Abstract
Multiple myeloma (MM) is preceded by monoclonal gammopathy of undetermined significance (MGUS). Up to date, it is difficult to predict an individual's time to disease progression and the treatment response. To examine whether the nuclear telomeric architecture will unravel some of these questions, we carried out. Three-dimensional (3D) telomere analysis on samples from patients diagnosed with MGUS and MM, as well as from patients who went into relapse. Telomere signal intensity, number of telomere aggregates, nuclear volume, and the overall nuclear telomere distribution (a/c ratio) were analyzed. The telomeric profiles allowed for the differentiation of the disease stages. The telomeric profiles of myeloma cells obtained from blood and bone marrow aspirates were identical. Based on this study, we discuss the use of 3D telomere profiling as a potential future tool for risk stratification and personalized treatment decisions.
Introduction
Monoclonal gammopathies are a heterogeneous group of plasma cell disorders characterized by the proliferation of a single B-cell clone, encompassing malignancies such as multiple myeloma (MM) and monoclonal gammopathy of undetermined significance (MGUS), smoldering MM, solitary plasmacytoma, Waldenström macroglobulinemia, and osteosclerotic myeloma [1,2]. MM accounts for 13% of all hematological cancers [3,4] and for nearly 2% of deaths from cancer [5]. The incidence rate of MM in the United States is 5.9 of 100,000 [6], whereas in other developed countries, it is 4 of 100,000 [7]. For unknown reasons, myeloma is more common among men than women, and the prevalence in African Americans is two-fold higher than in Caucasian Americans [8–10]. MGUS is the precursor state of MM[11,12]; the rate of transformation from MGUS to MM is 1% per annum [13]. The cause of progression to MM has remained elusive, although reports have suggested that secondary genetic changes such as N-ras and K-ras mutations [14], altered cytokine profiles, and increased bone marrow (BM) neovascularization based on median microvascular density [15] may play a role.
Genetic changes frequently occur in MM, which include gross chromosomal rearrangements such as translocations, deletions, duplications, and amplifications of parts of a chromosome or entire chromosomes [16–18]. Furthermore, whole genome sequencing of 38 MM tumor genomes revealed changes in histone methylation patterns and mutations in genes involved in the nuclear factor kappa light chain enhancer of activated B cells (NF-κB) pathway [19]. Recently, the Multiple Myeloma Research Foundation launched the CoMMpassSM study to identify molecular motifs and variations associated with MM [20].
The International Staging System [21] and the Durie Salmon Staging System [18] have been used in staging myeloma; the International Staging System and Durie Salmon Staging System are based on serum markers such as β2-microglobulin, albumin, hemoglobin, calcium, monoclonal proteins, and creatinine, coupled with conventional radiography, computerized tomography, magnetic resonance imaging, and fludeoxyglucose (18F) positron emission tomography/computerized tomography [22]. However, the reliability of these systems is currently under scrutiny [23]. The International Myeloma Working Group has issued guidelines for the clinical risk stratification of patients, which consist of factors such as the type and size of the M protein [24], number of BM plasma cells (BMPCs), constitutional symptoms, anemia, hyperviscosity, lymphadenopathy, and hepatosplenomegaly [25]. On the other hand, the European System relies on the quantification of plasma cells based on their cluster of differentiation (CD)markers [26–32] using flow cytometry [31–34]. However, both guidelines are geared toward detecting existing symptoms and thus do not address the unmet need for a reliable method for risk evaluation of patients.
Telomeres, the highly repetitive (TTAGGG)n nucleotide sequences situated at the ends of chromosomes, have been strongly associated with tumorigenesis [35–40]. Changes in telomere structure result in the formation of aggregates [41], and studies assessing the three-dimensional (3D) nuclear architecture have facilitated monitoring the progression of glioblastoma [42], chronic lymphocytic leukemia [43–46], acute myeloid leukemia (AML) [47,48], and Hodgkin lymphoma [49]. Our current study aimed to apply this 3D telomeric structure-based imaging tool for plasma cells from patients with MGUS, MM, and relapsed MM (MMrel) to potentially serve as a complementary approach in the design of personalized monitoring regimens and in the assessment of individual risk for MM, MGUS, and MMrel.
Materials and Methods
Study Population
The study population consisted of a total of 86 patients, which were subdivided into three groups, namely, MM (N = 44), MGUS (N = 27), and MMrel (N = 15). Patient characteristics are presented as Supplementary materials (Table W1). Informed consent was obtained from all patients after a presentation and discussion with a collaborating research nurse from CancerCare Manitoba and Estonia, respectively. This study was approved by the Research Ethics Review Board on Human Studies of the University of Manitoba (Ethics Reference No. H2010:170) and the Ethics Review Committee on Human Research of the University of Tartu (Protocol No. 194T-11).
Isolation of Lymphocytes and Plasma Cells
Lymphocytes from blood samples and BM aspirates were overlaid in Ficoll-Paque (GE Healthcare Life Sciences, Baie d'Urfe, Quebec) and separated by centrifugation at 200g for 30 minutes. The cells was washed with 10 ml of RPMI (Gibco Life Technologies Inc, Burlington, Ontario) supplemented with 10% FBS (Gibco Life Technologies Inc) and collected by centrifugation, and the cell pellet was resuspended in 100 µl of RPMI with 10% FBS.
CD138 Immunostaining
Cell preparation for immunostaining is described elsewhere [50]. Briefly, 10 µl of the cell suspension was laid onto poly-l-lysine precoated slides and incubated at 37°C for 60 minutes in a humidified chamber. After incubation, the slides were incubated in 0.3x phosphate-buffered saline (PBS) for 40 seconds, fixed with 3.7% formaldehyde/1x PBS for 20 minutes, washed three times with 1x PBS for 5 minutes, and blocked with 4% BSA in 4x sodium chloride/sodium citrate (SSC) for 15 minutes.
For immunostaining, the cells were incubated with mouse anti-CD138 antibodies (fluorescein isothiocyanate-mouse anti-human CD138 antibody; BD Pharmingen, San Diego, CA), diluted in 1/20 in blocking buffer, for 60minutes. After three 5-minute washes with PBS, the cells were subjected to post-fixation using 3.7% formaldehyde/1x PBS for 20 minutes. The nuclei were counterstained with 25 µl of 0.1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes, followed by a brief rinse with distilled water. The slides were dehydrated through an ethanol series (75%, 95%, and 100%), air-dried, andmounted using VECTASHIELD (Vector Laboratories, Burlington, Ontario).
Three-dimensional quantitative fluorescence in situ hybridization was carried out as described elsewhere [38]. The isolated lymphocytes and plasma cells were incubated in 5 ml of a 75-mM KCl solution for 15 minutes and fixed in 3:1 methanol/acetic acid [51]. Approximately 10 µl of the fixed cells was loaded onto each slide, airdried, and fixed in 3.7% formaldehyde/PBS for 20 minutes, followed by three washes with PBS for 5 minutes. The slides were then incubated in 0.5% Triton X-100 in PBS for 10 minutes, then in 20% glycerol for 1 hour. After permeabilization, the cells were subjected to a series of four freeze-thaw cycles [50], followed by three washes with PBS, one 5-minute incubation in 0.1 N HCl, and two washes with PBS. The slides were again dehydrated through the ethanol series.
For hybridization, approximately 5 µl of cyanine 3 (Cy3)-labeled peptide nucleic acid probe (DAKO, Glostrup, Denmark) was applied to the target nuclei. Nuclear DNA was denatured using a HYBrite Denaturation and Hybridization System (Vysis; Abbott Diagnostics, Des Plains, IL) using the following conditions: denaturation at 80°C for 3 minutes, followed by probe annealing at 30°C for 120 minutes. The slides were subjected to a series of washes including two 15-minute washes in 70% formamide (Fluka; Sigma-Aldrich, St Louis, MO), 10 mM Tris (pH 7.4), a 5-minute wash in 0.1x SSC at 55°C, and two washes in 2x SSC/0.05% Tween-20 for 5 minutes. Finally, the nuclei were counterstained with 25 µl of 0.1 µg/ml DAPI, dehydrated through an ethanol series, and mounted in VECTASHIELD (Vector Laboratories). The slides were stored at -20°C until analysis.
Fluorescence Imaging
Fluorescence microscopy was performed using a Zeiss AxioImager Z1 microscope (Carl Zeiss, Toronto, Ontario), equipped with an AxioCam HRm camera and 63x/1.4 oil Plan apochromat objective. The data acquisition was performed using the AXIOVISION 4.8 software (Carl Zeiss). For 3D imaging, each cell was imaged using a series of 80 z-stacks at x, y and z steps of 102 and 200 nm, respectively. Approximately 40 interphase nuclei were imaged for analysis. The exposure time for Cy3 was 343 milliseconds, whereas for DAPI it was 2 milliseconds. Thirty nuclei per patient sample were deconvolved using a constrained iterative algorithm [52]. The deconvolved images were converted into TIFF files and exported for the analysis using the TeloView software [41], which is embedded in MATLAB (MathWorks, Torrance, CA).
Three-dimensional Image Analyses and Statistical Analyses
To determine telomere number, signal intensity, which is proportional to the telomere length [41], and spatial distribution (a/c ratio), as well as to quantify the occurrence of telomeric aggregates (TAs) and measure nuclear volume, TeloView was employed [41]. Statistical parameters considered for characterization lymphocytes were given as follows: 1) percentage of cells with aggregates, 2) average number of telomeres per cell (ANT), 3) average number of telomeric aggregates per cell (ANTA), and 4) average nuclear volume (ANV). For statistical analysis, the software package SAS 9.3 (SAS Institute Inc, Cary, NC) was used, performing the nested factorial analysis of variance. Statistical analysis was employed to compare the percentage of interphase telomeric signals at defined intensity, using bins at an interval of 1000. To compare signal intensities among myeloma stages (i.e., MM, MGUS, and MMrel), nested factorial analysis of variance was used. To compare nuclear and telomeric features of lymphocytes and plasmocytes from blood and BM specimens collected from various myeloma stages, the Breslow-Day test was performed.
Results
The average age of the patient cohort was 67.8 ± 10.9 years; the majority of the patients belonged to the IgG group (Table 1). The percentile of BMPCs and the amount of secreted M-protein increased with disease progression. Additional patient characteristics are presented in Table W1.
Table 1.
Clinical Characteristics of Patients Included in This Study.
| MGUS | MM | MMrel | |
| Sample number | 27 | 43 | 15 |
| Age | 68.1 ± 11.9 | 68.7 ± 10.3 | 64.9 ± 11.1 |
| BMPC (%) | 6.0 ± 7.5 | 45.8 ± 28.7 | 56.9 ± 26.7 |
| M-protein (g/l) | 8.3 ± 5.9 | 42.2 ± 25.2 | 52.8 ± 28.4 |
| IgG (%) | 81.5 | 72.2 | 73.3 |
| IgA (%) | 0 | 23.3 | 13.3 |
| IgM (%) | 11.1 | 0 | 0 |
| n/d | 7.4 | 4.7 | 13.4 |
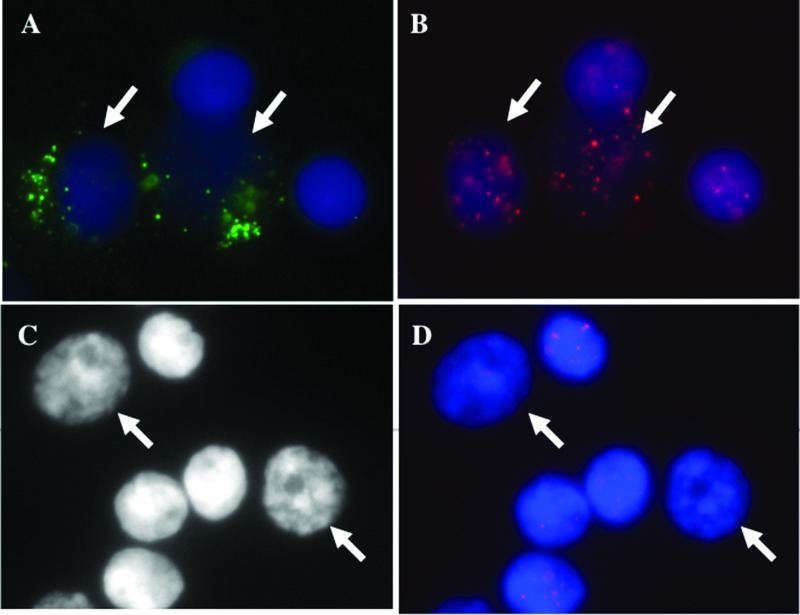
Normal lymphocytes were differentiated from myeloma cells and plasma cells based on the intensity of green fluorescence signals emitted by the fluorescein isothiocyanate-labeled CD138 antibody (Figure 1). The telomeres were visualized as red signals (Figure 1, B and D). The analysis of the telomere length of the CD138+ lymphocytes and plasmocytes and its CD138- counterpart revealed distinct differences. The telomeres of CD138+ cells are shorter, as indicated by lower telomere signal intensities (Figure W1A). No differences in telomere numbers were observed between CD138- and CD138+ lymphocytes and plasma cells. However, the CD138+ cells showed an increase in the number of TAs, a larger nuclear volume, as well as changes in the overall nuclear telomere organization that is expressed by the a/c ratio (Table 2) [41].
Figure 1.
Telomere signals in CD138+ plasma cells (A–D). (A) Myeloma cells fluoresce green, whereas normal cells remained unstained (see arrows). (B) The telomeres, hybridized with Cy3-labeled PNA probes, appear as red signals. The nuclei are counterstained with DAPI (blue). (C) Identification of 3D fixed nuclei in myeloma cells and normal lymphocytes based on size and intensity of the counterstain DAPI (see arrows). (D) Cy3-labeled PNA telomeres in 3D fixed lymphocytes.
Table 2.
Comparison of 3D Nuclear Architecture.
| (A) | ||||||||
| CD138+ | CD138- | Fold Difference | ||||||
| Mean | Standard Mean | Mean | Standard Mean | |||||
| ANT | 23.93 | 1.33 | 23.37 | 0.51 | 1 | |||
| ANTA | 2.6 | 0.38 | 1.93 | 0.24 | 1.3 | |||
| ANV (µm3) | 340.86 | 27.57 | 219.91 | 5.03 | 1.6 | |||
| a/c ratio | 1.97 | 0.23 | 1.49 | 0.06 | 1.3 | |||
| (B) | ||||||||
| Large | Small | Fold Difference | ||||||
| Mean | Standard Mean | Mean | Standard Mean | |||||
| ANT | 25.27 | 1.16 | 23.77 | 0.56 | 1.1 | |||
| ANTA | 2.6 | 0.87 | 1.93 | 0.27 | 1.3 | |||
| ANV (µm3) | 479.5 | 42.48 | 253.39 | 5.52 | 1.9 | |||
| a/c ratio | 4.55 | 0.3 | 3.16 | 0.11 | 1.4 | |||
In our previous studies involving Hodgkin lymphoma, we demonstrated an association between disease progression and the increase in the nuclear volume of tumor cells [53,54]. To determine whether this correlation also occurred in myeloma cells independent of the fixation methodology, we examined the telomeric features of malignant plasma cells and normal lymphocytes based on the size of the nuclei (Figure 1C). The nuclei of malignant plasma cells were larger than those of normal lymphocytes. Additionally, the former nuclei showed a lower intensity for the DAPI counterstain than the normal lymphocytes.
The comparison of normal plasma cells and neoplastic lymphocytes showed differences between CD138+ and CD138- cells with respect to the signal numbers within defined intensity bins (Figure W1A). The malignant plasma cells exhibited a higher number of smaller telomeric signals compared to those signals observed in the controls (Figure W1B). The ANV and a/c ratio were higher in myeloma cells compared to normal lymphocytes (Table 2, A and B). A graphical overlay of intensity readings of CD138+ cells and of 3D fixed large cells as well as the overlay of the intensity profiles of CD138- cells and 3D fixed small cells showed virtually identical results (Figure W2, A and B). We repeated this experiment using myeloma cells from blood and BM specimens (Figure W3, A–D). In either case, we obtained the same results when comparing the myeloma cells as described before.
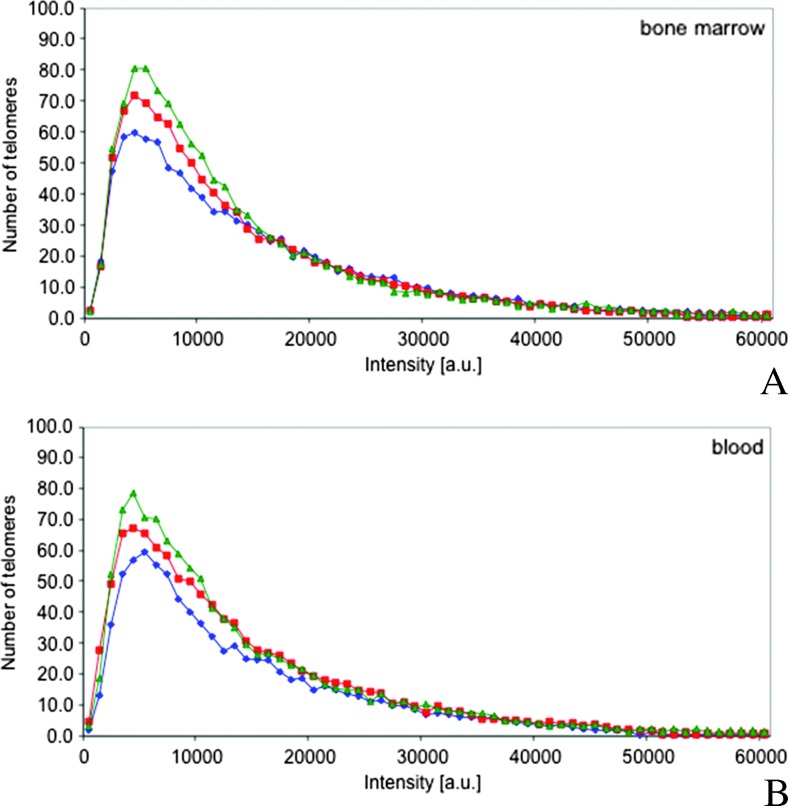
Comparison of the telomeric and nuclear architecture of B-cells in MM, MGUS, and MMrel revealed a distinct pattern involving an increase in the number of low-intensity signals and a decrease in the number of higher intensity range signals as the disease progressed (Figure 2A). For statistical analysis, the intensity range was subdivided into four groups based on signal intensities, <5000 (short telomeres), 5000 to 10,000 (short- to medium-sized telomeres), 10,001 to 19,000 (medium-sized telomeres), and >19,000 (large telomeres), as well as into two groups (≤19,000 and >19,000). Chi-square analysis of the intensity ranges of BM-derived plasma cells showed a significant difference between MGUS and MM (P < .0001), MM and MMrel (P < .0001), and MGUS and MMrel (P < .0064) based on the four-level and two-level intensities. A significant increase in the number of telomeric signals during MGUS to MM progression was also observed (P = .045).
Figure 2.
Telomere intensities change with disease progression. (A and B). MGUS (blue), MM (red), and MMrel (green) show distinct profiles with respect to the telomere intensities and number of telomeres in the low-intensity range. The profiles obtained with BM aspirates (A) are identical to the profiles obtained with blood samples (B).
The comparison of telomeric profiles of neoplastic lymphocytes from BM and blood specimens showed identical patterns (Figure 2, A and B). Differences in signal intensities were observed among the MGUS, MM, and MMrel groups (P < .0001). The Breslow-Day analysis did not detect any statistical differences in the telomere length between BM and blood samples (MGUS: P = .13, MM: P = .08, MMrel: P = .67).
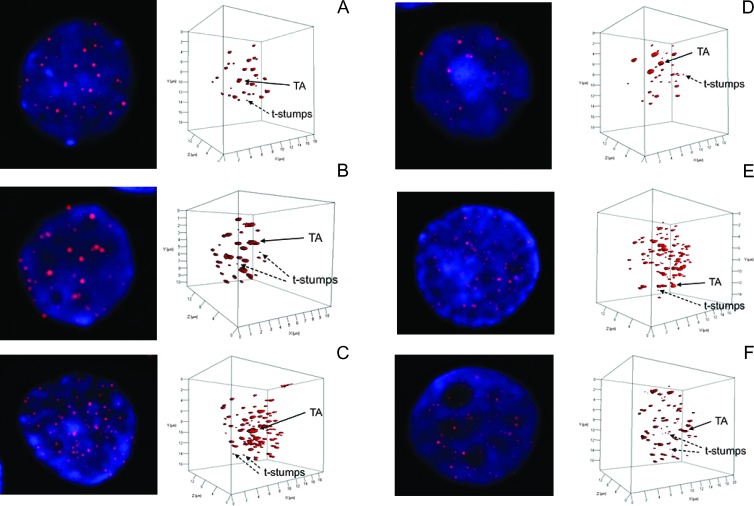
The analysis of the 3D nuclear architecture of the telomeres in MGUS, MM, and MMrel nuclei showed two distinct sizes of telomeres: very short telomeres, described as t-stumps [55] and often observed in normal cells, and very large telomeres or TAs (Figure 3) [39]. During disease progression from MGUS (Figure 3, A and E) to MM (Figure 3, B and F) and MMrel (Figure 3, C and G), the number of TAs as well as the number of t-stumps increased, accompanied by an increase in the nuclear volume. These features were also observed in the neoplastic lymphocytes from blood (Figure 3, A–C) as well as in BM (Figure 3, D–F).
Figure 3.
Evolution of 3D nuclear architecture in blood (A–C) and BM (D–F). The left side of the panel shows the telomere distribution (red) within the counterstained nucleus (blue). During evolution, the 3D nuclear architecture is undergoing changes (right-hand panels). The number of t-stumps and TAs and the nuclear volume are increasing (see scale).
Table 3 shows the changes in the ANT in both BM and blood samples. We also detected an increase in ANTA in MM compared to that in MGUS. Furthermore, we observed a statistically significant increase in the ANV in both blood (P = .0463) and BM (P = .0449) from MGUS with MM samples, although this was not observed between MM and MMrel.
Table 3.
Comparison of Telomeric Features of Myeloma Cells from Blood and BM Specimens.
| Blood | BM | |||||||
| ANT | ANTA | ANV (µm3) | a/c Ratio | ANT | ANTA | ANV (µm3) | a/c Ratio | |
| MGUS | 29.56 | 2.9 | 790.47 | 12.2 | 33.05 | 3.43 | 1163.5 | 13.87 |
| MM | 36.08 | 3.86 | 930.99 | 11.74 | 35.15 | 3.7 | 1178.5 | 11.51 |
| MMrel | 34.71 | 3.87 | 1083 | 10.61 | 39.14 | 4.46 | 1402.5 | 12.33 |
Comparison of MGUS with MM and MGUS with MMrel revealed statistically significant changes in the ANTA in blood samples (P = .039 and .045, respectively). However, we did not observe any statistically significant differences between MM and MMrel. Although we detected a tendency toward an increase in ANV, these differences were not significant among MGUS, MM, and MMrel in blood and BM samples. Finally, we compared the nuclear and telomeric features of cells among all groups. We observed statistically significant differences between MGUS and MM as well as between MGUS and MMrel with respect to ANT (.045 and .042, respectively) and ANTA (.045 and .011, respectively). In terms of the ANV, statistically significant differences were observed between MGUS and MMrel (P = .017). The results of this study thus indicate a correlation between changes in the 3D nuclear architecture and telomeric length of blood- and BM-derived lymphocytes and plasmocytes and disease progression, as supported by the measurements of the ANT, ANTA, and ANV.
Discussion
This study examined the 3D nuclear architecture and telomeric profile of plasma cells from patients with MGUS, MM, and MMrel. Recently, we have also classified patients with myelodysplastic syndrome (MDS) and AML into subgroups using their 3D telomeric architecture [48]. Other tumors that display alterations in their 3D telomeric architecture have been studied, including thyroid cancer [56], endometrial cancer [57], circulating tumor cells [58], chronic myeloid leukemia [43], plasmacytoma [59], cervical cancer [60], Burkitt lymphoma [61,62], head and neck cancer [35], Hodgkin lymphoma [49,53,54,63], and glioblastoma [42].
We identified the myeloma cells based on CD138, a transmembrane heparan sulfate proteoglycan, also called syndecan-1, that is overexpressed as soluble CD138 in myeloma cells [64] as well as on the cell surface [65]. CD138 expression has also been detected on circulating myeloma cells [66]. In our study, we observed that the telomere length of syndecan-1-overexpressing cells was shorter than that of CD138- lymphocytes, as reflected by the detected lower signal intensities, indicating accelerated telomere attrition in myeloma cells. Furthermore, we observed an increase in the ANTA, which is considered a hallmark of cancer cells [39]. We also detected an increase in the ANV, a characteristic feature of tumor cells [67], and changes in the spatial organization of the telomeres, which are reflected in changes of the a/c ratio [35,38,41]. These changes (Table 2) were also observed when comparing small nuclei of normal cells with larger nuclei of myeloma cells, confirming that both methods generated similar results.
This study showed changes in 3D nuclear architecture during disease progression from MGUS to MM, based on our findings of increased telomere attrition, resulting in shorter telomeres in MM, as well as in MMrel, compared to MGUS (Figure 2). Telomere dysfunction and disease progression has been associated with telomere attrition in various hematological malignancies [68] such as MDS/AML [47,48,69–71], chronic lymphocytic leukemia (CLL) [44,45, 72–74], accompanied by telomeric deletions, chromosomal aberrations, and resistance to treatment, thus providing a novel method for differentiating disease stages and potentially serving as a reliable tool for disease monitoring and treatment response [42,44,45,48,53,72].
We also observed the presence of larger TAs, the occurrence of very short t-stumps (Figure 5), and an increase in nuclear volume (Table 3 and Figure 5). TAs initiate breakage-bridge-fusion cycles [38] and are considered a hallmark of cancer cells [39] and have been described for solid tumors [35,57,75] and various hematological malignancies [43,46,48,50,58]. We observed an increase in the ANTA as the disease progressed from MGUS to MM, as well as in myeloma cells that survived treatment, thus suggesting an association with relapsed myeloma. However, statistical analysis of MM and MMrel did not reveal significant differences, indicating that myeloma cells in MMrel are selected survivors that undergo cell expansion. Aggregate formation has been described for squamous cell carcinoma, basal cell carcinoma, keratoacanthoma, and MM [75]. These observations of telomere reorganization during disease progression are similar to that described in earlier studies on Hodgkin lymphoma [65] and MDS/AML [50]. We also detected very short t-stumps [55], which have been associated by previous studies as an indication of the role of telomerase in protecting this distinct class of extremely short telomeres [53,55]. Previous reports have shown that t-stumps accumulate in telomerase-containing cells that lack checkpoint pathways involving p53 and/or pRb, possibly circumventing the DNA damage checkpoint response [76,77] and protecting these nuclear substructures [55]. As previously mentioned, we did not detect any statistical differences between MM and MMrel based on the ANTA. However, we observed an increase in the number of t-stumps in MMrel (Figure 5), suggesting that the myeloma cells in MMrel may have escaped apoptosis. Furthermore, we observed an increase in the number of myeloma cells in blood and BM samples (data not shown). The occurrence of a higher number of t-stumps in relation to tumor aggressiveness [78] also supports this notion. Telomerase inhibition could lead to the loss of protection of t-stumps, resulting in the apoptosis of malignant cells.
The results of this study showed a correlation between an increase in the ANV and disease progression from MGUS to MM and from MM to MMrel. Changes in the nuclear volume have been described for other hematological cancers, such as MDS/AML [48] and Hodgkin lymphoma [53,54], also coinciding with disease progression. However, we did not find any statistical significance between changes in nuclear volume in MM and MMrel, suggesting that myeloma cells do not undergo an evolutionary process as described for cells in Hodgkin lymphoma. As anticipated, the nuclear architecture of myeloma cells derived from BM resembled that of myeloma cells in blood, confirming earlier observations [79–81].
MM is a highly heterogeneous disease at presentation [1,2], often impacting the treatment response of homogeneously treated patients. On the basis of the nature of disease heterogeneity, patients diagnosed with MGUS often remain untreated until additional symptoms develop, such as an increase in the level of M-protein and changes in free light chains. Furthermore, BM aspiration and biopsy are also performed to confirm the initial diagnosis and to assist in the design of an effective treatment regimen. The identification of patients who are at high-risk for a relapse after treatment has long been a dilemma to hematologists and oncologists. Recently, minor clones that could serve as a reservoir for MMrel have been identified using whole-genome analysis [82], thus confirming the multisubclonal nature of myeloma cells and the high heterogeneity of this malignancy. Telomere-based analysis of the 3D nuclear architecture of myeloma cells thus facilitates a patient classification based on their telomere profile.
The analysis of the 3D nuclear architecture and telomeric profile of myeloma cells has established a classification scheme for the disease stages of MGUS, MM, and MMrel. We have also differentiated MGUS from MM based on the ANT and TAs. In the future, a blood test may be used for the prognosis of disease progression and treatment response that may potentially alleviate the need for BM aspirations for analysis. On the basis of the studies involving circulating tumor cells [58] in conjunction with the automated analysis of telomeric features [83], it may also be possible to detect minimal residual disease, which has remained an elusive issue in myeloma [84]. We have recently initiated a longitudinal study that examines the correlation between changes in the 3D nuclear architecture and the risk of progression and relapse.
Supplementary Material
Acknowledgments
This study is dedicated to Morel Rubinger and Adebayo Olujohungbe who passed away this year. Their dedication and passion for the patients with MMdrove this study. In their honor, we will continue our search for the best possible individualized care of patients with MM. We thank Olujohungbe's daughter Olivia and her friends Deanna Mills and Siya Vij, who dedicated their summer 2013 school break to learn about MM and the analysis of patient samples. The authors thank the patients who contributed blood and BM samples to this study. We acknowledge Hans Knecht for a critical review of the paper, Donna Hewitt for obtaining patient consent, and Mary Cheang, senior system analyst, for statistical data analysis. Samples from Estonia were provided by the Hematology and Oncology Clinic, Tartu University Hospital, and we thank the technicians Peeter Toit, Riin Klade, and Mare Jürgenson for their support.
Footnotes
This study was supported by Myeloma Canada and CancerCare Manitoba, the Canada Foundation for Innovation, the Province of Manitoba, and the Canadian Institutes of Health Research.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W3 and are available online at www.transonc.com.
References
- 1.Therneau TM, Kyle RA, Melton LJ, III, Larson DR, Benson JT, Colby CL, Dispenzieri A, Kumar S, Katzmann JA, Cerhan JR, et al. Incidence of monoclonal gammopathy of undetermined significance and estimation of duration before first clinical recognition. Mayo Clin Proc. 2012;87:1071–1079. doi: 10.1016/j.mayocp.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle RA, Rajkumar SV. Monoclonal gammopathies of undetermined significance. Rev Clin Exp Hematol. 2002;6:225–252. doi: 10.1046/j.1468-0734.2002.00076.x. [DOI] [PubMed] [Google Scholar]
- 3.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111:2962–2972. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terpos E, Rahemtulla A. Myeloma. In: Hoffbrand AV, Catovsky D, Tuddenham EGD, editors. Postgraduate Haematology. 5th ed. Oxford, United Kingdom: Blackwell Publishing Ltd; 2007. pp. 681–702. [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 6. http://seer.cancer.gov/statfacts/html/mulmy.html#incidence-mortality.
- 7.Graham-Rowe D. Overview: multiple lines of attack. Nature. 2011;14:S34–S35. doi: 10.1038/480S34a. [DOI] [PubMed] [Google Scholar]
- 8.Landgren O, Weiss BM. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia. 2009;23:1691–1697. doi: 10.1038/leu.2009.134. [DOI] [PubMed] [Google Scholar]
- 9.Alexander DD, Mink PJ, Adami HO, Cole P, Mandel JS, Oken MM, Trichulos D. Multiple myeloma: a review of the epidemiologic literature. Int J Cancer. 2007;120:40–61. doi: 10.1002/ijc.22718. [DOI] [PubMed] [Google Scholar]
- 10.Landgren O, Gridley G, Turesson I, Caporaso NE, Goldin LR, Baris D, Fears TR, Hoover RN, Linet MS. Risk of monoclonal gammopathy of undetermined significance (MGUS) and subsequent multiple myeloma among African American and white veterans in the United States. Blood. 2006;107:904–906. doi: 10.1182/blood-2005-08-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landgren O, Waxman AJ. Multiple myeloma precursor disease. JAMA. 2010;304:2397–2404. doi: 10.1001/jama.2010.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgren O. Multiple myeloma precursor disease: current clinical dilemma and future opportunities. Semin Hematol. 2011;48:1–3. doi: 10.1053/j.seminhematol.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance and smouldering multiple myeloma: emphasis on risk factors for progression. Br J Haematol. 2007;139:730–743. doi: 10.1111/j.1365-2141.2007.06873.x. [DOI] [PubMed] [Google Scholar]
- 14.Steinbrunn T, Stühmer T, Gattenlöhner S, Rosenwald A, Mottok A, Unzicker C, Einsele H, Chatterjee M, Bargou RC. Mutated RAS and constitutively activated Akt delineate distinct oncogenic pathways, which independently contribute to multiple myeloma cell survival. Blood. 2011;117:1998–2004. doi: 10.1182/blood-2010-05-284422. [DOI] [PubMed] [Google Scholar]
- 15.Rajkumar SV, Mesa RA, Tefferi A. A review of angiogenesis and anti-angiogenic therapy in hematologic malignancies. J Hematother Stem Cell Res. 2002;11:33–47. doi: 10.1089/152581602753448522. [DOI] [PubMed] [Google Scholar]
- 16.Bergsagel PL, Chesi M. V. Molecular classification and risk stratification of myeloma. Hematol Oncol. 2013;31:38–41. doi: 10.1002/hon.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadji M, Vallente R, Klewes L, Righolt C, Wark L, Kongruttanachok N, Knecht H, Mai S. Nuclear remodeling as a mechanism for genomic instability in cancer. Adv Cancer Res. 2011;112:77–126. doi: 10.1016/B978-0-12-387688-1.00004-1. [DOI] [PubMed] [Google Scholar]
- 18.Chng WC, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20:571–596. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://www.themmrf.org/research-programs/commpass-study/#sthash. apOWszeO.dpuf. [Google Scholar]
- 21.Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Bladé J, Boccadoro M, Child JA, Avet-Loiseau H, Kyle RA, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 22.Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975;36:842–854. doi: 10.1002/1097-0142(197509)36:3<842::aid-cncr2820360303>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 23.Durie BG. The role of anatomic and functional staging in myeloma: description of Durie/Salmon plus staging system. Eur J Cancer. 2006;42:1539–1543. doi: 10.1016/j.ejca.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Hari PN, Zhang MJ, Roy V, Pérez WS, Bashey A, To LB, Elfenbein G, Freytes CO, Gale RP, Gibson J, et al. Is the International Staging System superior to the Durie-Salmon staging system? A comparison in multiple myeloma patients undergoing autologous transplant. Leukemia. 2009;23:1528–1534. doi: 10.1038/leu.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansell SM, Kyle RA, Reeder CB, Fonseca R, Mikhael JR, Morice WG, Bergsagel PL, Buadi FK, Colgan JP, Dingli D, et al. Diagnosis and management of Waldenström macroglobulinemia: Mayo stratification of macroglobulinemia and risk-adapted therapy (mSMART) guidelines. Mayo Clin Proc. 2010;85:824–833. doi: 10.4065/mcp.2010.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greco A, Tedeschi A, Varettoni M, Nichelatti M, Paris L, Ricci F, Vismara E, Morra E. Factors predicting transformation of asymptomatic IgM monoclonal gammopathy. Clin Lymphoma Myeloma Leuk. 2011;11:77–79. doi: 10.3816/CLML.2011.n.012. [DOI] [PubMed] [Google Scholar]
- 27.Pellat-Deceunynck C, Bataille R, Robillard N, Harousseau JL, Rapp MJ, Juge-Morineau N, Wijdenes J, Amiot M. Expression of CD28 and CD40 in human myeloma cells: a comparative study with normal plasma cells. Blood. 1994;84:2597–2603. [PubMed] [Google Scholar]
- 28.Robillard N, Jego G, Pellat-Deceunynck C, Pineau D, Puthier D, Mellerin MP, Barillé S, Rapp MJ, Harousseau JL, Amiot M, et al. CD28, a marker associated with tumoral expansion in multiple myeloma. Clin Cancer Res. 1998;4:1521–1526. [PubMed] [Google Scholar]
- 29.Dahl IM, Rasmussen T, Kauric G, Husebekk A. Differential expression of CD56 and CD44 in the evolution of extramedullary myeloma. Br J Haematol. 2002;116:273–277. doi: 10.1046/j.1365-2141.2002.03258.x. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Andres M, Almeida J, Martin-Ayuso M, Moro MJ, Martin-Nuñez G, Galende J, Hernandez J, Mateo G, San Miguel JF, Orfao A, et al. Characterization of bone marrow T cells in monoclonal gammopathy of undetermined significance, multiple myeloma, and plasma cell leukemia demonstrates increased infiltration by cytotoxic/Th1 T cells demonstrating a squed TCR-Vβ repertoire. Cancer. 2006;106:1296–1305. doi: 10.1002/cncr.21746. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Andrés M, Almeida J, Martín-Ayuso M, Moro MJ, Martin-Nuñez G, Galende J, Borrego D, Rodríguez MJ, Ortega F, Hernandez J, et al. Clonal plasma cells from monoclonal gammopathy of undetermined significance, multiple myeloma and plasma cell leukemia show different expression profiles of molecules involved in the interaction with the immunological bone marrow microenvironment. Leukemia. 2005;19:449–455. doi: 10.1038/sj.leu.2403647. [DOI] [PubMed] [Google Scholar]
- 32.San Miguel JF, Gutiérrez NC, Mateo G, Orfao A. Conventional diagnostics in multiple myeloma. Eur J Cancer. 2006;42:1510–1519. doi: 10.1016/j.ejca.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 33.Rawstron AC, Orfao A, Beksac M, Bezdickova L, Brooimans RA, Bumbea H, Dalva K, Fuhler G, Gratama J, Hose D, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica. 2008;93:431–438. doi: 10.3324/haematol.11080. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Andrés M, Santiago M, Almeida J, Mateo G, Porwit-MacDonald A, Bjorklund E, Valet G, Kraan J, Gratama JW, D'Hautcourt JL, et al. Immunophenotypic approach to the identification and characterization of clonal plasma cells from patients with monoclonal gammopathies. J Biol Regul Homeost Agents. 2004;18(3–4):392–398. [PubMed] [Google Scholar]
- 35.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle. 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. [DOI] [PubMed] [Google Scholar]
- 37.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 38.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mai S. Initiation of telomere-mediated chromosomal rearrangements in cancer. J Cell Biochem. 2010;109:1095–1102. doi: 10.1002/jcb.22501. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Li S, Stohr BA. The role of telomere biology in cancer. Annu Rev Pathol. 2013;24:49–78. doi: 10.1146/annurev-pathol-020712-164030. [DOI] [PubMed] [Google Scholar]
- 41.Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- 42.Gadji M, Fortin D, Tsanaclis AM, Garini Y, Katzir N, Wienburg Y, Yan J, Klewes L, Klonisch T, Drouin R, et al. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia. 2010;12:183–191. doi: 10.1593/neo.91752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samassekou O, Hébert J, Mai S, Yan J. Nuclear remodeling of telomeres in chronic myeloid leukemia. Genes Chromosomes Cancer. 2013;52:495–502. doi: 10.1002/gcc.22046. [DOI] [PubMed] [Google Scholar]
- 44.Sellmann L, de Beer D, Bartels M, Opalka B, Nückel H, Dührsen U, Dürig J, Seifert M, Siemer D, Küppers R, et al. Telomeres and prognosis in patients with chronic lymphocytic leukaemia. Int J Hematol. 2011;93:74–82. doi: 10.1007/s12185-010-0750-2. [DOI] [PubMed] [Google Scholar]
- 45.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, Fegan C, Pepper C, Baird DM. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 46.Ramsay AJ, Quesada V, Foronda M, Conde L, Martínez-Trillos A, Villamor N, Rodríguez D, Kwarciak A, Garabaya C, Gallardo M, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 47.Zou YS, Ouahchi K, Lu Y, Liu W, Christensen T, Schappert J, Saleki R. Jumping translocations of 3q21 in an acute monocytic leukemia (M5) patient reveal mechanisms of multistage telomere shortening in pathogenesis of AML. Leuk Res. 2012;36:e31–e33. doi: 10.1016/j.leukres.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Gadji M, Adebayo Awe J, Rodrigues P, Kumar R, Houston DS, Klewes L, Dièye TN, Rego EM, Passetto RF, de Oliveira FM, et al. Profiling three-dimensional nuclear telomeric architecture of myelodysplastic syndromes and acute myeloid leukemia defines patient subgroups. Clin Cancer Res. 2012;18:3293–3304. doi: 10.1158/1078-0432.CCR-12-0087. [DOI] [PubMed] [Google Scholar]
- 49.Knecht H, Brüderlein S, Mai S, Möller P, Sawan B. 3D structural and functional characterization of the transition from Hodgkin to Reed-Sternberg cells. Ann Anat. 2010;192:302–308. doi: 10.1016/j.aanat.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Cremer M, Grasser F, Lanctôt C, Müller S, Neusser M, Zinner R, Solovei I, Cremer T. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- 51.Guffei A, Sarkar R, Klewes L, Righolt C, Knecht H, Mai S. Dynamic chromosomal rearrangements in Hodgkin's lymphoma are due to ongoing three-dimensional nuclear remodeling and breakage-bridge-fusion cycles. Haematologica. 2010;95:2038–2046. doi: 10.3324/haematol.2010.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204:99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 53.Knecht H, Kongruttanachok N, Sawan B, Brossard J, Prévost S, Turcotte E, Lichtensztejn Z, Lichtensztejn D, Mai S. Three-dimensional telomere signatures of Hodgkin- and Reed-Sternberg cells at diagnosis identify patients with poor response to conventional chemotherapy. Transl Oncol. 2012;5:269–277. doi: 10.1593/tlo.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knecht H, Brüderlein S, Wegener S, Lichtensztejn D, Lichtensztejn Z, Lemieux B, Möller P, Mai S. 3D nuclear organization of telomeres in the Hodgkin cell lines U-HO1 and U-HO1-PTPN1: PTPN1 expression prevents the formation of very short telomeres including “t-stumps”. BMC Cell Biol. 2010;11:99. doi: 10.1186/1471-2121-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wark L, Danescu A, Natarajan S, Zhu X, Cheng SY, Hombach-Klonisch S, Mai S, Klonisch T. Three-dimensional telomere dynamics in follicular thyroid cancer. Thyroid. 2013 doi: 10.1089/thy.2013.0118. E-pub ahead of print September 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danescu A, Herrero Gonzalez S, Di Cristofano A, Mai S, Hombach-Klonisch S. Three-dimensional nuclear telomere architecture changes during endometrial carcinoma development. Genes Chromosomes Cancer. 2013;52:716–732. doi: 10.1002/gcc.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adebayo Awe J, Xu MC, Wechsler J, Benali-Furet N, Cayre YE, Saranchuk J, Drachenberg D, Mai S. Three-dimensional telomeric analysis of isolated circulating tumor cells (CTCs) defines CTC subpopulations. Transl Oncol. 2013;6:51–65. doi: 10.1593/tlo.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuzyk A, Mai S. Selected telomere length changes and aberrant three-dimensional nuclear telomere organization during fast-onset mouse plasmacytomas. Neoplasia. 2013;14:344–351. doi: 10.1593/neo.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guijon FB, Greulich-Bode K, Paraskevas M, Baker P, Mai S. Premalignant cervical lesions are characterized by dihydrofolate reductase gene amplification and c-Myc overexpression: possible biomarkers. J Low Genit Tract Dis. 2007;11:265–272. doi: 10.1097/LGT.0b013e31803c4df0. [DOI] [PubMed] [Google Scholar]
- 61.Fest T, Guffei A, Williams G, Silva S, Mai S. Uncoupling of genomic instability and tumorigenesis in a mouse model of Burkitt's lymphoma expressing a conditional box II-deleted Myc protein. Oncogene. 2005;24:2944–2953. doi: 10.1038/sj.onc.1208467. [DOI] [PubMed] [Google Scholar]
- 62.Fest T, Mougey V, Dalstein V, Hagerty M, Milette D, Silva S, Mai S. c-MYC overexpression in Ba/F3 cells simultaneously elicits genomic instability and apoptosis. Oncogene. 2002;21:2981–2990. doi: 10.1038/sj.onc.1205274. [DOI] [PubMed] [Google Scholar]
- 63.Knecht H, Sawan B, Lichtensztejn D, Lemieux B, Wellinger RJ, Mai S. The 3D nuclear organization of telomeres marks the transition from Hodgkin to Reed-Sternberg cells. Leukemia. 2009;23:565–573. doi: 10.1038/leu.2008.314. [DOI] [PubMed] [Google Scholar]
- 64.Lovell R, Dunn JA, Begum G, Barth NJ, Plant T, Moss PA, Drayson MT, Pratt G. Soluble syndecan-1 level at diagnosis is an independent prognostic factor in multiple myeloma and the extent of fall from diagnosis to plateau predicts for overall survival. Br J Haematol. 2005;130:542–548. doi: 10.1111/j.1365-2141.2005.05647.x. [DOI] [PubMed] [Google Scholar]
- 65.Wijdenes J, Vooijs WC, Clément C, Post J, Morard F, Vita N, Laurent P, Sun RX, Klein B, Dore JM. A plasmocyte selective monoclonal antibody (B-B4) recognizes syndecan-1. Br J Haematol. 1996;94:318–323. doi: 10.1046/j.1365-2141.1996.d01-1811.x. [DOI] [PubMed] [Google Scholar]
- 66.Merryweather A. CD138 expression on circulating myeloma cells: disease activity correlation. The Biomedical Scientist; IBMS Biomedical Sciences Congress; 28th-30th September; Birmingham, UK. 2009. pp. 952–953. [Google Scholar]
- 67.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 68.Jones CH, Pepper C, Baird DM. Telomere dysfunction and its role in haematological cancer. Br J Haematol. 2012;156:573–587. doi: 10.1111/j.1365-2141.2011.09022.x. [DOI] [PubMed] [Google Scholar]
- 69.Gancarcíková M, Zemanová Z, Brezinová J, Berková A, Vcelíková S, Smigová J, Michalová K. The role of telomeres and telomerase complex in haematological neoplasia: the length of telomeres as a marker of carcinogenesis and prognosis of disease. Prague Med Rep. 2010;111:91–105. [PubMed] [Google Scholar]
- 70.Lange K, Holm L, Vang Nielsen K, Hahn A, Hofmann W, Kreipe H, Schlegelberger B, Göhring G. Telomere shortening and chromosomal instability in myelodysplastic syndromes. Genes Chromosomes Cancer. 2010;49:260–269. doi: 10.1002/gcc.20737. [DOI] [PubMed] [Google Scholar]
- 71.Rollison DE, Epling-Burnette PK, Park JY, Lee JH, Park H, Jonathan K, Cole AL, Painter JS, Guerrier M, Meléndez-Santiago J, et al. Telomere length in myelodysplastic syndromes. Leuk Lymphoma. 2011;52:1528–1536. doi: 10.3109/10428194.2011.568648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brugat T, Nguyen-Khac F, Grelier A, Merle-Béral H, Delic J. Telomere dysfunction-induced foci arise with the onset of telomeric deletions and complex chromosomal aberrations in resistant chronic lymphocytic leukemia cells. Blood. 2010;116:239–249. doi: 10.1182/blood-2009-12-257618. [DOI] [PubMed] [Google Scholar]
- 73.Mansouri L, Grabowski P, Degerman S, Svenson U, Gunnarsson R, Cahill N, Smedby KE, Geisler C, Juliusson G, Roos G, et al. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88:647–651. doi: 10.1002/ajh.23466. [DOI] [PubMed] [Google Scholar]
- 74.Brugat T, Nguyen-Khac F, Merle-Béral H, Delic J. Concomitant telomere shortening, acquisition of multiple chromosomal aberrations and in vitro resistance to apoptosis in a single case of progressive CLL. Leuk Res. 2011;35:e37–e40. doi: 10.1016/j.leukres.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 75.Leufke C, Leykauf J, Krunic D, Jauch A, Holtgreve-Grez H, Böhm-Steuer B, Bröcker EB, Mauch C, Utikal J, Hartschuh W, et al. The telomere profile distinguishes two classes of genetically distinct cutaneous squamous cell carcinomas. Oncogene. 2013 doi: 10.1038/onc.2013.323. E-pub ahead of print August 19. [DOI] [PubMed] [Google Scholar]
- 76.d'Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 77.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 78.Pai RB, Pai SB, Yang L, Joshi HC. Abundance of a distinct cluster of telomere t-stumps in advanced breast cancer cell line. Oncol Lett. 2010;1:339–343. doi: 10.3892/ol_00000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thiago LS, Perez-Andres M, Balanzategui A, Sarasquete ME, Paiva B, Jara-Acevedo M, Barcena P, Sanchez ML, Almeida J, Gonzalez M, et al. Circulating clonotypic B-cells in multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2013 doi: 10.3324/haematol.2013.092817. E-pub ahead of print July 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szczepek AJ, Seeberger K, Wizniak J, Mant MJ, Belch AR, Pilarski LM. A high frequency of circulating B cells share clonotypic Ig heavy-chain VDJ rearrangements with autologous bone marrow plasma cells in multiple myeloma, as measured by single-cell and in situ reverse transcriptase-polymerase chain reaction. Blood. 1998;92:2844–2855. [PubMed] [Google Scholar]
- 81.Bergsagel PL, Masellis Smith A, Belch AR, Pilarski LM. The blood B-cells and bone marrow plasma cells in patients with multiple myeloma share identical IgH rearrangements. Blood. 1995;85:436–447. doi: 10.1007/978-3-642-79275-5_3. [DOI] [PubMed] [Google Scholar]
- 82.Magrangeas F, Avet-Loiseau H, Gouraud W, Lodé L, Decaux O, Godmer P, Garderet L, Voillat L, Facon T, Stoppa AM, et al. Minor clone provides a reservoir for relapse in multiple myeloma. Leukemia. 2013;27:473–481. doi: 10.1038/leu.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klewes L, Höbsch C, Katzir N, Rourke D, Garini Y, Mai S. Novel automated three-dimensional genome scanning based on the nuclear architecture of telomeres. Cytometry A. 2011;79:159–166. doi: 10.1002/cyto.a.21012. [DOI] [PubMed] [Google Scholar]
- 84.Hart AJ, Jagasia MH, Kim AS, Mosse CA, Savani BN, Kassim A. Minimal residual disease in myeloma: are we there yet? Biol Blood Marrow Transplant. 2012;18:1790–1799. doi: 10.1016/j.bbmt.2012.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.