Abstract
Studies pertaining to Salmonella enterica serovar Typhimurium infection by utilizing model systems failed to mimic the essential aspects of immunity induced by Salmonella enterica serovar Typhi, as the determinants of innate immunity are distinct. The present study investigated the physiological and innate immune responses of S. Typhi infected Caenorhabditis elegans and also explored the Ty21a mediated immune enhancement in C. elegans. Ty21a is a known live vaccine for typhoidal infection in human beings. Physiological responses of C. elegans infected with S. Typhi assessed by survival and behavioral assays revealed that S. Typhi caused host mortality by persistent infection. However, Ty21a exposure to C. elegans was not harmful. Ty21a pre-exposed C. elegans, exhibited significant resistance against S. Typhi infection. Elevated accumulation of S. Typhi inside the infected host was observed when compared to Ty21a exposures. Transcript analysis of candidate innate immune gene (clec-60, clec-87, lys-7, ilys-3, scl-2, cpr-2, F08G5.6, atf-7, age-1, bec-1 and daf-16) regulations in the host during S. Typhi infection have been assessed through qPCR analysis to understand the activation of immune signaling pathways during S. Typhi infections. Gene silencing approaches confirmed that clec-60 and clec-87 has a major role in the defense system of C. elegans during S. Typhi infection. In conclusion, the study revealed that preconditioning of host with Ty21a protects against subsequent S. Typhi infection.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-013-0424-x) contains supplementary material, which is available to authorized users.
Keywords: Caenorhabditis elegans, Innate immune response, Pre-exposed host, RNAi, Salmonella enterica serovar Typhi
Introduction
Typhoid fever is a severe infection in the reticulo-endothelial system caused by the etiological agent Salmonella enterica serovar Typhi. Due to indiscriminate use of antibiotics to fight against the disease, it has led to the emergence of multidrug-resistant strains of S. Typhi. Invasive S. Typhi is able to overcome the stomach acid barrier and causes disease by initially colonizing the intestinal tract and entering the lymphatic system through the passage of lumen. However, the induction of a defensive immune response is reliant upon the bacteria possessing an entire lipopolysaccharide (LPS) [1]. In order to prevent and adopt immune to the disease, vaccines were developed [2, 3] like S. enterica serovar Typhi Ty21a, which is the licensed live attenuated oral bacterial vaccine against typhoid fever [4]. Ty21a is an attenuated strain of S. Typhi Ty2 isolated in the year 1970, which has been attenuated by an irreversible genetic defect, achieved by multiple mutations induced by chemical mutagenesis [1]. Multiple mutations made Ty21a as an avirulent strain and its inability of the reversion was confirmed through in vitro and in vivo studies [5].
Earlier studies have demonstrated that certain genes were induced against S. Typhimurium infection in Caenorhabditis elegans [6, 7]. These studies fail to mimic the essential aspects of immunity induced by S. Typhi [8]. Moreover, the determinants of host defense system to S. Typhimurium and S. Typhi are basically different [9–12] and earlier studies designate that the host immune interaction in response to S. Typhimurium and S. Typhi are qualitatively diverse. The lack of a small model has been a foremost hindrance in understanding the virulence mechanisms of S. Typhi [8]. In contrast, there are no reports available on physiological and molecular response of host altered by S. Typhi and S. Typhi Ty21a. Caenorhabditis elegans as a suitable model for studying human pathogen due to its functional similarity with human [13]. Therefore this study aimed to evaluate the role of candidate innate immune genes against S. Typhi infection and also demonstrated the Ty21a mediated immune enhancement against S. Typhi using the simple tractable model system, C. elegans.
Materials and Methods
Bacterial Strains, Nematode, Media and Culture Conditions
Salmonella enterica serovar Typhi Ty21a was generously provided by Dr. S. Krishnaswamy (Madurai Kamaraj University, Madurai). The wild type strain of S. enterica Typhi (MTCC 733) was obtained from MTCC. Escherichia coli OP50 was provided by CGC. Bacterial cultures were maintained on Luria–Bertani (LB) medium at 37 °C, with aeration. Media was solidified by the addition of agar (18 g/l). Caenorhabditis elegans wild type strain N2 Bristol was provided by CGC, and maintained on nematode growth medium [14]. All the experiments were carried out using age-synchronized L4 stage worms. Synchronous populations were acquired as described by Sivamaruthi et al. [15].
Chemotaxis Assay
The cultures (0.5 OD) of S. Typhi or S. Typhi Ty21a and E. coli OP50 were spotted at a distance of 3 cm from the centre of NGM plates (90 mm) and denoted as zone A and zone B, respectively. Wild-type C. elegans were thoroughly washed from E. coli OP50 lawn and 25 worms were placed at the center of the culture spotted NGM plate. The number of worms in zone A and zone B were counted every 4 h. Similarly in control plates, both zones A and B were spotted with E. coli OP50.
Liquid Survival Assay
Immune conditioning of the host was done by treating the naive host with Ty21a for 12 h at 20 °C. Liquid killing assay was performed with naive or Ty21a pre-exposed (conditioned) C. elegans against S. Typhi and S. Typhi Ty21a as described by Sivamaruthi et al. [15]. Worms treated with E. coli OP50 served as a control and all the experiments were conducted at least three independent occasions. Statistical analysis was performed using SPSS Statistics 17.0. P values were generated by ANOVA followed by a Dunnett’s T3 post hoc test. P < 0.05 was considered to be significant.
Short-Time Exposure Assays on C. elegans
L4 staged hermaphrodites were exposed to S. Typhi or S. Typhi Ty21a for short durations (1, 3, 5, and 14 h) and assessed the physiological changes as described by Sivamaruthi et al. [15].
Bacterial Accumulation
Accumulation of bacterial load (S. Typhi or S. Typhi Ty21a) inside the host system was assessed through the procedure prescribed by Sivamaruthi et al. [15]. Salmonella Shigella Agar (Himedia) medium was used for the plating purpose.
Brood Size and Pharyngeal Pumping Assay
To determine the pumping rate, naive and conditioned worms were placed on NGM plates seeded with E. coli OP50 (control) and S. Typhi. The number of progeny of the worms was counted and the pharyngeal pumping rate also was observed using a stereomicroscope (Nikon SMZ1000, Japan) for 30 consecutive seconds [15].
Aldicarb Sensitivity (Acetylcholine-Specific) Assay
Approximately, 20 N2 (control/S. Typhi infected) worms transferred onto 0.5 mM aldicarb plates. Every 10 min, worms were scored for paralysis for 2 h. The worms were allowed to stay on the aldicarb plates and the number of paralyzed worms was counted.
Osmosensation Assay
Infected worms were assessed for their ASH osmosensory function. High osmolarity ring with 1 cm radius was created on NGM plates using 4 M NaCl constitute of phenol-red (non-toxic dye to observe the solution and act as an indicator). Approximately, 20 worms (control/S. Typhi exposed) were washed with M9 buffer, allowed to crawl on an unseeded NGM agar plate, and then transferred to the center of high (4 M NaCl) osmolarity rings or normal NGM plate (control plate). The worms that escaped from the barrier ring were counted.
qPCR
Salmonella enterica serovar Typhi or E. coli OP50 infected worms were collected after each 24 h of post-infection and washed thrice with DEPC treated M9 buffer. Washed worms were incubated at −80 °C after treating with TRIzol (Invitrogen) reagent for 24–48 h. Total RNA was isolated according to the manufacturer’s instructions. Total RNA was reverse-transcribed using Superscript III (Invitrogen) after the verification of RNA purity (BioSpec-nano). Real-time PCR was carried out to analyze the expression pattern of candidate antimicrobial genes (clec-60, clec-87, lys-7, ilys-3, scl-2, cpr-2, F08G5.6, atf-7, age-1, bec-1 and daf-16) using gene specific primers (online resource Table S1). The conditions for qPCR were 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 40 s. The fold difference in the expression was calculated by comparative Ct value analysis with act-2 (housekeeping gene) normalization.
RNAi
The mRNA sequences of target genes (clec-60 and clec-87) were retrieved from Wormbase and the spliced sequences were used as input for the siRNA designing tool, Target Finder (AmbionR life technologies). Two targets were selected for each gene from the software suggested silencing targets. The selection of the silencing target is based on the GC content and the position in the mRNA sequence. Specificity of selected targets was analyzed by NCBI–BLAST. The BLAST results revealed that the selected targets were 100 % specific to targeted mRNA. The siRNA duplexes were chemically synthesized from VBC biotech, Austria. The delivery of siRNA to the worms was done successfully by soaking method [16].
Results and Discussion
C. elegans Does Not Avoid S. Typhi and S. Typhi Ty21a
Bacterial pathogens are detected by C. elegans through its olfactory neurons and they respond against the pathogen by simply avoiding the bacteria. Hence, it is necessary to analyze the chemotaxis behavior of C. elegans against the bacteria to be studied intensively. In the present study, the chemotaxis assay results revealed that C. elegans does not avoid S. Typhi as well as S. Typhi Ty21a. C. elegans moved freely without any hindrance over the NGM plates spread with S. Typhi and E. coli OP50 or S. Typhi Ty21a and E. coli OP50. Both S. Typhi and S. Typhi Ty21a plates showed an equal distribution of worms (online resource Fig. S1).
S. Typhi Ty21a Enhanced the Survivability of Pre-exposed C. elegans
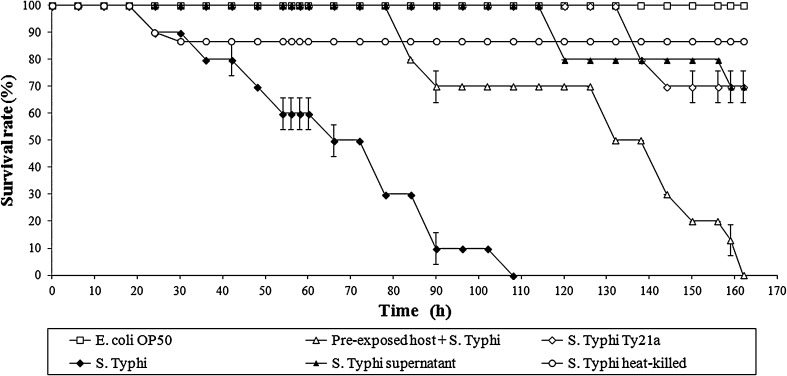
Even though the chemotaxis assay results indicated that C. elegans can uptake S. Typhi and S. Typhi Ty21a in solid condition, all the assays were performed in liquid medium to enhance the rate of intake of bacteria by the host. In liquid assays, C. elegans were exposed to S. Typhi or S. Typhi Ty21a continuously and monitored for their survival rate. Results of liquid assay revealed that the continuous exposure of S. Typhi to C. elegans caused host mortality with LT50 of 64 ± 3.4 h and complete death at 100 ± 6.9 h, whereas null mortality of the worms exposed to E. coli OP50 and reduced mortality (20 %) of S. Typhi Ty21a exposed worms were observed. S. Typhi Ty21a is a vaccine for typhoid. Typically, the function of a vaccine is stimulation of the host immune system against the respective infection. The previous study suggested that food and folate supplementation enhances the survivability of S. Typhi infected C. elegans [17]. The present study evaluated the Ty21a mediated induction of C. elegans immune system against S. Typhi infection. Interestingly, conditioned (12 h pre-exposed to S. Typhi Ty21a) worms showed an increased survival rate (LT50 of 132 ± 2.1 h) against S. Typhi, compared to naive C. elegans (LT50 of 64 ± 3.4 h), which revealed that the pre-exposed C. elegans survived longer (about ~54 %) than naive worms upon subsequent infection with S. Typhi. Neither heat-killed S. Typhi nor cell free supernatants caused significant mortality, suggesting that S. Typhi pathogenesis required live infection (Fig. 1). The above results supported the notion that Ty21a can be used as an oral vaccine for a multicellular system infected with S. Typhi. In addition, it appears that Ty21a mediated enhancement of innate immunity is confined to S. Typhi mediated infection and appeared to be not significantly effective for other Gram-negative bacterial pathogens (Data not shown).
Fig. 1.
Survival of C. elegans in liquid assays. Pre-exposed C. elegans (wild-type) exhibited resistance against S. Typhi infection during liquid infection assay. Age synchronized young adult worms were pre-exposed with S. Typhi Ty21a (pre-treatment for 12 h) as described in “Materials and Methods” section. Escherichia coli OP50 exposed worms were used as a control (open square). The naive C. elegans exposed to pathogenic S. Typhi (closed rhombus) showed a significant difference in their survival rate when compared with the pre-exposed C. elegans exposed to pathogenic S. Typhi (open triangle). The differences between the survival rate of E. coli OP50 exposed naive C. elegans and S. Typhi challenged pre-exposed C. elegans (P = 0.006) or naive C. elegans (P = 0.004), were significant. Ty21a exposed worms survived significantly different from naive C. elegans exposed to S. Typhi (P = 0.012). P values were generated by ANOVA (Dunnett’s T3 post hoc test). P < 0.05 was considered to be significant
Five-Hour Exposure of S. Typhi is Enough to Cause Infection
In short-time exposure study, the life span of C. elegans exposed to S. Typhi/S. Typhi Ty21a was kinetically monitored after placing them in E. coli OP50 seeded NGM plates. Ty21a exposed worms (for about 1–14 h) were active and produced progenies. Worms exposed to S. Typhi for 1–3 h were active and produced the next generation in the presence of a food source (E. coli OP50). However, worms exposed for 5 h were dead after 144 h with no progeny. Worms exposed to S. Typhi for 14 h were dead after 98 h even in the presence of a food source. These data indicated that S. Typhi required at-least 5 h of interaction to significantly infect and cause mortality in C. elegans, whereas Ty21a did not cause any lethal effect to the host system (Table 1).
Table 1.
Status of C. elegans placed on E. coli OP50 seeded medium during the short time exposure assay
| Time (h) | S. Typhi | S. Typhi Ty21a |
|---|---|---|
| 1 | C. elegans were active and able to produce next generation | C. elegans were active and able to produce next generation |
| 3 | C. elegans were active and able to produce next generation | C. elegans were active and able to produce next generation |
| 5 | C. elegans were active for 72 h without producing progeny and died after 144 h | C. elegans were active and able to produce next generation |
| 14 | C. elegans were active for 72 h without producing progeny and died after 98 h | C. elegans were active and able to produce next generation |
Typhi Cause Persistent Infection in C. elegans
In order to demonstrate the presence and multiplication of S. Typhi inside the infected C. elegans, CFU assay was performed. Accumulation of S. Typhi or S. Typhi Ty21a in C. elegans after 24 h exposure was found to be 4.51 × 102 or 3.2 × 102 per worm, respectively. However, the multiplication of S. Typhi and S. Typhi Ty21a in exposed C. elegans was distinctly different in both the cases (after 24 h) (online resource Fig. S2). No live bacteria could be recovered from the control C. elegans. These results confirmed that accumulation of S. Typhi was gradually increased over the course of infection within the host compared to Ty21a (online resource Fig. S2). This result stated that wild-type S. Typhi is highly virulent to C. elegans when compared to Ty21a.
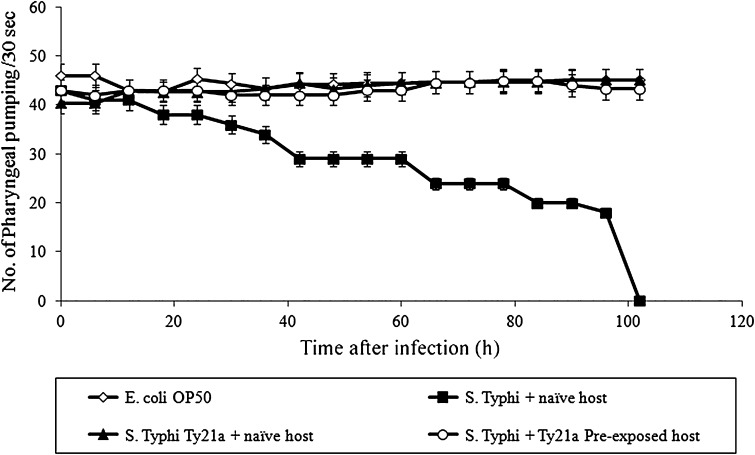
In egg laying assay, the wild-type worms exposed to S. Typhi produced 70 ± 9 progenies compared to the control worm (268 ± 10 progenies) which revealed that S. Typhi infection affects the reproductive mechanism of C. elegans. In pharyngeal pumping assay, the pumping rate of naive wild-type C. elegans infected with S. Typhi have been reduced in a time dependent manner compared to control (Fig. 2). This result indicated that the bacterial infection damages the pharyngeal region of the host.
Fig. 2.
Impact of infection in pharyngeal region of the host. Pharyngeal pumping rate (number of flings per 30 s) of control (open rhombus), S. Typhi infected naive (closed square), pre-exposed C. elegans (open circle) and S. Typhi Ty21a (closed triangle) infected naive host during infection
Behavioral Changes in C. elegans Exposed to S. Typhi
Additionally, behavioral assays (Ach-mediated behavioral assay and osmosensation assay) were performed to study the possible defects in specific classes of neurons within the control and pathogen exposed worms. Aldicarb blocks acetylcholine esterase (AchE) and that leads to paralysis. Infection mediated alteration in the expression of AchE has an impact on the time duration required for the aldicarb induced paralysis in naive C. elegans [18]. Uninfected wild-type C. elegans in aldicarb incorporated plate showed paralysis after 2 h from the time of transfer and its pharyngeal pumping rate (no. of flings per 30 s) decreased from about 51 to 39. The wild-type worms exposed to S. Typhi for 24 h were also paralyzed after 2 h with a decrease in pharyngeal pumping (online resource Table S2). This result suggested that S. Typhi infection appears to not affect the expression of AchE in C. elegans.
Worms avoid the high osmotic strength areas and this behavior is controlled by several neurons located in the head of the worm. The defects associated with specific neurons of worms exposed with OP50 and the pathogen was analyzed based on this avoidance behavior of high osmolarity barrier. When N2 control worms are placed at the midpoint of a high osmolarity ring, they tend to withdraw from the boundary and are trapped inside the ring. However, wild-type C. elegans exposed to S. Typhi for 48 h crossed the high osmolarity ring and the uninfected C. elegans sensed the boundary of the high osmolarity ring remains trapped inside the ring (online resource Fig. S3).
Regulation of Candidate Innate Immune Genes in C. elegans Upon S. Typhi Infection
Regulations of candidate innate immune genes (clec-60, clec-87, lys-7, ilys-3, scl-2, cpr-2, F08G5.6, atf-7, age-1, bec-1 and daf-16) were analyzed in C. elegans during S. Typhi infection. C-type lectin-like domain containing proteins (CTLD protein) are secreted as transmembrane proteins in the intestine and as soluble proteins. In C. elegans, the potential contribution of CTLD proteins to immune specificity is demonstrated by their differential up-regulation towards pathogens [19–24].
In the present study, the level of expression of clec-87 and clec-60 was evaluated. During the initial hours of infection, both clec-60 and clec-87 were up-regulated compared to the control. After 48 h of post-infection, reduction in the expression of clec-60 was observed, whereas a higher level of expression of clec-87 was observed after 72 h of infection. The reason for the elevated expression of clec-87 is may be the necessity to recognize the proliferating bacterial cells within the host, which suggested that clec-60 and clec-87 are required for the recognition of invading pathogen, thus host can provoke the active defense mechanism against infection (online resource Fig. S4).
Lysozymes play a significant role in both vertebrate and invertebrate immunity. In C. elegans, lys-7 expression was up-regulated in response to infection with the Gram-negative bacterium Serratia marcescens, indicating that lys-7 likely plays a role in the innate immune response [20]. Invertebrate lysozyme (ilys-3) and lys-7 were gradually up-regulated upon infection, which indicated the degradation of intracellular S. Typhi in C. elegans. After 96 h of post-infection, lys-7 and ilys-3 expression was observed to the maximum level. This elevation may be the resultant of survived worms even after 72 h of continuous exposure to S. Typhi (online resource Fig. S5).
Gradual reduction of daf-16 was observed in infected N2 worms. daf-16 was up-regulated in C. elegans after 72 h of S. Typhi infection (up-regulation of daf-16 leads to the longevity in C. elegans) indicating its involvement in extending the life of the host. However, the virulence mechanism of bacterial pathogen masked the expression of daf-16 by suppressing the expression during the later hours of infection (online resource Fig. S4).
Previous reports revealed that decreased level of age-1 increases the lifespan in C. elegans [25, 26]. During the initial hour of exposure, age-1 was found to be up-regulated to lead the normal life cycle, whereas after 72 h of post-infection, age-1 expression was suppressed to facilitate the host to extend the lifespan to tackle the pathogenic condition. Salmonella enterica serovar Typhi infected host illustrated gradual up-regulation of bec-1 except 96 h. Previous findings revealed that BEC-1 has role in C. elegans dauer formation and plays a role in autophagy pathway. The higher expression of this gene during S. Typhi infection revealed that host struggled to resist the infection by attaining the dauer phenomena and lysosomal degradation of the pathogen.
Level of cpr-2 was up-regulated with higher fold induction at 24 h and sudden reduction at the latter hours of S. Typhi infection. Drastic reduction of cpr-2 (CPR-2 is a Cysteine protease related family member known to involve in development and apoptosis) might have favoured the multiplication of pathogen within the host system. F08G5.6 is an unnamed CUB domain containing protein involved in both MAPK and daf-16 regulated insulin signaling pathway of C. elegans. Expression of this gene is greatly regulated in C. elegans during S. Typhi infection. The previous study revealed that F08G5.6 has been involved in defense mechanism and its expression can be altered by the pesticides. ATF-7 plays a key role in the innate immune response by regulating the expression of immune genes downstream of PMK-1/p38 MAPK. During S. Typhi infection, F08G5.6 and atf-7 was up-regulated with gradual reduction except 96 h of post-infection, respectively (online resource Fig. S5). These results suggested that S. Typhi infection alters the defense mechanism of host by altering the regulatory genes of aging, defense and apoptotic process.
C-type Lectins (clec-60, clec-87) are the Essential Players in the Defense System of C. elegans
Physiological observation of silenced C. elegans indicated that RNAi of clec-60 affects the normal development of worm tail. Tail swelling was observed in notable number of clec-60 silenced worms (online resource Fig. S6) whereas, there is no physiological alternations observed in clec-87 silenced worms. Both clec-60 and clec-87 silenced worms exhibited normal life span and silencing was confirmed through semi-qPCR analysis (online resource Fig. S7).
Survival assays were performed using silenced C. elegans to analyze the role of clec-60 and clec-87.Salmonella enterica serovar Typhi infected clec-60 and clec-87 RNAi worms died with complete mortality at 41 ± 1.4 and 50 ± 1.4 h, respectively (online resource Figs. S8, S9). Compared to the mortality of wild type worms during S. Typhi infection (Fig. 1), the silenced worms showed a huge difference by causing infection followed by earlier death, which revealed that clec-60 and clec-87 are necessary players of C. elegans immunity during S. Typhi infection. Although, the qPCR data indicated that clec-87 is up-regulated than clec-60, the silencing study results indicated that clec-60 is more important than clec-87. It appears that though the level of regulation of clec-60 is comparatively lower than that of clec-87, the siRNA data suggested that among the pattern recognition receptors analyzed, clec-60 appeared to have more regulatory role during the immune response against S. Typhi infection. These results revealed that the pattern recognition receptors of a host system probably have major impact in C. elegans immunity during S. Typhi infection.
In conclusion, the present study confirmed that S. Typhi causes infection and leads C. elegans mortality by an active process that required live infection, which correlated with the bacterial accumulation and persistent infection in the intestine. Merely 5-h exposure of S. Typhi is enough to cause infection in C. elegans and the S. Typhi infection affects the neuron system of the host. Surprisingly, Ty21a pre-exposed host exhibited enhanced immunity merely specific against subsequent S. Typhi infection. This findings have supported the theories [27] of special adaptive immune response in C. elegans. The immune system of the host was altered in response to S. Typhi infection. The C-type lectins, clec-60 and clec-87 are the major immune players essential for the dynamic defense system of C. elegans against S. Typhi infection. Finally, the present study explores the simple nematode model, C. elegans for the study of human specific pathogen S. Typhi.
Electronic supplementary material
Fig. S1 Olfactory responses of C. elegans against S. Typhi and S. Typhi Ty21a. a NGM plate spotted with E. coli OP50 (Zone: A) and S. Typhi (Zone: B). b NGM plate spotted with E. coli OP50 (Zone: A) and S. Typhi Ty21a (Zone: B). c NGM plate spotted with S. Typhi (Zone: A) and S. Typhi Ty21a (Zone: B). The bacterial spots were indicated as a black circle. In all the combinations, worms were freely crawling over the bacterial spots which indicated that C. elegans does not exhibit the bacterial avoidance behavior against both S. Typhi and S. Typhi Ty21a. Supplementary material 1 (TIFF 592 kb)
Fig. S2 Bacterial accumulations in C. elegans. The proliferation of Salmonella strains inside C. elegans was analyzed as described in “Materials and Methods” section. Bacterial load of S. Typhi and S. Typhi Ty21a over the course of infection. (TIFF 127 kb)
Fig. S3 Diagrammatic representation of osmosensation assay. Salmonella enterica serovar Typhi infected or control worms were placed on NGM plate (a) and High osmolarity region of medium (b) was created around the worms. A Control animals recognized the high osmolarity zone and they have avoided the crossing of the high osmolarity area, whereas S. Typhi infected C. elegans B has lost their osmosensation and crossed the zone. (TIFF 443 kb)
Fig. S4 Regulation of candidate immune genes. Expression patterns of clec-60, clec-87, age-1, bec-1, daf-16 during S. Typhi infection. Fold difference (2−ΔΔCt) of the expression of candidate genes were calculated after normalization with act-2 expression (housekeeping gene) and basal level expression of corresponding gene in control worms. The regulations of these genes indicated that infection also altered the aging related genes. (TIFF 337 kb)
Fig. S5 Regulation of candidate immune genes. Expression patterns of lys-7, ilys-3, scl-2, cpr-2, F08G5.6, atf-7 during S. Typhi infection in naive host. Fold difference (2−ΔΔCt) of the expression of candidate genes were calculated after normalization with act-2 expression (housekeeping gene) and basal level expression of corresponding gene in control worms. (TIFF 387 kb)
Fig. S6 Representative images of C. elegans depicting the phenotypic variation in clec-60 silenced C. elegans. Status of C. elegans after 36 h from the treatment with siRNA specific to clec-60 for 24 h and the tail swelling was highlighted by a circle (a, b) when compared to control (f) and the swelling phenotype of C. elegans after 72 h (c, d). The swelling at the posterior end is due to the vacuole formation (e, clearly indicated with arrows). The reason for the vacuole formation is not known. (TIFF 1969 kb)
Fig. S7 Expression pattern of clec-60 and clec-87 in silenced and control C. elegans. Lane 1 shows the 100 bp ladder. No amplification was observed in clec-60 and clec-87 silenced worms for their respective gene. This results suggested that worms treated with RNAi were silenced for respective genes and also indicated the specificity of siRNA. (TIFF 184 kb)
Fig. S8 Survival rate of clec-60 silenced C. elegans during S. Typhi infection. Rapid mortality was observed in S. Typhi infected clec-60 RNAi worms compared to the mortality of naïve worms exposed to S. Typhi. (TIFF 102 kb)
Fig. S9 Survival rate of clec-87 silenced C. elegans during S. Typhi infection. Rapid mortality was observed in S. Typhi infected clec-87 RNAi worms compared to the mortality of naïve worms exposed to S. Typhi. (TIFF 103 kb)
Abbreviations
- Ach
Acetylcholine
- ASH
Amphied sensilla neuron of head
- ATCC
American Type Culture Collection
- CFU
Colony forming unit
- CGC
Caenorhabditis Genetic Centre
- DEPC
Diethylpyrocarbonate
- IMTECH
Institute of Microbial Technology
- LB
Luria–Bertani
- LPS
Lipopolysaccharide
- MAPK
Mitogen activated protein kinase
- MTCC
Microbial type culture collection
- NGM
Nematode growth medium
- PAMP(s)
Pathogen associated molecular pattern(s)
- PMK
p38 MAP kinase
- SCP/TAPS
Sperm-coating protein/Tpx-1/Ag5/PR-1/Sc7
References
- 1.Germanier R, Fuer E. Isolation and characterization of Gal E mutant Ty21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131(5):553–558. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 2.Deng W, Liou SR, Plunkett G, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle EC, Bishop JL, Grassl GA, Finlay BB. Salmonella: from pathogenesis to therapeutics. J Bacteriol. 2007;189:1489–1495. doi: 10.1128/JB.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotz C, Fensterle J, Goebel W, Meyer SR, Kirchgraber G, Heisig M, Fürer A, Dietrich G, Rapp UR, Gentschev I. Improvement of the live vaccine strain Salmonella enterica serovar typhi Ty21a for antigen delivery via the hemolysin secretion system of Escherichia coli. Int J Med Microbiol. 2009;299:109–119. doi: 10.1016/j.ijmm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Kopecko DJ, Sieber H, Ures JA, Fürer A, Schlup J, Knof U, Collioud A, Xu D, Colburn K, Dietrich G. Genetic stability of vaccine strain Salmonella typhi Ty21a over 25 years. Int J Med Microbiol. 2009;299:233–246. doi: 10.1016/j.ijmm.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Aballay A, Ausubel FM. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc Natl Acad Sci USA. 2001;98:2735–2739. doi: 10.1073/pnas.041613098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. doi: 10.1016/S0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- 8.Libby SJ, Brehm MA, Greiner DL, Shultz LD, McClelland M, Smith KD, Cookson BT, Karlinsey JE, Kinkel TL, Porwollik S, Canals R, Cummings LA, Fang FC. Humanized nonobese diabetic-scid IL2rγnull mice are susceptible to lethal Salmonella Typhi infection. Proc Natl Acad Sci USA. 2008;107:15589–15594. doi: 10.1073/pnas.1005566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghjoo E, Galan JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci USA. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickard D, Wain J, Baker S, Line A, Chohan S, Fookes M, Barron A, Gaora PO, Chabalgoity JA, Thanky N, Scholes C, Thomson N, Quail M, Parkhill J, Dougan G. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J Bacteriol. 2003;185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtiss R, Kelly SM. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun. 1987;55:3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hone DM, Attridge SR, Forrest B, Morona R, Daniels D, LaBrooy JT, Bartholomeusz RC, Shearman DJ, Hackett J. A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect Immun. 1988;56:1326–1333. doi: 10.1128/iai.56.5.1326-1333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balamurugan k. Bacterial infection alters proteome of Caenorhabditis elegans. J Protein Proteomic. 2011;2:49–53. [Google Scholar]
- 14.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivamaruthi BS, Ganguli A, Kumar M, Bhaviya S, Pandian SK, Balamurugan K. Caenorhabditis elegans as a model for studying Cronobacter sakazakii ATCC BAA-894 pathogenesis. J Basic Microbiol. 2011;51:540–549. doi: 10.1002/jobm.201000377. [DOI] [PubMed] [Google Scholar]
- 16.Ausubel FM (2008) Current protocols in molecular biology. Wiley Interscience, New york, pp 26.0.1-26.0.2
- 17.Sivamaruthi BS, Balamurugan K. Impact of food and folate supplementation during Salmonella typhi infection in Caenorhabditis elegans. J Protein Proteomic. 2012;3:9–14. [Google Scholar]
- 18.McMullan R, Anderson A, Nurrish S. Behavioral and immune responses to infection require Gaq-RhoA signaling in C. elegans. PLoS Pathog. 2012;8:e1002530. doi: 10.1371/journal.ppat.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/S0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 21.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewbank JJ (2006) Signaling in the immune response. In: WormBook (ed) The C. elegans research community. WormBook, doi: 10.1895/wormbook.1.83.1 [DOI] [PMC free article] [PubMed]
- 25.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhyaya R. The roles that age-1 and daf-2 genes play in aging. Eukaryon. 2010;6:93–95. [Google Scholar]
- 27.Zhao YL, Wang DY. Formation and regulation of adaptive response in nematode Caenorhabditis elegans. Oxid Med Cell Longev. 2012;2012:564093. doi: 10.1155/2012/564093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Olfactory responses of C. elegans against S. Typhi and S. Typhi Ty21a. a NGM plate spotted with E. coli OP50 (Zone: A) and S. Typhi (Zone: B). b NGM plate spotted with E. coli OP50 (Zone: A) and S. Typhi Ty21a (Zone: B). c NGM plate spotted with S. Typhi (Zone: A) and S. Typhi Ty21a (Zone: B). The bacterial spots were indicated as a black circle. In all the combinations, worms were freely crawling over the bacterial spots which indicated that C. elegans does not exhibit the bacterial avoidance behavior against both S. Typhi and S. Typhi Ty21a. Supplementary material 1 (TIFF 592 kb)
Fig. S2 Bacterial accumulations in C. elegans. The proliferation of Salmonella strains inside C. elegans was analyzed as described in “Materials and Methods” section. Bacterial load of S. Typhi and S. Typhi Ty21a over the course of infection. (TIFF 127 kb)
Fig. S3 Diagrammatic representation of osmosensation assay. Salmonella enterica serovar Typhi infected or control worms were placed on NGM plate (a) and High osmolarity region of medium (b) was created around the worms. A Control animals recognized the high osmolarity zone and they have avoided the crossing of the high osmolarity area, whereas S. Typhi infected C. elegans B has lost their osmosensation and crossed the zone. (TIFF 443 kb)
Fig. S4 Regulation of candidate immune genes. Expression patterns of clec-60, clec-87, age-1, bec-1, daf-16 during S. Typhi infection. Fold difference (2−ΔΔCt) of the expression of candidate genes were calculated after normalization with act-2 expression (housekeeping gene) and basal level expression of corresponding gene in control worms. The regulations of these genes indicated that infection also altered the aging related genes. (TIFF 337 kb)
Fig. S5 Regulation of candidate immune genes. Expression patterns of lys-7, ilys-3, scl-2, cpr-2, F08G5.6, atf-7 during S. Typhi infection in naive host. Fold difference (2−ΔΔCt) of the expression of candidate genes were calculated after normalization with act-2 expression (housekeeping gene) and basal level expression of corresponding gene in control worms. (TIFF 387 kb)
Fig. S6 Representative images of C. elegans depicting the phenotypic variation in clec-60 silenced C. elegans. Status of C. elegans after 36 h from the treatment with siRNA specific to clec-60 for 24 h and the tail swelling was highlighted by a circle (a, b) when compared to control (f) and the swelling phenotype of C. elegans after 72 h (c, d). The swelling at the posterior end is due to the vacuole formation (e, clearly indicated with arrows). The reason for the vacuole formation is not known. (TIFF 1969 kb)
Fig. S7 Expression pattern of clec-60 and clec-87 in silenced and control C. elegans. Lane 1 shows the 100 bp ladder. No amplification was observed in clec-60 and clec-87 silenced worms for their respective gene. This results suggested that worms treated with RNAi were silenced for respective genes and also indicated the specificity of siRNA. (TIFF 184 kb)
Fig. S8 Survival rate of clec-60 silenced C. elegans during S. Typhi infection. Rapid mortality was observed in S. Typhi infected clec-60 RNAi worms compared to the mortality of naïve worms exposed to S. Typhi. (TIFF 102 kb)
Fig. S9 Survival rate of clec-87 silenced C. elegans during S. Typhi infection. Rapid mortality was observed in S. Typhi infected clec-87 RNAi worms compared to the mortality of naïve worms exposed to S. Typhi. (TIFF 103 kb)