Significance
Microbial pathogens use complex secretion systems to deliver virulence factors into host cells, where they disrupt host cell function. Understanding these systems is essential to the development of new treatments for infectious disease. A challenge in such studies arises from the abundance of host cell proteins, which interfere with detection of microbial effectors. Here we describe a metabolic labeling strategy that allows selective enrichment of microbial proteins from the host cell cytoplasm. The method enables efficient identification of microbial proteins that have been delivered to the host, identifies distinct secretion profiles for intracellular and extracellular bacteria, and allows for determination of the order of injection of microbial proteins into host cells.
Keywords: proteomics, click chemistry, BONCAT, Yop
Abstract
Pathogenic microbes have evolved complex secretion systems to deliver virulence factors into host cells. Identification of these factors is critical for understanding the infection process. We report a powerful and versatile approach to the selective labeling and identification of secreted pathogen proteins. Selective labeling of microbial proteins is accomplished via translational incorporation of azidonorleucine (Anl), a methionine surrogate that requires a mutant form of the methionyl-tRNA synthetase for activation. Secreted pathogen proteins containing Anl can be tagged by azide-alkyne cycloaddition and enriched by affinity purification. Application of the method to analysis of the type III secretion system of the human pathogen Yersinia enterocolitica enabled efficient identification of secreted proteins, identification of distinct secretion profiles for intracellular and extracellular bacteria, and determination of the order of substrate injection into host cells. This approach should be widely useful for the identification of virulence factors in microbial pathogens and the development of potential new targets for antimicrobial therapy.
Many bacterial pathogens use elaborate secretion systems to transfer effector proteins into target cells (1). The injected proteins disrupt host cell functions, including cytoskeletal assembly and cytokine production, to promote infection (2). An important step in understanding virulence mechanisms is the identification of injected and secreted bacterial proteins. Traditional methods have included genetic screens and candidate protein approaches, which can be laborious and noncomprehensive. Proteome-wide labeling strategies offer the potential to rapidly identify secreted pathogen proteins without bias and with limited previous knowledge of host–pathogen interactions (3).
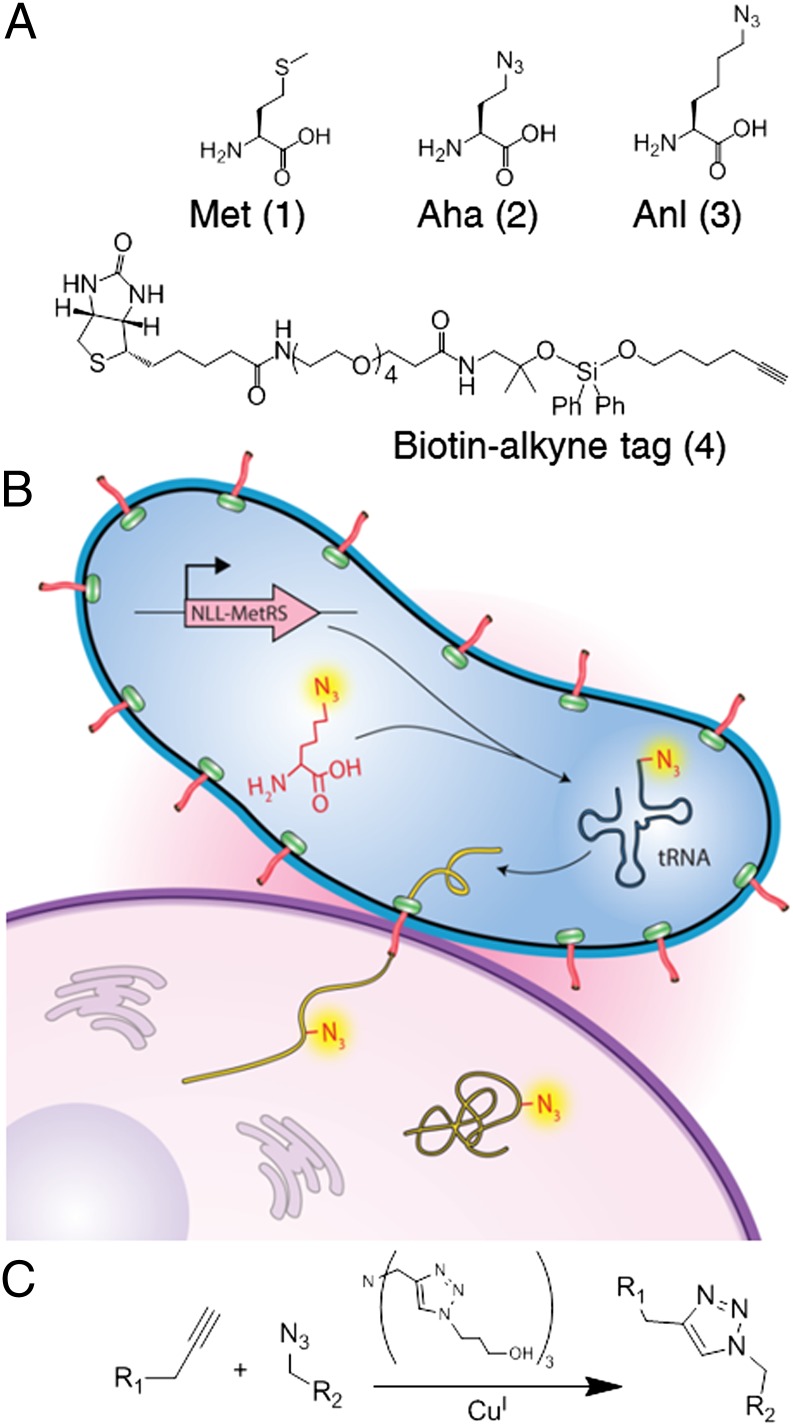
We have developed a method, termed bio-orthogonal noncanonical amino acid tagging (BONCAT), for incorporating azide functional groups into proteins as a general strategy for the enrichment of newly synthesized cellular proteins, making it possible to elucidate the spatial and temporal character of proteomic changes (4, 5). Our initial studies used the noncanonical amino acid azidohomoalanine (Aha) (structure 2, Fig. 1A), a methionine (Met) surrogate, to label newly synthesized proteins (4, 5). The azide side chain of Aha allows newly synthesized proteins to be tagged with alkyne-functionalized affinity reagents and separated from preexisting proteins by affinity chromatography. After separation, proteins are identified by tandem MS. Enrichment of newly synthesized proteins reduces the complexity of the sample and facilitates identification of the proteins of interest.
Fig. 1.
Incorporation of Anl into injected pathogen proteins and enrichment of labeled proteins. (A) Structures of amino acids Met (1) and Met analogs Aha (2) and Anl (3). The alkyne-functionalized biotin affinity probe (4) contains an acid-cleavable silane linker. The probe can be appended to the azide side chain of Anl. Cleavage with formic acid transfers a small mass tag to each modified Anl residue. (B) NLL-MetRS charges the tRNAMet with Anl; its expression in the pathogen allows cell-selective labeling of pathogen proteins during infection. Injected pathogen proteins are enriched after labeling with 4 and identified by tandem MS. (C) Copper-catalyzed azide-alkyne cycloaddition yields a triazole linkage.
We recently showed that introduction of a mutant form of the methionyl-tRNA synthetase (designated NLL-MetRS) into Escherichia coli enables incorporation of the noncanonical amino acid azidonorleucine (Anl) (structure 3, Fig. 1A) into the bacterial proteome (6). Because Anl is not activated to any significant extent by any of the WT synthetases (6), labeling is restricted to cells in which NLL-MetRS is expressed (7). This approach has prompted recent efforts to study proteomic changes in pathogens during infection (8). At the same time, there has been considerable interest in protein labeling strategies to study secreted pathogen proteins, most notably through the use of stable-isotope labeling of amino acids in cell culture (SILAC) (9, 10). Isotopic labeling does not allow enrichment of secreted pathogen proteins, however, and enrichment is important for identification of virulence factors that otherwise would go undetected among abundant host proteins. Here, using a shotgun, bottom-up proteomics approach, we show that noncanonical amino acid labeling enables enrichment of secreted virulence factors and identification of injected proteins from host cell lysates.
Pathogenic bacteria secrete proteins through various mechanisms. Secretion via type III, type IV, and type VI systems occurs by direct injection of proteins into host cells, whereas type II and type V secretion systems use a two-step passage through the inner and outer membranes of the pathogen. Secreted outer membrane vesicles also mediate export of a complex array of proteins (11). We focus here on the well-characterized type III secretion system (T3SS) of Yersinia enterocolitica, a Gram-negative bacterium. In Yersinia, the majority of secreted proteins, designated Yersinia outer proteins (Yops), are encoded on the 70-kb virulence plasmid pYV (2, 12). In addition to encoding Yops, the plasmid encodes machinery consisting of needle-shaped structures that assemble on the bacterial surface and inject proteins into the cytoplasm of host cells. The T3SS is activated by a temperature shift from 26 °C to the host temperature (37 °C); injection is initiated on surface contact with target cells (13, 14). The pYV virulence plasmid also encodes a low calcium response (LCR) that enables secretion of T3SS substrates into the medium in the absence of host cells (15). As a control for type III secretion, we used a YscU mutant strain (designated T3SS-Mut), which is unable to secrete Yops (16). YscU is an inner membrane protein required for T3SS assembly and recruitment of substrates (17).
In this study, NLL-MetRS was introduced to both WT and mutant Yersinia strains to enable selective Anl labeling of bacterial proteins (Fig. 1B). Because host cells do not express NLL-MetRS, host cell proteins are not labeled with Anl. After Anl was added to the infection medium, Anl-labeled proteins were tagged by copper-catalyzed cycloaddition (18) (Fig. 1C) with alkyne-functionalized dyes and detected by in-gel fluorescence or by confocal fluorescence imaging of infected host cells. Similarly, enrichment of Anl-labeled proteins was performed after attachment of a cleavable affinity tag (structure 4, Fig. 1A) that permits binding of labeled proteins to immobilized streptavidin resin and removal of unlabeled proteins. The small mass modification resulting from tagging of Anl residues is readily detected by MS, thereby facilitating identification of enriched proteins (SI Appendix, Figs. S1 and S2).
In a HeLa cell infection model, we identified the Yersinia proteins that were secreted into the medium and injected into HeLa cells. In addition to identifying known Yops, we identified secreted proteins that may play important roles in Yersinia infection. An extension of this approach allowed us to selectively label proteins secreted by Yersinia that had invaded HeLa cells and to reveal secretion of distinct subsets of virulence factors. Pulse-labeling with Anl was used to investigate the order of injection of type III substrates into HeLa cells, providing a simple method to determine the hierarchy of injection of virulence factors. The approach described here is not limited to the study of T3SS substrates, but can be used to examine the many different secretion systems of microbial pathogens.
Results
Labeling of the Yersinia Proteome and T3SS Substrates.
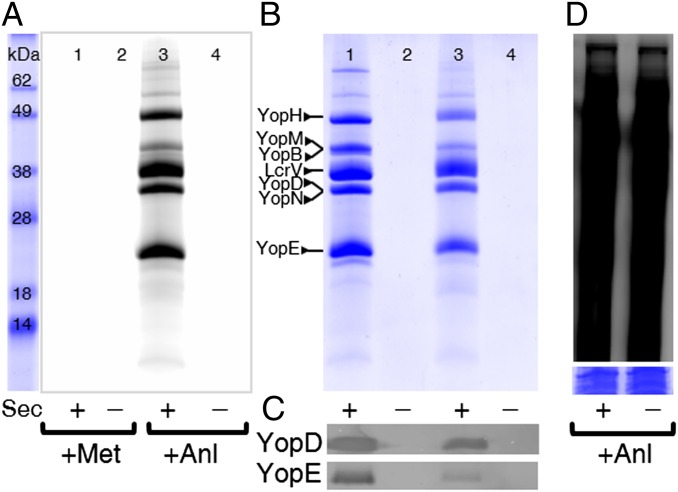
E. coli NLL-MetRS was constitutively expressed in Y. enterocolitica under control of its natural promoter to enable incorporation of Anl into bacterial proteins (SI Appendix, Fig. S3). Proteins secreted under LCR conditions were tagged with an alkyne-functionalized tetramethylrhodamine (TAMRA) dye (SI Appendix, Fig. S4) and detected by in-gel fluorescence imaging (Fig. 2A). Labeling was observed only in samples treated with Anl (Fig. 2A, lane 3); nonspecific labeling in the absence of Anl was negligible (Fig. 2A, lane 1). Lack of TAMRA labeling in the absence of Anl was not related to the absence of secreted proteins, because these proteins were detected by colloidal blue staining (Fig. 2B, lane 1). These results confirm the chemoselectivity of the copper-catalyzed click reaction. As expected, the T3SS-Mut strain did not secrete any labeled proteins (Fig. 2B, lanes 2 and 4). The similarity of the protein secretion profiles in the Met- and Anl-treated samples (Fig. 2B, lanes 1 and 3), along with the lack of secretion by the mutant strain (lanes 2 and 4), indicate that Anl incorporation does not interfere with type III secretion. Western blot analysis with antibodies specific for YopD and YopE confirmed Yop secretion by the T3SS-Wt strain (Fig. 2C). Analysis of Yersinia lysates showed proteome-wide incorporation of Anl into Yersinia proteins (Fig. 2D).
Fig. 2.
Labeling of secreted T3SS substrates. (A) Secretion competent (+) and secretion mutant (−) Y. enterocolitica strains harboring the NLL-MetRS were induced to secrete T3SS substrates under LCR conditions. Secreted proteins were labeled with alkyne-TAMRA and detected by in-gel fluorescence. (B) Detection of all secreted proteins in A by colloidal blue staining. (C) Western blot detection of YopD and YopE in samples from A. (D) In-gel fluorescence detection of alkyne-TAMRA labeling of Y. enterocolitica lysates from conditions corresponding to lanes 3 and 4 in A shows proteome-wide incorporation of Anl. (Inset) Colloidal blue staining of the same samples.
Detection of Injected Proteins in Host Cells by Fluorescence Imaging.
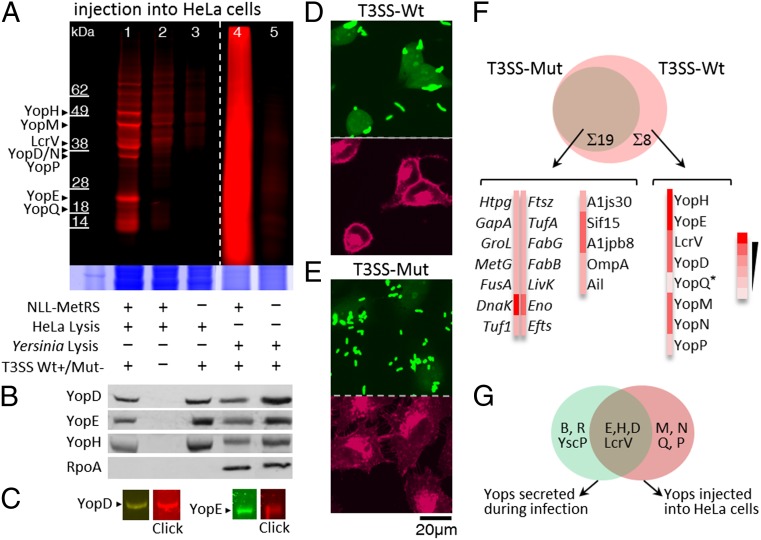
We examined the injection of Yops into HeLa cells, a widely used in vitro model for Yersinia pathogenesis (17). Yop injection results in a characteristic rounded HeLa cell morphology that can be used to track injection (SI Appendix, Fig. S5) (19, 20). Infections were performed with 1 mM Anl; incubation of Yersinia at 37 °C before infection increased the efficiency of injection (SI Appendix, Fig. S6). Digitonin was used to lyse HeLa cells selectively without causing significant disruption of Yersinia membranes (SI Appendix, Fig. S7) (19, 21). Injected T3SS substrates were labeled with alkyne-TAMRA and detected by in-gel fluorescence imaging. We observed distinct bands of labeled proteins corresponding to molecular weights of known YopD, YopE, YopH, YopM, YopN, YopP, YopQ, and low calcium response virulence (LcrV) protein (Fig. 3A, lane 1). These bands were not observed in infections with the T3SS-Mut strain [although a low level of background labeling was observed (Fig. 3A, lane 2)], and proteins injected by Yersinia lacking NLL-MetRS were not labeled (Fig. 3A, lane 3). Cell-specific proteome-wide incorporation of Anl in Yersinia was confirmed in these cocultures (Fig. 3A, lanes 4 and 5). Injected proteins were readily detected by this chemical labeling approach despite the much greater abundance of host cell proteins. Western blot analysis of the same samples with antibodies for YopD, YopE, and YopH showed the presence of these Yops in infected HeLa cells; antibody staining for Yersinia bacterial RNA polymerase A (RpoA) confirmed the absence of significant Yersinia cell lysis (Fig. 3B). Transference of HeLa cell lysates treated with alkyne-TAMRA to nitrocellulose membranes and probing of the membranes with antibodies for YopD and YopE revealed that the protein bands detected by these antibodies were also labeled with the TAMRA dye (Fig. 3C).
Fig. 3.
Detection and identification of injected Y. enterocolitica virulence factors in HeLa cells. (A) HeLa cells were infected with Y. enterocolitica in media containing 1 mM Anl. Proteins were labeled with alkyne-TAMRA and detected by in-gel fluorescence. (Inset) Colloidal blue staining of the same gels. (B) Western blots of HeLa cell lysates with antibodies specific to YopD, YopE, YopH, and RpoA. The legend at the bottom of A applies to B as well. (C) Lysates of infected HeLa cells were treated with alkyne-TAMRA (click) and transferred to nitrocellulose membranes after SDS/PAGE. The same membranes were probed with antibodies specific to YopD and YopE. (D) Detection of injected Yersinia proteins in HeLa cells by fluorescence confocal microscopy. Anl-tagged proteins were labeled with alkyne-Alexa Fluor 488 (green). An Alexa Fluor 633-WGA conjugate was used to label membranes of HeLa cells (red). (E) The same analysis was performed for infections with secretion mutant Y. enterocolitica. (F) Shotgun proteomic identification of virulence factors injected into HeLa cells. Color code indicates the number of independent experiments in which each protein was detected. *YopQ was detected in only one infection. (G) Venn diagram showing Yops injected into HeLa cells or secreted into medium during infection.
To visualize injection, we used fluorescence confocal microscopy to detect Anl-labeled T3SS substrates in the HeLa cell cytoplasm. Anl-labeled proteins were tagged with alkyne-Alexa Fluor 488. We observed increased fluorescence in the cytoplasm of HeLa cells infected with the T3SS-Wt strain compared with infections with T3SS-Mut Yersinia (Fig. 3 D and E), although a low level of background labeling was observed in experiments with the mutant strain (Fig. 3E). As expected, infections with Yersinia that lacked NLL-MetRS resulted in HeLa cell rounding, but no evidence of labeled proteins in the cytoplasm (SI Appendix, Fig. S8). These results indicate that Anl labeling can be used to detect injected virulence factors inside host cells.
Identification of Virulence Factors.
As a first step toward identifying injected virulence factors, we performed a directed MS search for Anl-labeled Yops secreted under LCR conditions. We were able to detect incorporation of Anl at Met positions distributed throughout the secreted proteins (SI Appendix, Figs. S9–S11). We next sought to enrich and identify injected T3SS substrates from HeLa cells using a shotgun MS approach. HeLa cells were infected with Y. enterocolitica in Anl-supplemented medium and selectively lysed with digitonin after the infection. Cell lysates were treated with probe 4 for affinity enrichment of Anl-labeled proteins. Biotinylation was detectable by Western blot analysis (SI Appendix, Fig. S12), and labeled proteins were affinity-enriched on streptavidin resin (SI Appendix, Fig. S13). In-gel tryptic digestion was performed on these samples, and the resulting peptide mixtures were analyzed on a nanoliquid chromatography, linear trap quadrupole, Fourier transform (nano-LC-LTQ-FT) mass spectrometer (SI Appendix, Figs. S14 and S15). Type III-specific virulence factors were determined by comparing lysates of HeLa cells infected by the T3SS-Wt and T3SS-Mut strains (Fig. 3F). This analysis identified previously reported T3SS-specific substrates, including YopD, YopE, YopH, YopM, YopN, YopP, YopQ, and LcrV (17, 22). We did not detect YopT or YopO, perhaps because these proteins are associated with host membranes. YopT was previously observed in the insoluble fraction of HeLa cell lysates (23, 24), and YpkA (the YopO counterpart in Yersinia pseudotuberculosis) has been reported to associate with the plasma membrane after injection (25). It is also possible that YopT and YopO are made or secreted at levels below our limit of detection.
In addition to identifying proteins injected into HeLa cells, we investigated proteins that were secreted into the medium during infection (SI Appendix, Fig. S16). This was done by precipitating the proteins from the infection medium and enriching Anl-labeled proteins. Comparison of T3SS substrates that were injected into HeLa cells with those secreted into medium during infection (Fig. 3G) showed that YopB, YscP, and YscH/YopR were found only in the medium and not inside infected HeLa cells. This finding supports a previously proposed mechanism of action of these Yops, in which secretion of YscP is followed by secretion of YopR into the extracellular medium, resulting in injection of YopN and other effector Yops into host cells (19).
Nineteen Yersinia proteins were found both in lysates prepared from HeLa cells infected with the secretion-competent strain and in lysates prepared from cells infected with the secretion mutant strain (Fig. 3F). These proteins, which are not T3SS substrates, fall into two classes: highly abundant bacterial proteins and proteins associated with the bacterial cell surface. The highly abundant proteins, defined as those ranked among the top 10% of bacterial proteins in terms of abundance according to the PaxDb database (26), are listed in italic type in Fig. 3F and include Tuf1 (ranked as the first of 3,163 proteins in terms of abundance), TufA (ranked fourth), GapA (ranked fifth), Eno (ranked sixth), GroL (ranked seventh), and DnaK (ranked tenth). We have also listed LivK in this group, as it is in the top 10.8% of bacterial proteins in terms of abundance, as well as MetG (MetRS), because it is overexpressed in both Yersinia strains. It seems likely that these highly abundant proteins are found in the HeLa cell lysate as a result of a low level of adventitious bacterial cell lysis, although we cannot rule out other mechanisms of transfer. Previous proteomic studies of factors secreted by Yersinia found subsets of these proteins, including HtpG, OmpA, GroL, and several elongation factors (27), as well as DnaK and Eno (28).
The second class of proteins found in experiments conducted with the secretion mutant strain includes the membrane-associated proteins A1js30, Sif15, A1jpb8, OmpA, and Ail. Yersinia surface proteins invasin, Ail, and YadA mediate binding to host cells (29), and it has been shown that Ail mediates the attachment and uptake of bacterially secreted outer membrane vesicles (11). Outer membrane protein A (OmpA), also identified in our analysis, is known to be present in such vesicles and released by Gram-negative bacteria (11, 30). OmpA has been detected in monocyte cell lysates after infection with Y. pestis (27), is known to bind scavenger receptors (31), and is considered a potent Yersinia virulence factor (30). Sif15, also known as systemic factor protein-a (Sfpa), is involved in systemic infection of Y. enterocolitica, is induced at 37 °C, and is necessary for colonization of mesenteric lymph nodes in a mouse Peyer’s patch infection model (32). We also found the putative exported protein A1jpb8 and the outer membrane porin A1js30 in lysates prepared from HeLa cells infected with the secretion-mutant strain. Transfer of these proteins to the HeLa cell lysate could occur via various mechanisms, including regulated release of outer membrane vesicles. Further work is needed to establish the mode of transfer of each protein.
Identification of Virulence Factors Secreted by Internalized Bacterial Cells.
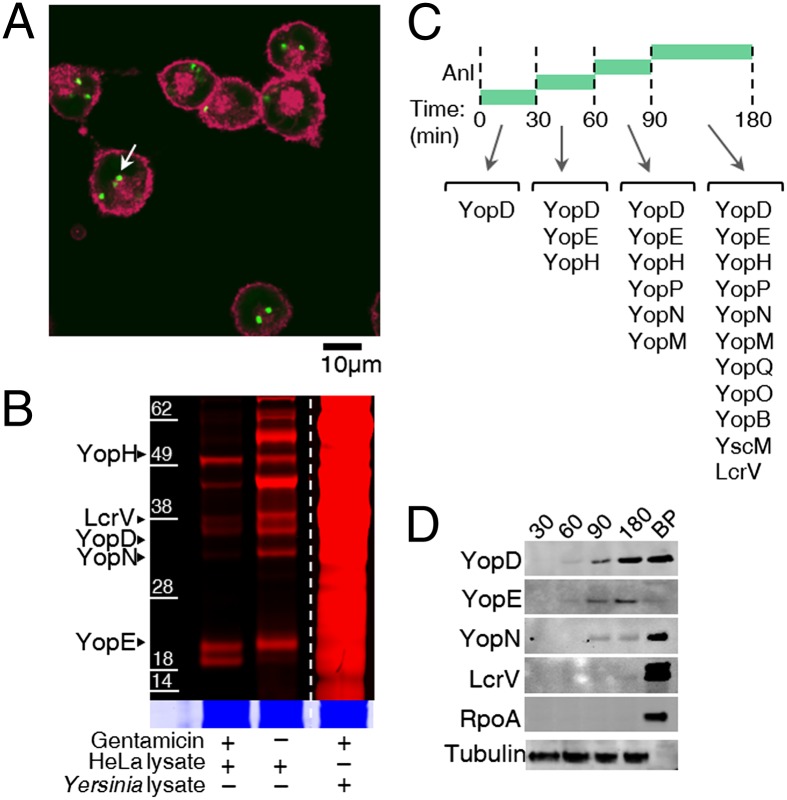
Many pathogens, including Yersinia strains, invade host cells during infection. Internalized pathogens may secrete virulence factors that are distinct from those released by extracellular bacteria (33). We used the gentamicin protection assay with pulsed Anl labeling to compare the type III secretion profiles of internalized and extracellular Yersinia cells. Two parallel infections were initiated with the T3SS-Wt strain. After 1 h, to allow internalization of the pathogen by HeLa cells, gentamicin was added to one of the samples, thereby inhibiting protein synthesis in the extracellular bacteria in this sample. Thereafter, Anl was introduced into both samples for identical labeling times of 3 h. Confocal microscopy verified selective labeling of Yersinia proteins inside infected HeLa cells (Fig. 4A). In the absence of gentamicin, both extracellular and intracellular Yersinia were labeled (SI Appendix, Figs. S17 and S18 and Movies S1 and S2).
Fig. 4.
Labeling of proteins injected into HeLa cells by internalized Y. enterocolitica and identification of the order of Yop injection into HeLa cells. (A) Confocal fluorescence microscopy showed Anl incorporation into the proteome of internalized Y. enterocolitica. HeLa cell membranes were labeled with Alexa Fluor 633-WGA conjugate (red), and Anl residues were labeled with alkyne-Alexa Fluor 488 (green). The arrow indicates labeled Y. enterocolitica inside HeLa cells. (B) Infected HeLa cells were selectively lysed with digitonin and treated with alkyne-TAMRA to detect the proteins injected by internalized Y. enterocolitica. (Inset) Colloidal blue staining of the same gel. In the presence of gentamicin only, internalized Yersinia can inject proteins into HeLa cells. (C) The order of injection of Yops was determined by pulsed-Anl labeling and shotgun MS. Anl was added only during the indicated times for each infection, and HeLa cells were lysed with digitonin at the end of each time interval. (D) Western blot analysis detected Yops in pulsed-Anl labeling experiments. RpoA served as a control for bacterial lysis; antibody for α-tubulin was used as a loading control for HeLa lysates. BP, bacterial pellet.
Comparison of Anl-labeled proteins in HeLa cell lysates by in-gel fluorescence detection revealed a distinct pattern of proteins secreted by internalized Yersinia (Fig. 4B); internalized cells appear to secrete a subset of virulence factors. MS analysis, after enrichment of injected proteins, also indicated that a subset of Yops is secreted by internalized Yersinia (SI Appendix, Fig. S19); YopM, YopP, and YopQ were not detected, whereas YopD, YopN, LcrV, and effectors YopE and YopH were injected by the intracellular subpopulation.
Yops Are Injected in a Temporally Distinct Manner.
The BONCAT method is ideally suited to the study of time-dependent cellular phenomena. To analyze the order in which Yops are injected into host cells, we used an Anl pulse-labeling strategy in which Anl was added to the medium at specified times after infection (Fig. 4C). MS analysis revealed that injection of YopD, which is part of the type III needle complex that inserts into the host cell membrane, is followed by injection of effector YopE and YopH. Identification of YopD as the earliest injected substrate is supported by previous reports indicating that its injection is required to establish translocation of other Yops (34). YopD, YopE, and YopH have previously been detected on the surface of bacteria before contact with host cells, potentially allowing rapid injection of these substrates to stop phagocytosis (35). YopE disrupts the host cell cytoskeleton and can interfere with phagocytosis, and YopE and YopH are thought to control injection of effector Yops (36).
Our finding that YopN and YopM are injected after YopE is supported by the fact that impassable YopE-DHFR fusion substrates can be used to block injection of YopN and YopM (37). YopP was first detected at 60–90 min after initiation of infection, in agreement with previous findings that its cytotoxic effect is not detected until 60 min postinfection and that its inhibition of NF-κB signaling in dendritic cells is detected starting at 90 min after infection (38). Detection of YscM and YopO may indicate that Anl pulse-labeling may be particularly good for identifying low-abundance and transiently injected proteins that would otherwise be undetected. Taken together, these results demonstrate the capacity of the BONCAT method to elucidate the hierarchy of secretion of virulence factors.
Discussion
Identification of effector proteins that are secreted or injected by pathogenic bacteria offers new opportunities for understanding mechanisms of pathogenesis and developing novel therapeutics. Here we show that cell-selective, noncanonical amino acid tagging enables labeling, enrichment, and identification of virulence factors secreted by pathogenic bacteria. Cell-selective proteomic labeling was achieved by outfitting Yersinia cells with E. coli NLL-MetRS, which charges Yersinia tRNAMet with the azide-functionalized noncanonical amino acid Anl (Fig. 1B). Treatment of Anl-labeled proteins with alkyne affinity reagents provided a selective chemical tagging method and enabled enrichment of secreted virulence factors from abundant host proteins. Yersinia proteins isolated from HeLa cell lysates included eight T3SS substrates and 19 proteins that were transferred via type III-independent mechanisms (Fig. 3F). Because some (or perhaps all) of the latter proteins may have been released via adventitious bacterial cell lysis, the overall selectivity of the method might be enhanced by further improvements in the removal of bacterially shed proteins and better host cell lysis techniques.
For live cell applications, cyclooctyne-functionalized reagents can be used to tag the azide side chain of Anl residues in a copper-free manner (39, 40). Our chemical tagging strategy is compatible with routine MS sample preparation methods such as gel electrophoresis liquid chromatography-mass spectrometry (GeLC-MS), filter-aided sample preparation (FASP) (41, 42), and multidimensional protein identification technology (MudPIT) (43), and is easily combined with SILAC, isobaric tag for relative and absolute quantification (iTRAQ), and multiple-reaction monitoring (MRM) quantitative MS methods (44, 45). This approach can be complemented with candidate protein methods, such as expression of tagged substrates, to verify the secretion and identify the location of newly identified substrates inside host cells.
Pulsed Anl labeling was combined with a gentamicin protection assay to identify proteins injected into HeLa cells by internalized Yersinia (Fig. 4 A and B). The results demonstrate that the method can be used in different compartments of the host and should be applicable to studies of functional redundancy, in which multiple effectors perform similar functions (46). Pulsed Anl labeling was used to study the hierarchy of Yop injection, enabling direct elucidation of the order of injection of T3SS substrates (Fig. 4 C and D). As an alternative to pulsed Anl labeling, spatial and temporal resolution may be achieved in future studies by placing NLL-MetRS under the control of specific promoters that are stage-specific or spatiotemporally regulated (47). This approach may allow for Anl labeling at different stages of infection, particularly host cells, or subcellular compartments. Straightforward extensions of the technology will enable investigation of both pathogen proteins and host proteins during infections in animals. The simplicity of the approach makes it suitable for the study of numerous host–microbe interactions.
Materials and Methods
Expression of NLL-MetRS in Y. enterocolitica.
DH10B strains were used for genetic manipulations. E. coli NLL-MetRS with mutations L13N, Y360L, and H301L was isolated from pJTN1 (7) by Nhe1 digestion and inserted into pQE80 (Qiagen). Kanamycin resistance was used for selection, because Y. enterocolitica is resistant to ampicillin (48) and nalidixic acid. The resulting plasmid, which carries NLL-MetRS under control of the endogenous E. coli MetG promoter, is termed pAM1 and has been deposited in Addgene. The plasmid was transformed into electrocompetent Y. enterocolitica, and transformants were grown at 26 °C on agar plates or in LB medium, both containing 50 µg/mL kanamycin.
Secretion of T3SS Substrates Under LCR Conditions.
Y. enterocolitica W2273 was diluted 1:50 from an overnight LB culture into M9 medium at 26 °C with agitation at 250 rpm. At OD600 = 0.5, protein secretion was initiated by a temperature shift to 37 °C. Labeling with Anl was performed in M9 medium lacking calcium and containing 1 mM Anl. After 2.5 h, bacteria were sedimented for 15 min at a relative centrifugal force of 15,000 × g at 4 °C. The medium was passed through a 0.2-µm filter, and proteins were precipitated with chloroform/methanol.
HeLa Cell Infection and Anl Labeling of T3SS Substrates.
HeLa cells (American Type Culture Collection) were routinely cultured in DMEM supplemented with 10% FBS, trypsinized (Gibco), and expanded every 72 h. Before infection, cells were washed twice with PBS and resuspended in Opti-MEM medium (Gibco). Y. enterocolitica was diluted 1:25 from overnight cultures in LB and incubated at 26 °C with agitation at 250 rpm until an OD600 = 0.5 was reached. Yersinia were preincubated at 37 °C for 3 h before the start of infection. The preincubation time was determined by tracking T3SS injection of Anl-labeled proteins (SI Appendix, Fig. S6). Labeling was performed at a multiplicity of infection of 100, with 107 HeLa cells per condition, at 1 mM Anl and 50 µg/mL kanamycin. Infections were carried out for 3.5 h. Infected HeLa cells were lysed with digitonin as described below for analysis of injected Yersinia proteins in HeLa cells (Fig. 3F).
Selective Lysis of HeLa Cells After Infection.
After infection, HeLa cells were washed five times with PBS to remove surface-bound proteins. Cells were incubated with 0.1% (wt/vol) digitonin in PBS for 20 min at room temperature with agitation at 100 rpm. EDTA-free protease inhibitor (Roche) was added to the lysis buffer. Bacterial cells were removed from the lysates by centrifugation at 15,000 × g for 15 min at 4 °C and filtration through a 0.2-µm filter. Western blot analysis with an antibody for RpoA was used to confirm the absence of Yersinia lysis.
Enrichment of Anl-Labeled Proteins.
Probe 4 was appended to Anl-labeled proteins by copper-catalyzed azide-alkyne cycloaddition (SI Appendix, Fig. S1). Proteins were precipitated with acetone, dissolved in 250 µL of 4% SDS in PBS, and diluted to 0.1% SDS by the addition of PBS supplemented with EDTA-free protease inhibitor (Roche). Proteins were incubated with 400 µL of Streptavidin Plus Ultralink resin (Pierce) for 1.5 h at room temperature. Affinity purification was performed according to a previously published protocol (49). Elution fractions were combined with Amicon Ultra 0.5 centrifuge filters (3 kDa molecular weight cutoff; Millipore). Enrichment was also performed with Click-iT alkyne-agarose resin (Invitrogen).
Detection of Proteins in Gels and Western Blots.
Bicinchoninic acid protein quantification (Pierce) was used to equalize the amounts of proteins analyzed under different conditions. After dye labeling via the copper-catalyzed click reaction described above, proteins were washed with methanol to remove unreacted dye and then electrophoresed on a Novex 12% Bis-Tris polyacrylamide gel (Invitrogen). Colloidal blue dye (Invitrogen) was used for nonspecific protein detection. Antibodies were used at the following dilutions: YopD, 1:20,000; YopE, 1:40,000; YopH, 1:4,000; RpoA, 1:40,000, α-tubulin (Abcam) and secondary antibody anti-rabbit IgG-Alexa Fluor 488 conjugate (Cell Signal Technologies), 1:1,000. Fluorescence imaging of Western blots and gels was performed with a Typhoon 9400 molecular imager (GE Healthcare).
Fluorescence Confocal Microscopy.
Adherent HeLa cells were infected as described above and fixed with 3.7% formaldehyde in PBS before labeling with 10 µg/mL Alexa Fluor 633-wheat germ agglutinin (WGA) conjugate (Invitrogen) in PBS for 30 min. Cells were permeabilized with ice-cold methanol for 3 min. Labeling with alkyne-TAMRA (Invitrogen) was performed as described above. Fluorescence confocal images were obtained on a Zeiss LSM 510 microscope.
Comparison of Yops Injected by Extracellular and Internalized Y. enterocolitica.
Y. enterocolitica (T3SS-Wt) was diluted 1:25 from overnight cultures in LB and incubated at 26 °C with agitation until OD600 = 0.5 was reached. Two parallel infections of 5 × 107 HeLa cells each were initiated at a multiplicity of infection of 100 in Opti-MEM without Phenol Red (Invitrogen). After 1 h of infection, 80 µg/mL gentamicin was added to a sample of infected HeLa cells; the other sample did not contain the antibiotic. After 1 h, the medium was changed to Opti-MEM without gentamicin, and 1 mM Anl was added to both samples. The infection that was initially treated with gentamicin was supplemented with 4 µg/mL gentamicin to maintain inhibition of protein synthesis by extracellular bacteria. After 3 h of labeling, HeLa cells in both samples were lysed with 0.1% digitonin for enrichment and MS analysis or fixed with 3.7% formaldehyde for fluorescence confocal microscopy as described above.
Determination of the Order of Yop Injection.
T3SS-Wt Y. enterocolitica was diluted 1:25 from overnight cultures in LB and incubated at 26 °C with agitation up to an OD600 = 0.5. Cells were pelleted at 5,000 × g and washed with PBS. Infection of four parallel samples, corresponding to the four time windows of interest (Fig. 4C), was initiated as described above, with no preincubation at 37 °C. Anl was added to the infection medium at 1 mM for the indicated times (Fig. 4C), and HeLa cells were lysed with digitonin at the end of each interval. HeLa cell lysates were treated with probe 4 as described above for enrichment and identification of injected proteins by MS.
Mass Spectrometry.
Analyses were performed with either a hybrid LTQ-Orbitrap or LTQ-FT Ultra (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source connected to an EASY-nLC II instrument (Thermo Fisher Scientific). Details of instrument setup and data analysis are provided in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Geoff Smith (Proteome Exploration Laboratory, Beckman Institute, California Institute of Technology) for technical assistance. This work was supported by the National Institutes of Health (Grant R01 GM062523), the Institute for Collaborative Biotechnologies (Grant W911NF-09-0001 from the US Army Research Office), the Burroughs Wellcome Fund in the Pathogenesis of Infectious Disease, and the Gordon and Betty Moore Foundation. A.M. is the recipient of a Natural Sciences and Engineering Reasearch Council of Canada scholarship and a Donna and Benjamin M. Rosen postgraduate scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301740111/-/DCSupplemental.
References
- 1.Cornelis GR. The Yersinia Ysc-Yop “type III” weaponry. Nat Rev Mol Cell Biol. 2002;3(10):742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4(11):811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt F, Völker U. Proteome analysis of host–pathogen interactions: Investigation of pathogen responses to the host cell environment. Proteomics. 2011;11(15):3203–3211. doi: 10.1002/pmic.201100158. [DOI] [PubMed] [Google Scholar]
- 4.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103(25):9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieterich DC, et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2(3):532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 6.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, 3rd, Tirrell DA. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci USA. 2009;106(36):15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5(10):715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grammel M, Zhang MM, Hang HC. Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection. Angew Chem Int Ed Engl. 2010;49(34):5970–5974. doi: 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W, et al. A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J Biol Chem. 2010;285(9):6790–6800. doi: 10.1074/jbc.M109.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rechavi O, et al. Trans-SILAC: Sorting out the non–cell-autonomous proteome. Nat Methods. 2010;7(11):923–927. doi: 10.1038/nmeth.1513. [DOI] [PubMed] [Google Scholar]
- 11.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host–pathogen interaction. Genes Dev. 2005;19(22):2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular “toxin” delivery. Biol Chem. 2010;391(7):745–751. doi: 10.1515/BC.2010.079. [DOI] [PubMed] [Google Scholar]
- 13.Lee VT, Mazmanian SK, Schneewind O. A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J Bacteriol. 2001;183(17):4970–4978. doi: 10.1128/JB.183.17.4970-4978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBord KL, Galanopoulos NS, Schneewind O. The ttsA gene is required for low-calcium–induced type III secretion of Yop proteins and virulence of Yersinia enterocolitica W22703. J Bacteriol. 2003;185(12):3499–3507. doi: 10.1128/JB.185.12.3499-3507.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snellings NJ, Popek M, Lindler LE. Complete DNA sequence of Yersinia enterocolitica serotype 0:8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect Immun. 2001;69(7):4627–4638. doi: 10.1128/IAI.69.7.4627-4638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allaoui A, Woestyn S, Sluiters C, Cornelis GR. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176(15):4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008;68(6):1485–1501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48(52):9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaylock B, Berube BJ, Schneewind O. YopR impacts type III needle polymerization in Yersinia species. Mol Microbiol. 2010;75(1):221–229. doi: 10.1111/j.1365-2958.2009.06988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aepfelbacher M, Trasak C, Ruckdeschel K. Effector functions of pathogenic Yersinia species. Thromb Haemost. 2007;98(3):521–529. [PubMed] [Google Scholar]
- 21.Sorg JA, Blaylock B, Schneewind O. Secretion signal recognition by YscN, the Yersinia type III secretion ATPase. Proc Natl Acad Sci USA. 2006;103(44):16490–16495. doi: 10.1073/pnas.0605974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson DM, Ramamurthi KS, Tam C, Schneewind O. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol. 2002;184(5):1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Håkansson S, Galyov EE, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20(3):593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt G. Yersinia enterocolitica outer protein T (YopT) Eur J Cell Biol. 2011;90(11):955–958. doi: 10.1016/j.ejcb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Navarro L, Alto NM, Dixon JE. Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr Opin Microbiol. 2005;8(1):21–27. doi: 10.1016/j.mib.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang M, et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11(8):492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chromy BA, et al. Proteomic characterization of Yersinia pestis virulence. J Bacteriol. 2005;187(23):8172–8180. doi: 10.1128/JB.187.23.8172-8180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponnusamy D, Hartson SD, Clinkenbeard KD. Intracellular Yersinia pestis expresses general stress response and tellurite resistance proteins in mouse macrophages. Vet Microbiol. 2011;150(1-2):146–151. doi: 10.1016/j.vetmic.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Miller VL, Bliska JB, Falkow S. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J Bacteriol. 1990;172(2):1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellman J, Warren HS. Outer membrane protein A (OmpA), peptidoglycan-associated lipoprotein (PAL), and murein lipoprotein (MLP) are released in experimental Gram-negative sepsis. J Endotoxin Res. 2001;7(1):69–72. [PubMed] [Google Scholar]
- 31.Jeannin P, et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 2005;22(5):551–560. doi: 10.1016/j.immuni.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Mildiner-Earley S, Miller VL. Characterization of a novel porin involved in systemic Yersinia enterocolitica infection. Infect Immun. 2006;74(7):4361–4365. doi: 10.1128/IAI.00154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steele-Mortimer O, et al. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4(1):43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- 34.Francis MS, Wolf-Watz H. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: Evidence of a structural domain necessary for translocation. Mol Microbiol. 1998;29(3):799–813. doi: 10.1046/j.1365-2958.1998.00973.x. [DOI] [PubMed] [Google Scholar]
- 35.Akopyan K, et al. Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci USA. 2011;108(4):1639–1644. doi: 10.1073/pnas.1013888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wulff-Strobel CR, Williams AW, Straley SC. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol Microbiol. 2002;43(2):411–423. doi: 10.1046/j.1365-2958.2002.02752.x. [DOI] [PubMed] [Google Scholar]
- 37.Sorg JA, Miller NC, Marketon MM, Schneewind O. Rejection of impassable substrates by Yersinia type III secretion machines. J Bacteriol. 2005;187(20):7090–7102. doi: 10.1128/JB.187.20.7090-7102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adkins I, Schulz S, Borgmann S, Autenrieth IB, Gröbner S. Differential roles of Yersinia outer protein P-mediated inhibition of nuclear factor-kappa B in the induction of cell death in dendritic cells and macrophages. J Med Microbiol. 2008;57(Pt 2):139–144. doi: 10.1099/jmm.0.47437-0. [DOI] [PubMed] [Google Scholar]
- 39.Beatty KE, et al. Live-cell imaging of cellular proteins by a strain-promoted azide-alkyne cycloaddition. ChemBioChem. 2010;11(15):2092–2095. doi: 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beatty KE, Szychowski J, Fisk JD, Tirrell DA. A BODIPY-cyclooctyne for protein imaging in live cells. ChemBioChem. 2011;12(14):2137–2139. doi: 10.1002/cbic.201100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiśniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8(12):5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 42.Manza LL, Stamer SL, Ham AJ, Codreanu SG, Liebler DC. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 43.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 44.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7(12):952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 45.Howden AJ, et al. QuaNCAT: Quantitating proteome dynamics in primary cells. Nat Methods. 2013;10(4):343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngo JT, Babin BM, Champion JA, Schuman EM, Tirrell DA. State-selective metabolic labeling of cellular proteins. ACS Chem Biol. 2012;7(8):1326–1330. doi: 10.1021/cb300238w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornelis G. Distribution of beta-lactamases A and B in some groups of Yersinia enterocolitica and their role in resistance. J Gen Microbiol. 1975;91(2):391–402. doi: 10.1099/00221287-91-2-391. [DOI] [PubMed] [Google Scholar]
- 49.Szychowski J, et al. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J Am Chem Soc. 2010;132(51):18351–18360. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.