation.
Fig 1.
Changes in the genetic complexity of the HCV genome before and after liver transplantation. (A) Mean Shannon entropy index values for the overall HCV genome in 5 LDLT recipients before and after liver transplantation. (B) Mean Shannon entropy index values for each HCV genomic region before (black bars) and after (white bars) liver transplantation are shown. The error bars in panels A and B represent the standard deviation. *, P < 0.05; ns, nonsignificant.
Defective HCV clones became undetectable immediately after liver transplantation.
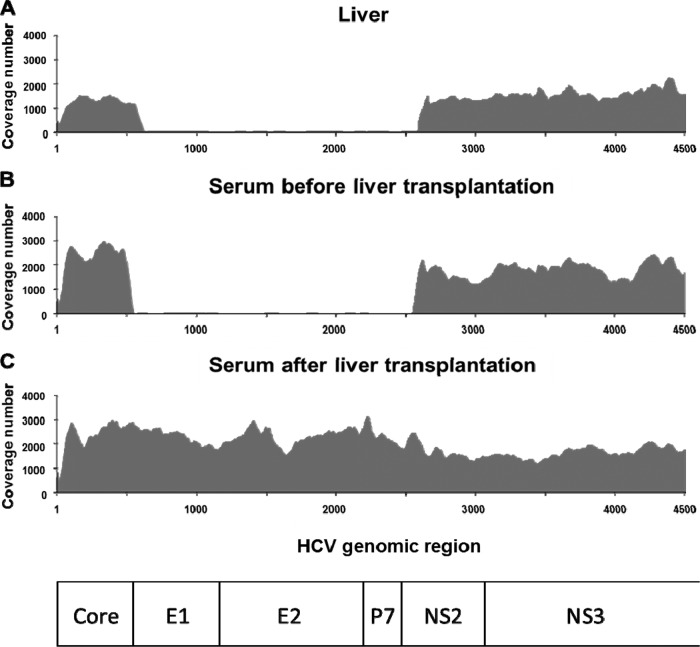
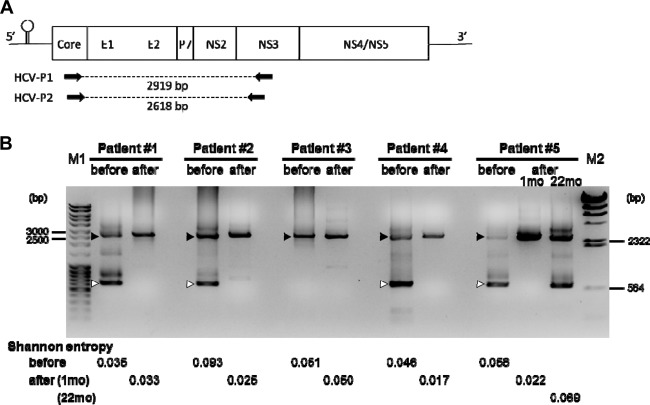
Using the ultradeep sequencing analyses, we found that the sequence coverage of viral genomic regions spanning from the end of the core to the middle of NS2 was smaller than those of the other regions in several liver and serum samples before liver transplantation, but this tendency was not observed in the samples after liver transplantation (Fig. 2). As we previously identified the defective HCV clones lacking the structural regions of the HCV genome in the serum samples of HCV-positive liver transplant recipients (8), we speculated that the presence of the defective HCV clones would result in the smaller coverage of E1-NS2 before transplantation, and the population of the defective clones would change after liver transplantation. Therefore, we next analyzed the population change of the defective HCV clones between before and after liver transplantation. Using RT-PCR analysis with the primers HCV-P1 and HCV-P2 (Fig. 3A), we detected both defective HCV clones and full-genome HCV clones before liver transplantation at various ratios in each sample, except for in patient 3 (Fig. 3B). The defective HCV clones became undetectable and the full-genome HCV clones became dominant in the serum samples 1 month after liver transplantation, indicating that the defective HCV clones have less of a replication advantage than the full-genome clones. In patient 3, the defective HCV clones were undetectable both before and after liver transplantation.
Fig 2.
Dynamics of defective HCV clones indicated by coverage numbers of ultradeep sequence of HCV genome. Coverage of ultradeep sequence of HCV genome in liver (A), serum samples before liver transplantation (B), and serum samples after liver transplantation (C) for patient 1. The degree of coverage (fold) at each nucleotide site of the HCV sequence is shown. Nucleotide 1 indicates the first nucleotide of the core region of HCV RNA. Similar results were obtained in the samples from patients 2, 4, and 5.
Fig 3.
Dynamics of defective HCV clones based on RT-PCR analysis. (A) Schematic presentation of the HCV genome and the primer sets used in this study. (B) Results of RT-PCR analysis by using RNA samples as a template, which were extracted from blood serum before and 1 month after liver transplantation in all patients and 22 months after transplantation in patient 5. HCV-P1 and HCV-P2 (panel A) were used as primers. Lanes M1 and M2, the molecular weight markers MassRuler DNA ladder mix (Fermentas, Canada) and Lambda DNA-HindIII Digest (New England BioLabs, USA), respectively. The values shown indicate the sizes of the band in the molecular weight markers. Black arrowheads, full-length PCR fragment of 2,618 bp; white arrowheads, defective HCV clones that were confirmed by sequencing analysis. The Shannon entropy index values of these HCV specimens in the serum (before and after liver transplantation) are shown at the bottom.
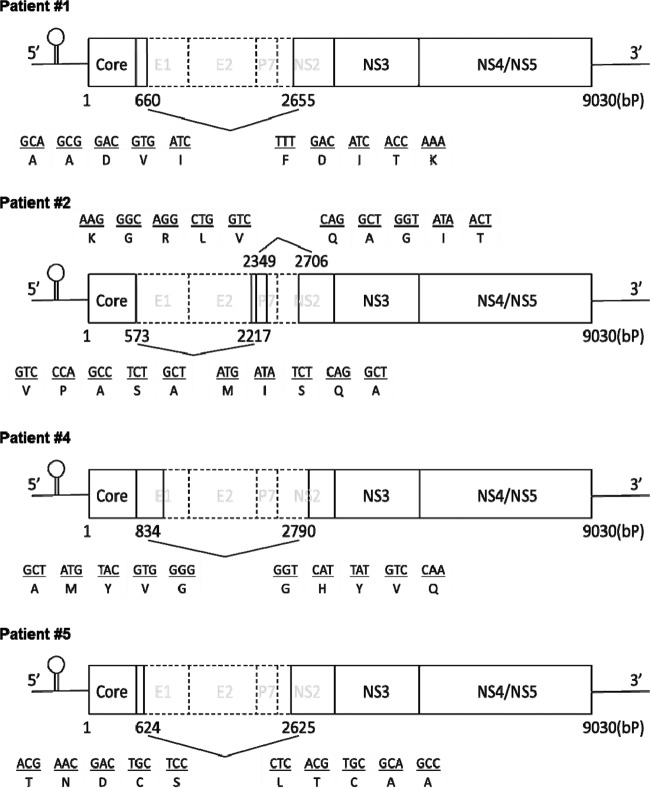
To determine the internal structure of the deletions in the defective HCV genomes, major amplified fragments from each of the four patients with defective HCV clones before transplantation were subcloned for further sequence analyses. Schematic representations of the defective HCV RNA detected in the blood serum specimens of these patients are shown in Fig. 4. Sequence analyses revealed that the structural region was widely deleted in all of the defective HCV clones. The 3′-boundaries of the deletions were quite diverse in the clones, while the 5′-untranslated region and core regions were preserved in all four clones, as reported previously (8). Two distinct defective clones were found in patient 2. All of the deletions identified were in frame, implying that these defective HCV genomes have the potential for translation from the core to the authentic end of NS5B without a frameshift.
Fig 4.
Schematic presentation of major defective HCV clones in 4 patients before liver transplantation. The values in the schema indicate the nucleotide numbers from the first ATG of the core region in HCV RNA. Nucleotide and amino acid sequences before and after the deleted region of the HCV genome are shown. E1, envelope glycoprotein 1; E2, envelope glycoprotein 2; NS, nonstructural protein.
We then analyzed the dynamics of the defective HCV clones at the chronic hepatitis phase after liver transplantation in patient 5. As shown in the Patient 5 column in Fig. 3B, RT-PCR from a serum sample collected at 22 months after liver transplantation, when a liver biopsy specimen demonstrated findings of chronic hepatitis C with fibrosis (METAVIR score, A1 F1), showed that a defective HCV clone had reappeared. The size of the defective clone was the same as that found in the serum before transplantation, and we confirmed by sequence analysis that the deleted region of the defective HCV clone was identical to that in the pretransplant serum sample. The viral complexity analyzed by calculating the Shannon entropy index from ultradeep sequencing data also returned to the pretransplantation level at the chronic hepatitis phase (Shannon entropy values, 0.056 before transplantation, 0.022 at 1 month posttransplantation, and 0.069 at 22 months after liver transplantation). These findings indicated that the reconstitution of HCV heterogeneity occurs at the chronic hepatitis phase after liver transplantation, and the same defective HCV clone present before liver transplantation reappears at the chronic hepatitis phase after liver transplantation.
DISCUSSION
The present study revealed two major findings from ultradeep sequencing analyses of the HCV genome sequence in liver transplant recipients before and after liver transplantation. First, the viral heterogeneity of HCV significantly decreased after liver transplantation, indicating that the clones with advantages for infection and/or replication in hepatocytes rapidly increased after liver transplantation. Second, full-genome HCV clones selectively increased, while the defective clones did not increase in number during the period immediately after liver transplantation.
The discovery of differences in the populations of HCV quasispecies between the liver and serum of the same individuals has been controversial. Most previous studies examined the HCV sequencing mainly for the hypervariable region in E2 using the Sanger sequencing method (11–13) or single-strand conformation polymorphism (12, 14, 15), but the findings were conflicting. In the present study, we obtained full-genome HCV sequences using ultradeep sequencing analysis. Our results suggested that a similar HCV population exists in the liver and blood serum, at least at the specific sites related to interferon sensitivity and drug resistance. These results are clinically important because we confirmed that the serum samples, which are easily obtained from patients, reflect the HCV population in the liver and are thus useful for analyses of resistance and sensitivity to treatment.
Differences in the HCV population between individuals can be determined by multiple factors, such as the duration of hospital visit, route of HCV infection, fibrosis progression, degree of inflammation, and the presence of hepatocellular carcinoma. In our analysis, we could not find an association between these clinical characteristics and the nature of the HCV population between patients. However, we speculated that undetectable defective HCV clones present before liver transplantation in patient 3 might be associated with a shorter duration of HCV infection. In patients 1 and 3, the difference in viral complexity as measured by the Shannon entropy index values between before and after liver transplantation was small. The reason is unclear at present, but differences in the clinical features of infected patients might affect the results. Further large-scale investigations may reveal the relationship between clinical features in patients and the nature of a specific HCV population.
Our large-scale analysis using ultradeep sequencing demonstrated that the complexity of all regions of the HCV genome was dramatically reduced 1 month after liver transplantation compared with the pretransplantation level of complexity. This finding is consistent with findings from previous reports using Sanger sequencing methods that showed that heterogeneity is decreased in the hypervariable region of E2 of HCV after liver transplantation (16, 17). Gretch et al. (16) analyzed HCV quasispecies before and after liver transplantation by comparing the differences in the hypervariable region of the HCV genome in 5 transplant recipients. They found that different HCV clones were present in pretransplant blood serum and relatively homogeneous quasispecies variants emerged after liver transplantation in all 5 cases. Hughes et al. (17) demonstrated that the viral complexity of the hypervariable region 1 in postperfusion liver tissue at 2.5 h after liver transplantation was significantly lower than that in explanted liver and in pretransplant serum, although there was no significant difference in the complexity between the explanted liver and pretransplant serum. Our present data confirmed the results of these previous studies and added new information from the full-genome ultradeep sequencing. In particular, our data demonstrated a new aspect of the analyses of full-genome and defective HCV clones, because the defective HCV clones lack hypervariable regions that were analyzed in previous studies. Interestingly, our analysis revealed significant decreases in complexity in the NS4A, NS4B, NS5A, and NS5B regions after transplantation, although a decreasing trend was detected in all regions of the HCV genome. Because the region from NS4A to NS5B has important roles in HCV replication (18–20), a decrease in the complexity of the NS4A to NS5B sequence after liver transplantation might indicate the presence of the specific NS4A to NS5B sequence in the HCV genome that confers advantages in the reinfection and/or replication processes. Therefore, we attempted to identify the specific HCV genome sequences with such advantages. However, we could not identify a common feature of the HCV genomic changes in amplified HCV clones after liver transplantation among the 5 cases tested. This may be due to differences between individuals in the relative fitness of a viral subpopulation in its host, which is determined by multiple factors, including infection capacity, replication ability, and mechanisms by which to escape from immune pressure.
We previously identified defective HCV clones in the blood serum of patients after liver transplantation (8). Other groups also reported that defective HCV clones exist in the liver and serum of patients with chronic hepatitis C and patients with immunosilent infections (21–25). These reports demonstrated that deletions in the HCV genome were present mainly in the structural region, while the 5′-untranslated region, the core, and NS3 to NS5B regions were preserved, and that most of the deletions were in frame, indicating that the preserved regions can be translated to the authentic terminus. Indeed, Sugiyama et al. (24) recently demonstrated that the defective genome can be translated, self-replicated, and encapsidated as an infectious particle by trans-complementation of the structural proteins in vitro. Pacini et al. (23) also reported that defective HCV clones show robust replication, efficient trans-packaging, and infection of cultured cells. These data suggest that the abilities of defective HCV genomes to infect, replicate, and be encapsidated do not differ from those of full-genome HCV. The in vivo data reported here, however, clearly reveal that the amount of defective HCV clones was lower than that of full-genome HCV after liver transplantation, although the reason for this remains unknown. One possibility is that the capabilities to infect, replicate, or be encapsidated differ between defective HCV and full-genome HCV in vivo. It is noteworthy that an identical defective HCV clone that was detected before transplantation reappeared during the chronic hepatitis phase after transplantation in patient 5. This finding suggests that the defective clone in the blood serum also infected the graft liver, replicated, and was encapsidated in the graft liver after liver transplantation. Therefore, the speed of these steps would differ between defective HCV clones and full-genome HCV clones.
The present study revealed a limitation of the massively parallel ultradeep sequencing technology in the analyses of viral quasispecies. Because the massively parallel ultradeep sequencing platform is based on multitudinous short reads, it is difficult to separately evaluate the association between nucleotide sites that are mapped to different viral genome regions in a single viral clone. Indeed, it is difficult to clarify the potential mutational linkage between different viral genomic regions because of the short read lengths of the shotgun sequencing approach.
In conclusion, after liver transplantation, viral heterogeneity decreased significantly and the number of full-genome HCV clones increased immediately, whereas the defective HCV clones began to increase in number over a longer period. Further analysis will reveal the significance of the changes in defective HCV clones after liver transplantation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research no. 21229009 and 23590972 and Health and Labor Sciences research grants for research on intractable diseases and research on hepatitis from the Ministry of Health, Labor, and Welfare, Japan.
Footnotes
Published ahead of print 28 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00676-13.
REFERENCES
- 1. Shimotohno K. 1995. Hepatitis C virus as a causative agent of hepatocellular carcinoma. Intervirology 38:162–169 [DOI] [PubMed] [Google Scholar]
- 2. Mardis ER. 2009. New strategies and emerging technologies for massively parallel sequencing: applications in medical research. Genome Med. 1:40. 10.1186/gm40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ueda Y, Marusawa H, Kaido T, Ogura Y, Ogawa K, Yoshizawa A, Hata K, Fujimoto Y, Nishijima N, Chiba T, Uemoto S. 2012. Efficacy and safety of prophylaxis with entecavir and hepatitis B immunoglobulin in preventing hepatitis B recurrence after living-donor liver transplantation. Hepatol Res. 43:67–71 [DOI] [PubMed] [Google Scholar]
- 5. Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. 1997. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J. Clin. Microbiol. 35:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nasu A, Marusawa H, Ueda Y, Nishijima N, Takahashi K, Osaki Y, Yamashita Y, Inokuma T, Tamada T, Fujiwara T, Sato F, Shimizu K, Chiba T. 2011. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One 6:e24907. 10.1371/journal.pone.0024907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishijima N, Marusawa H, Ueda Y, Takahashi K, Nasu A, Osaki Y, Kou T, Yazumi S, Fujiwara T, Tsuchiya S, Shimizu K, Uemoto S, Chiba T. 2012. Dynamics of hepatitis B virus quasispecies in association with nucleos(t)ide analogue treatment determined by ultra-deep sequencing. PLoS One 7:e35052. 10.1371/journal.pone.0035052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iwai A, Marusawa H, Takada Y, Egawa H, Ikeda K, Nabeshima M, Uemoto S, Chiba T. 2006. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J. Viral Hepat. 13:523–531 [DOI] [PubMed] [Google Scholar]
- 9. Fishman SL, Branch AD. 2009. The quasispecies nature and biological implications of the hepatitis C virus. Infect. Genet. Evol. 9:1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537–542 [DOI] [PubMed] [Google Scholar]
- 11. Cabot B, Martell M, Esteban JI, Sauleda S, Otero T, Esteban R, Guàrdia J, Gómez J. 2000. Nucleotide and amino acid complexity of hepatitis C virus quasispecies in serum and liver. J. Virol. 74:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jang SJ, Wang LF, Radkowski M, Rakela J, Laskus T. 1999. Differences between hepatitis C virus 5′ untranslated region quasispecies in serum and liver. J. Gen. Virol. 80(Pt 3):711–716 [DOI] [PubMed] [Google Scholar]
- 13. Sakai A, Kaneko S, Honda M, Matsushita E, Kobayashi K. 1999. Quasispecies of hepatitis C virus in serum and in three different parts of the liver of patients with chronic hepatitis. Hepatology 30:556–561 [DOI] [PubMed] [Google Scholar]
- 14. De Mitri MS, Mele L, Chen CH, Piccinini A, Chianese R, D'Errico A, Alberti A, Pisi E. 1998. Comparison of serum and liver hepatitis C virus quasispecies in HCV-related hepatocellular carcinoma. J. Hepatol. 29:887–892 [DOI] [PubMed] [Google Scholar]
- 15. Sakamoto N, Enomoto N, Kurosaki M, Asahina Y, Maekawa S, Koizumi K, Sakuma I, Murakami T, Marumo F, Sato C. 1995. Comparison of the hypervariable region of hepatitis C virus genomes in plasma and liver. J. Med. Virol. 46:7–11 [DOI] [PubMed] [Google Scholar]
- 16. Gretch DR, Polyak SJ, Wilson JJ, Carithers RL, Jr, Perkins JD, Corey L. 1996. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J. Virol. 70:7622–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes MG, Jr, Rudy CK, Chong TW, Smith RL, Evans HL, Iezzoni JC, Sawyer RG, Pruett TL. 2004. E2 quasispecies specificity of hepatitis C virus association with allografts immediately after liver transplantation. Liver Transpl. 10:208–216 [DOI] [PubMed] [Google Scholar]
- 18. Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moradpour D, Brass V, Bieck E, Friebe P, Gosert R, Blum HE, Bartenschlager R, Penin F, Lohmann V. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278–13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shimakami T, Hijikata M, Luo H, Ma YY, Kaneko S, Shimotohno K, Murakami S. 2004. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78:2738–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernardin F, Stramer SL, Rehermann B, Page-Shafer K, Cooper S, Bangsberg DR, Hahn J, Tobler L, Busch M, Delwart E. 2007. High levels of subgenomic HCV plasma RNA in immunosilent infections. Virology 365:446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noppornpanth S, Smits SL, Lien TX, Poovorawan Y, Osterhaus ADME, Haagmans BL. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 81:12496–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pacini L, Graziani R, Bartholomew L, De Francesco R, Paonessa G. 2009. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J. Virol. 83:9079–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sugiyama K, Suzuki K, Nakazawa T, Funami K, Hishiki T, Ogawa K, Saito S, Shimotohno KW, Suzuki T, Shimizu Y, Tobita R, Hijikata M, Takaku H, Shimotohno K. 2009. Genetic analysis of hepatitis C virus with defective genome and its infectivity in vitro. J. Virol. 83:6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yagi S, Mori K, Tanaka E, Matsumoto A, Sunaga F, Kiyosawa K, Yamaguchi K. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 77:399–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.