Significance
Several different populations of T lymphocytes develop in the thymus from a common precursor. Each population plays a unique and critical role in the mounting and resolution of an immune response. The mechanisms responsible for the emergence of these different populations remain incompletely understood. We demonstrate that strict “Goldilocks” conditions of affinity for self-lipids by the T-cell antigen receptor expressed on T-cell precursors are necessary for imprinting the proper developmental program toward the invariant NK T-cell lineage. Our results establish a direct link between the affinity of the T-cell receptor for self-antigens and the proper development of a unique population of lymphocytes that has been implicated in the modulation of a multitude of immune responses in mice and humans.
Keywords: thymus, deletion
Abstract
The self-reactivity of their T-cell antigen receptor (TCR) is thought to contribute to the development of immune regulatory cells, such as invariant NK T cells (iNKT). In the mouse, iNKT cells express TCRs composed of a unique Vα14-Jα18 rearrangement and recognize lipid antigens presented by CD1d molecules. We created mice expressing a transgenic TCR-β chain that confers high affinity for self-lipid/CD1d complexes when randomly paired with the mouse iNKT Vα14-Jα18 rearrangement to study their development. We show that although iNKT cells undergo agonist selection, their development is also shaped by negative selection in vivo. In addition, iNKT cells that avoid negative selection in these mice express natural sequence variants of the canonical TCR-α and decreased affinity for self/CD1d. However, limiting the affinity of the iNKT TCRs for “self” leads to inefficient Egr2 induction, poor expression of the iNKT lineage-specific zinc-finger transcription factor PLZF, inadequate proliferation of iNKT cell precursors, defects in trafficking, and impaired effector functions. Thus, proper development of fully functional iNKT cells is constrained by a limited range of TCR affinity that plays a key role in triggering the iNKT cell-differentiation pathway. These results provide a direct link between the affinity of the TCR expressed by T-cell precursors for self-antigens and the proper development of a unique population of lymphocytes essential to immune responses.
Invariant NK T cells (iNKT) cells are a conserved population of innate-like T cells that recognize lipid antigens complexed to the antigen-presenting molecule CD1d, and rapidly produce large quantities of various cytokines and chemokines upon stimulation (1–3). In this way, iNKT cells have been shown to modulate various immunological conditions, including infection, cancer, autoimmunity, allergy, and metabolism (3, 4). In mice, the vast majority of iNKT cells, albeit not all (5), express a canonical T-cell receptor-α (TCR-α) chain composed of a Vα14-Jα18 gene rearrangement that is created randomly during thymic development at the CD4+CD8+ (double-positive, DP) stage (6, 7). When paired with certain TCR-β chains using the Vβ8, Vβ7, or Vβ2 gene segments, the resulting TCRs confer specificity for self-lipids presented by CD1d at the surface of cortical thymocytes, and this recognition event is thought to instruct differentiation into the iNKT cell lineage. Furthermore, this iNKT cell self-reactivity appears to be an intrinsic component of their immune functions (8–10).
Following positive selection at the DP stage, a few iNKT cell precursors can readily be characterized as CD4dimCD8dim CD69+ CD24hi cells (stage 0) (11). Cells at stage 1 down-regulate CD24 and appear otherwise phenotypically equivalent to other naïve mature CD4+ single-positive thymocytes. However, these cells are already committed to the iNKT cell lineage as exemplified by their high expression of the promyelocytic zinc finger transcription factor (PLZF) and their active proliferation. Soon after these stage 1 cells acquire a memory-like CD44hi phenotype (stage 2), they are ready to leave the thymus and colonize the peripheral tissues (12, 13). Progression to stage 3 is manifest by cessation of proliferation and acquisition of the NK-like program (CD44hi, NK1.1+, CD122hi). This terminal differentiation can occur either in peripheral tissues or in the thymus itself, where a small population of iNKT cells become long-term residents (14).
During iNKT cell positive selection, agonistic TCR signals are associated with high expression of the Ras- (15) and Ca2+-dependent early growth-response transcription factors early growth-response protein (Egr)-1 and, especially, Egr-2 (16, 17) that, in tandem with costimulation with the signaling lymphocyte activation-molecule family receptor Ly108 (18), leads to high PLZF expression. In turn, PLZF directs the acquisition of the iNKT cell effector program, including the cytokine and migratory properties of iNKT cells (19, 20). As such, poor integration of TCR stimulation and costimulation is expected to affect the required threshold for agonist selection and should have important consequences for the development of iNKT cells. However, the extent to which TCR affinity for agonist self-ligands controls entry into the iNKT lineage remains to be fully investigated.
We have previously constructed a TCR-β chain that, when paired with the canonical Vα14-Jα18 iNKT TCR-α chain increases the affinity of the αβ TCR for the antigen/CD1d complex, regardless of the antigen involved (21, 22). To study the effects of this TCR-β chain, we generated mice expressing the unusual TCR-β as a transgene driven by the CD2 promoter (23), in which the TCR-α locus remained unaltered so that T-cell development could proceed normally. iNKT cell precursors that expressed the high-affinity TCR for “self” in these mice did not fully mature, thus providing direct in vivo evidence that iNKT cells, like their “conventional” T-cell counterparts, are subject to clonal deletion during development. Unexpectedly, we also found that a small fraction of cells with iNKT cell properties avoided negative selection in these mice. These cells expressed variants of the canonical Vα14-Jα18 rearrangement in which the aspartic acid that appears at position 94 in the Vα-Jα junctional region of the canonical iNKT CDR3α is replaced by other amino acids. This aspartic acid has been shown in crystal structures to make extensive contacts with the α1 α-helix of CD1d (24, 25). We showed that an iNKT TCR with one of the CDR3α position 94 variants has about 40-fold lower affinity for self-ligands presented by CD1d than the wild-type TCR-α, a phenomenon that presumably allows thymocytes bearing the variant TCR-α to avoid clonal deletion. However, although these CDR3α 94 variant TCRs are still dependent upon CD1d for positive selection, their lower affinity for self leads to inefficient Egr2 induction, poor expression of the iNKT-lineage-specific transcription factor PLZF, inadequate proliferation of iNKT cell precursors, defect in trafficking, and impaired effector functions. These results demonstrate that proper iNKT cell development is not only dependent on TCR-mediated signals (26) but is also carefully tuned for proper iNKT cell differentiation and subsequent function.
Results
Normal Development of Conventional T Cells in 2A3-D TCR-β Transgenic Mice.
TCRs made up of the canonical iNKT TCR-α chain (Vα14-Jα18) and TCR-βs that include Vβ6 usually react poorly, if at all, with iNKT ligands, such as CD1d/PBS57 (27). However, randomization of CDR3β allowed us to identify CDR3β sequences that allowed TCRs that contained the canonical TCR-α and Vβ6 to react with CD1d/PBS57. Moreover, when the CDR2 Vβ6 sequence in this TCR was replaced with the CDR2 sequence of a Vβ, Vβ8.2 [which usually contributes well to reaction with CD1d plus glycolipid ligands (27)], the resulting TCRs reacted not only with CD1d bound to a strong ligand, PBS57, but also with CD1d bound to very weak ligands, such as those naturally found within CD1d in unmanipulated antigen presenting cells (APCs) (21). Thus, the combination of the new “hybrid” TCR-β chains with the canonical iNKT TCR-α created TCRs with high affinity for CD1d plus various ligands including those of self (21, 22, 28).
Crystal structure studies of one of these TCRs (2A3-D) in complex with various lipids presented by mouse CD1d molecules (21, 22) demonstrated that the 2A3-D TCR binds to CD1d-Ag in an identical manner to human and mouse wild-type iNKT TCRs (24). Furthermore, the results demonstrated that the autoreactivty of these TCRs resulted from increased affinity for the CD1d molecule itself, in which the CDR3β loop determines the threshold of CD1d autoreactivity in an antigen-independent manner (21, 28).
The cDNA coding for the hybrid 2A3-D TCR-β chain was cloned into the human CD2 promoter transgene cassette for T-cell–specific expression and the construct injected in C57BL/6 oocytes. We obtained three independent founder lines, one of which did not transmit the transgene. The other two founder lines had identical phenotypes. One line was selected for further use and bred to C57BL/6 TCR-β−/− mice to ensure that no other endogenous TCR-β chains would be expressed in the animals. This line has been used for all subsequent experiments and will be referred as 2A3-D Tg for the rest of the article. Importantly, the TCR-α locus in these mice was unmanipulated, thus allowing normal rearrangement and expression of TCR-α genes.
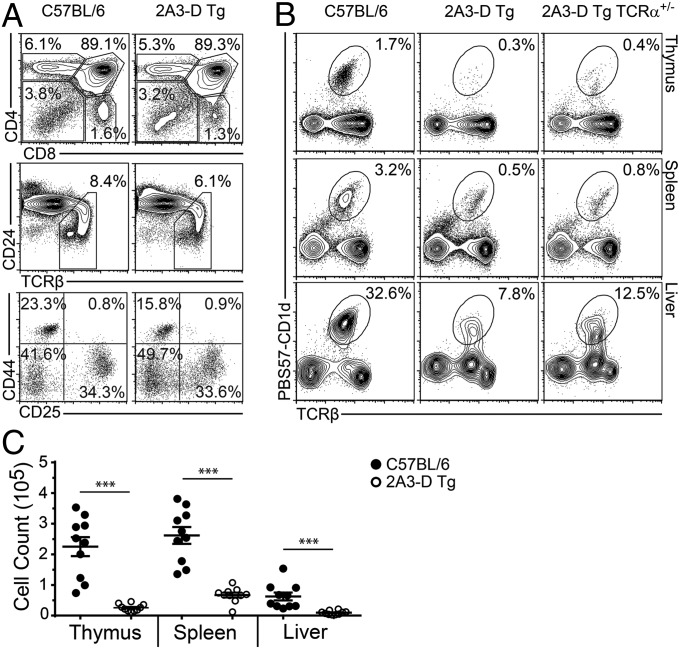
It was possible that the hybrid TCR-β in 2A3-D Tg mice (i.e., Vβ6 chain with a Vβ8.2-derived CDR2β) might affect the development of all T cells. To show that this was not so, we first checked whether or not the development of conventional T cells was normal. As seen in Fig. 1A, the proportions of CD4−CD8− double-negative, DP, and single positive CD4+ and CD8+ T cells was essentially identical in 2A3-D Tg and normal wild-type mice. The proportion of mature T cells, as measured by down-regulation of CD24 and up-regulation of the TCR was also normal. Finally, early stages of thymocyte development within the double-negative population were also normal. Taken together, the results demonstrate that expression of the TCR-β transgene did not significantly affect the development of conventional T cells, and that the hybrid nature of the TCR-β chain allowed for positive selection of conventional T cells on MHC class I and class II molecules.
Fig. 1.
Reduced number of iNKT cells in 2A3-D Tg mice. (A) Total thymocytes from C57BL/6 or 2A3-D Tg mice were stained for indicated markers to characterize specific stages of T-cell development (data representative of n = 3). (B) Total lymphocytes from indicated organs of C57BL/6, 2A3-D Tg or 2A3-D Tg TCR-α+/− mice were assessed for the presence of iNKT cells (data representative of n = 10). (C) Mean absolute numbers ± SEM of PBS57-CD1d tetramer-positive TCR-β+ iNKT cells were calculated for the thymus, spleen, and liver of C57BL/6 (●) and 2A3-D Tg (○) mice. Each dot represents a mouse (n = 10). ***P < 0.001.
Selective Reduction of iNKT Cells in 2A3-D TCR-β Transgenic Mice.
Because the rearrangement of the TCR-α chains occurs randomly at the DP stage, we anticipated that, if a thymocyte in these mice rearranged its TCR-α locus to create the canonical Vα14-Jα18 TCR-α chain, the TCR-α would pair with the TCR-β transgene and thereby generate an iNKT TCR of high affinity for self, potentially leading to deletion of the relevant thymocyte. Analysis with PBS57-loaded CD1 tetramers of the cells in 2A3-D Tg mice revealed a large reduction in, but not complete loss of, the proportion and total number of iNKT cells in the thymus, spleen, and liver compared with the numbers of such cells in wild-type mice (Fig. 1 B and C). However, no staining at all was detected in any organs when CD1d tetramers that had not been deliberately loaded with any ligand were used (Fig. S1A). These results suggest that cells bearing the transgenic TCR-β/canonical iNKT TCR-α were missing in the mice, because we have previously shown that cells bearing this combination of TCR-β/TCR-α stain well, even with self-CD1d tetramers (21). Therefore, it appears that the few remaining cells that can bind CD1d/PBS57 tetramers (hereinafter called iNKT-like) found in the 2A3-D Tg animals cannot interact with self-antigens.
To ascertain whether the few cells detected with the PBS57-CD1d tetramer represent “real” iNKT cells and not conventional T cells selected on MHC class I or MHC class II that had, by chance, rearranged Vα14-Jα18 on their second allele, we generated 2A3-D Tg TCR-β−/− TCR-α+/− mice, in which only one allele of the TCR-α locus can be expressed. Analysis of these mice revealed comparable frequencies and numbers of iNKT-like cells compared with TCR-α+/+ mice (Fig. 1B). We also crossed the 2A3-D Tg TCR-β−/− animals with CD1d−/− mice to determine whether the remaining iNKT-like cells were positively selected by CD1d. In the absence of CD1d, no iNKT cells of any sort were detected in any tissue of the 2A3-D transgenic mice (Fig. S1B). Therefore, forced expression of the 2A3-D transgene selectively reduces the iNKT cell population but a few iNKT-like cells continue to develop in a CD1d-dependent manner.
iNKT Cell Development in 2A3-D TCR-β Transgenic Mice.
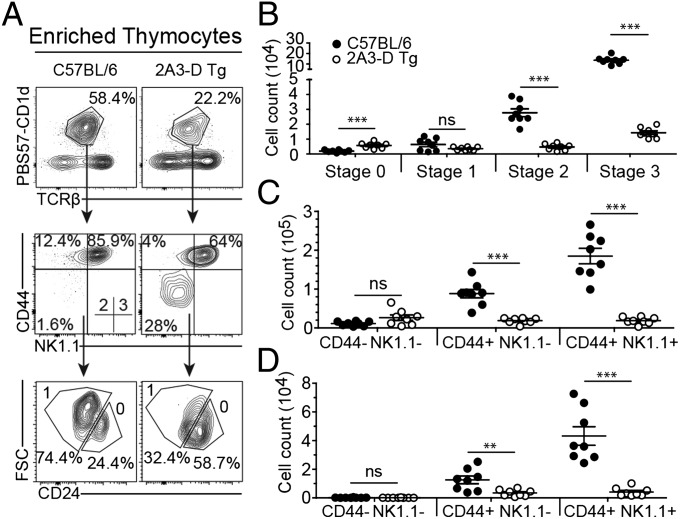
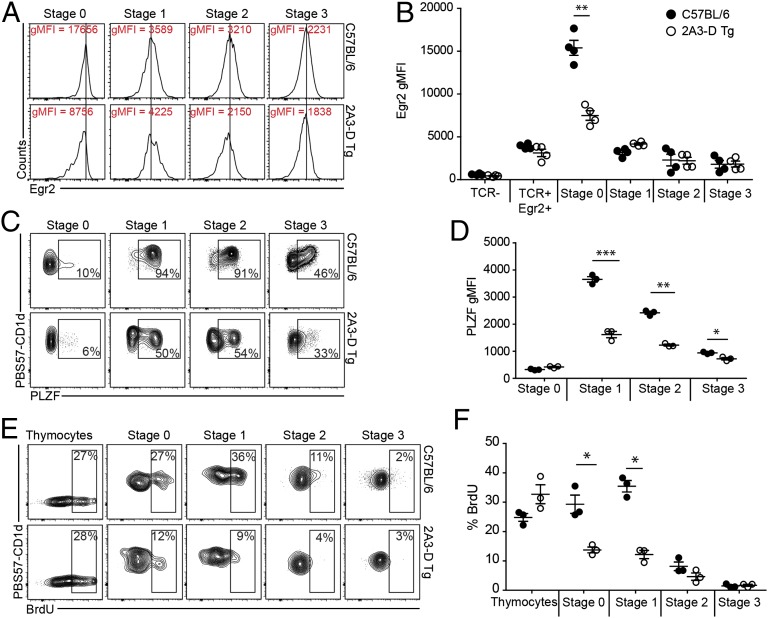
We enriched PBS57-CD1d tetramer-positive thymocytes from wild-type and 2A3-D Tg mice using magnetic beads and examined their expression of CD24, to identify the earliest stage 0 (CD24high) cells that are the immediate product of positive selection at the DP stage, and for the expression of CD44 and NK1.1 to follow the progression from stages 1–3 among the CD24low cells. Although we did not observe quantitative differences in the number of stage 0 and stage 1 iNKT cells between wild-type and transgenic animals, a large decrease in stage 2 and 3 iNKT cell numbers was evident (Fig. 2 A and B). This decrease was not because of a survival defect, as we could not detect any significant increase in apoptosis at any of the differentiation stages between wild-type and transgenic mice (Fig. S2). Furthermore, levels of CD1, signalling lymphocyte activation molecule (SLAM), and Ly108 expression on thymocytes were identical between the wild-type and transgenic animals (Fig. S3). Thus, the residual population of iNKT-like cells in the 2A3-D Tg mice is predominantly composed of stage 0 (CD24high) and stage 1 (CD44low) iNKT cells. This finding is further exemplified by the enrichment for CD44low NK1.1− cells in the spleen and for NK1.1low cells in the liver of 2A3-D mice (Fig. 2 C and D). This phenotype suggests that iNKT-like cells in 2A3-D Tg mice are blocked in their development at the transition from stage 1 to stage 2. However, this block is not absolute, as a small fraction of apparently mature iNKT-like cells (CD44high, NK1.1high) were detected in all organs. This phenotype is reminiscent of that previously reported for mice deficient in the transcription factor PLZF (19, 20), which upon its induction after positive selection has been shown to direct the effector program of the iNKT cell lineage.
Fig. 2.
iNKT cell development in 2A3-D Tg mice. (A) The gating strategy to identify the various iNKT cell developmental stages is depicted. Total thymocytes were MACS bead-enriched for the PBS57-CD1d tetramer-positive population and then stained for CD24, CD44, and NK1.1 to characterize the developmental stages of iNKT cells. The stages are defined as follows: stage 0 (CD24+CD44−NK1.1−), stage 1 (CD24−CD44−NK1.1−), stage 2 (CD24−CD44+NK1.1−), and stage 3 (CD24−CD44+NK1.1+) (data representative of n = 4). (B) Mean absolute numbers ± SEM of PBS57-CD1d tetramer-positive TCR-β+ NKT cell developmental stages were calculated in the thymus of C57BL/6 (●) and 2A3-D Tg (white dot) mice. Absolute numbers were calculated using population percentages from samples that were not subjected to MACS-enrichment. In these instances, large numbers of events were collected to clearly define the different populations. Each dot represents an independent mouse (n = 8). (C and D) Purified lymphocytes from the spleen (C) or the liver (D) of C57BL/6 or 2A3-D Tg mice were stained for CD44 and NK1.1. Mean absolute numbers ± SEM of iNKT cell maturation stages in the spleen (C) or liver (D) of C57BL/6 (black dot) and 2A3-D Tg (white dot) mice. Each dot represents a mouse (n = 8). Ns, not significant, **P < 0.01, and ***P < 0.001.
iNKT Cells Are Redistributed in the Peripheral Lymph Nodes of 2A3-D TCR-β Transgenic Mice.
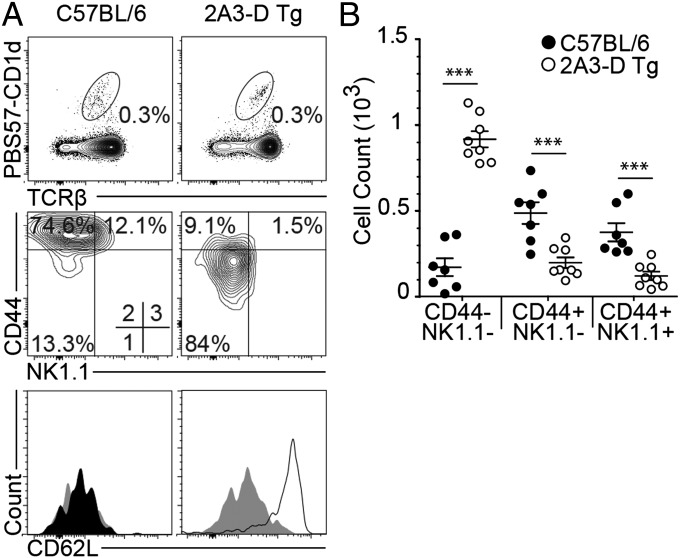
In the absence of PLZF, iNKT cell development is severely impaired, with a large reduction in the number of αGC/CD1d tetramer-positive cells and the preferential export of immature stage 1 iNKT cells to the peripheral tissues (19, 20). Furthermore, these naïve, stage 1-arrested, iNKT cells also tend to redistribute in the peripheral lymph nodes, a location where iNKT cells are normally poorly represented. We examined the numbers of iNKT-like cells in the peripheral organs of 2A3-D Tg mice. The cells were at low levels compared with wild-types in all organs examined, except in peripheral lymph nodes, where the proportion and total number of CD1d/PBS57 tetramer-positive cells was identical between wild-type and 2A3-D Tg mice (Fig. 3). However, on closer examination we found that the iNKT-like cells in the lymph nodes of 2A3-D Tg mice were essentially all CD44low NK1.1− and continued to express the homing receptor for high endothelial venules in lymph nodes, CD62L (Fig. 3). These results extend the previous findings of the thymic developmental defect and demonstrate that the majority of iNKT cells found in 2A3-D Tg mice maintain a naïve phenotype identical to that of conventional naïve CD4 T cells (CD24low, CD44low, NK1.1−), suggesting that they had not turned on the iNKT cell differentiation program.
Fig. 3.
iNKT cells are redistributed in the pLN of 2A3-D Tg mice. (A) Lymphocytes from peripheral lymph nodes (pLN) were pooled and stained for CD44, NK1.1, and CD62L to characterize iNKT cells. Histogram legend: isotype (gray filled), C57BL/6 (black filled) and 2A3-D Tg (black line) (data representative of n = 8 and histogram representative of n = 3). (B) Mean absolute numbers ± SEM of the different phenotypic PBS57-CD1d tetramer-positive TCR-β+ NKT cell populations in the pLN of C57BL/6 (black dot) and 2A3-D Tg (white dot) mice. Each dot represents a mouse (n = 8). ***P < 0.001.
The 2A3-D TCR-β Transgenic iNKT Cells Are Poorly Responsive in Vivo.
We next tested the functionality of the CD1d/PBS57-binding iNKT-like cells in 2A3-D Tg mice by injecting in vivo the strong iNKT cell agonist, αGC. Unlike in wild-type mice, serum cytokines appeared at very low levels in 2A3-D Tg mice in response to this treatment (Fig. 4A). This result could have been either because of a failure of the iNKT-like cells to respond or because of their very low numbers in the animals. To distinguish the two possibilities, we used intracellular staining to examine cytokine production by the iNKT-like cells. In agreement with their immature and naïve phenotype, iNKT-like cells in the spleen of 2A3-D mice were poor producers of both IFN-γ and IL-4 cytokines, compared with their wild-type counterparts (Fig. 4B).
Fig. 4.
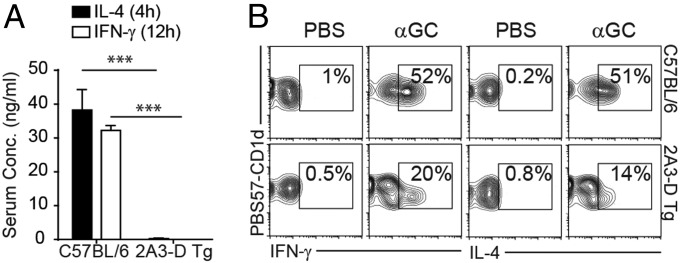
iNKT cells in the 2A3-D Tg mice are poorly responsive. C57BL/6 and 2A3-D Tg mice received an intraperitoneal injection of 2 μg of αGC or PBS. (A) Serum was collected at 4 h and 12 h after injection to quantify by ELISA IL-4 and IFN-γ concentration, respectively (n = 3). (B) Ninety minutes after injection, mice were killed and purified splenocytes were stained for PBS57-CD1d tetramer-positive TCR-β+ NKT cells and for intracellular IL-4 and IFN-γ (data representative of n = 3). ***P < 0.001.
Repertoire of Peripheral iNKT Cells in 2A3-D TCR-β Transgenic Mice.
The results described above demonstrated that transgenic expression of the 2A3-D TCR-β chain led to a striking decrease in iNKT cell number and that the remaining iNKT-like cells were developmentally arrested, functionally deficient, and did not seed the peripheral tissues appropriately. These results led us to question whether the “escapee” iNKT cells had the same TCR repertoire as wild-type iNKT cells. Although the TCR-β chain is fixed in the transgenic animals, the TCR-α chains rearrange normally, and thus can vary. We sorted total iNKT cells from the spleen of wild-type and 2A3-D Tg animals and analyzed their Vα chain use by PCR using Vα- and Cα-specific primers. Similar to what is found in wild-type mice, the iNKT cell populations in 2A3-D Tg mice also used the Vα14 gene segment (Fig. S4A).
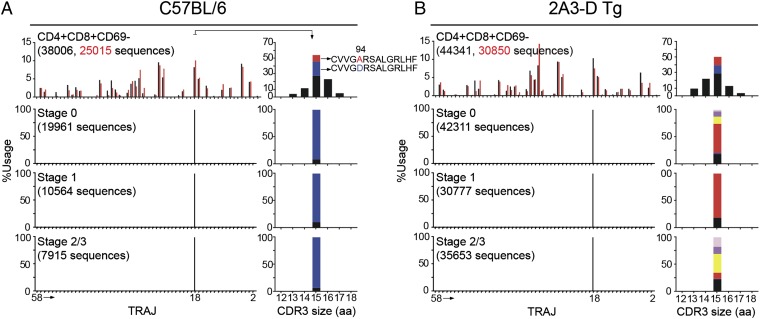
The PCR products were sequenced using the 454 platform. Essentially all in-frame productive Vα14 sequences (99.6%) in both types of mice rearranged to the Jα18 gene segment (Fig. 5A). In wild-type mice, most of the sequences (>89%) encoded the expected 15-aa canonical CDR3α, CVVGDRGSALGRLHF, which contains an aspartic acid residue at position 94 (Fig. 5B). In agreement with previous results (6), and in support of the idea that iNKT cells are selected to express exactly this TCR-α amino acid sequence, this protein sequence was encoded by four different nucleotide sequences (Fig. S4B). In contrast, this canonical CDR3α sequence was essentially absent (0.07% of total sequences) in 2A3-D iNKT-like cells. Although their CDR3αs were, like those in wild-type cells, 15-aa long, most sequences (>77%) encoded for a variant CDR3α, in which the D94 residue was replaced by an alanine (Fig. 5B). Analysis of nucleotide sequences encoding for this variant CDR3α also strongly implied that it was subject to selection pressure at the protein level (Fig. S4B). These results suggested that the association of the 2A3-D TCR-β chain with the canonical D94 Vα14-Jα18 rearrangement led to clonal deletion of iNKT cells bearing this TCR, thus explaining the conspicuous absence of the TCR-α D94 sequence from the peripheral repertoire.
Fig. 5.
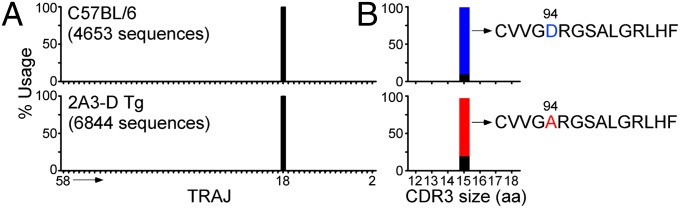
TCR-α repertoire of iNKT cells in the periphery of 2A3-D Tg mice. Splenocytes from C57BL/6 and 2A3-D Tg mice were MACS bead-enriched for PBS57-CD1d tetramer-positive cells and PBS57-CD1d tetramer-positive TCR-β+ iNKT cells were subsequently FACS sorted from the positive fraction. (A) Vα14-Cα PCR products were sequenced using the 454 platform and the frequency use of T-cell receptor-α joining (TRAJ) genes encoding productive rearrangements with T-cell receptor alpha variable gene (TRAV) TRAV11 is shown (data representative of n = 2). The TRAJ number, from 58 to 2, follow the order that the genes are found in the TCR-α locus. (B) Frequency and CDR3 size (in amino acids) of the Vα14-Jα18 rearrangements in C57BL/6 or 2A3-D Tg iNKT splenocytes (data representative of n = 2). The canonical D94 rearrangement is depicted in blue and the A94 variant sequence is depicted in red. Other sequences (i.e., other 94 variants as well as other position variants) are depicted in black.
To explore this hypothesis further, we sorted iNKT-like cells from the liver and the lymph nodes of the 2A3-D Tg mice and sequenced their Vα14 rearrangements. Similar to what was observed in the spleen, the canonical D94 CDR3α sequence was completely absent (Fig. 2B) from the liver and lymph node of 2A3-D iNKT-like cells. Instead, a number of different amino acids were found at position 94 of CDR3α, with a predominance of A94, especially in the lymph nodes (Fig. S4D).
Tracking of Vα14 Rearrangements During iNKT Cell Development.
The absence of the canonical D94 TCR-α sequence from the repertoire of peripheral 2A3-D iNKT-like cells strongly suggests that because this TCR-αβ combination is self-reactive (21, 22), thymocytes bearing it are probably clonally deleted during their development. If this were true, one would assume that the D94 sequence should be present within the preselected repertoire. Therefore, we sorted CD4+CD8+CD69− preselection thymocytes from wild-type and 2A3-D Tg mice and analyzed Vα14 rearrangements using high-throughput sequencing. Furthermore, to identify the stage of development at which deletion might occur, we also sorted cells at stages 0, 1, and 2/3 combined from wild-type and 2A3-D Tg mice and performed the same analysis. As seen in Fig. 6, in preselected DP thymocytes, the Vα14 gene segment rearranges with 38 different Jα gene segments of the 42 open-reading frame sequences that can be used as J segments in the genome of C57BL/6 mice. No rearrangements involving Jα45, Jα30, Jα7, and Jα5 could be found in our analysis. Focusing on productive in-frame rearrangements, the frequency of Jα use was reproducible between independent samples and, importantly, comparable between wild-type and 2A3-D Tg mice. These results demonstrate that the TCR-β transgene did not noticeably affect the intrinsic rearrangement of the TCR-α locus (Fig. 6). In DP CD69− thymocytes, Vα14 was found rearranged with Jα18 on average 9% of the time. Analysis of the CDR3 size of these rearrangements demonstrated a Gaussian distribution of sizes, centered on 15 amino acids. Within the Vα14-Jα18 rearrangements, the canonical D94 sequence was found 11–18% of the time, whereas the A94 variant sequence represented 8–11% of the analyzed sequences (Fig. 6). Although we specifically excluded PBS57/CD1d tetramer-positive cells during the sort of DP CD69− cells, the frequency of D94 sequence in C57BL/6 mice was slightly higher than that in 2A3-D Tg mice. Nevertheless, both D94 and A94 sequences could be detected in preselection thymocytes in both wild-type and 2A3-D Tg mice, in striking contrast with the peripheral results (Fig. 5). These results strongly argue for clonal deletion of the cells bearing the iNKT TCR with D94 sequences in 2A3-D Tg mice.
Fig. 6.
Tracking of TCR-α repertoire during iNKT cell development in the thymus. DP CD4+CD8+CD69− PBS57-CD1d tetramer-negative thymocytes were FACS-sorted from C57BL/6 (A) or 2A3-D Tg mice (B) (first row). Alternatively, thymocytes from C57BL/6 (A) and 2A3-D Tg (B) mice were MACS bead-enriched for PBS57-CD1d tetramer-positive cells and the different iNKT cell developmental stages were FACS-sorted based on CD24 and CD44 staining. Stage 0 (PBS57-CD1d+CD24+CD44−, second row), stage 1 (PBS57-CD1d+CD24−CD44−, third row), and stage 2/3 (PBS57-CD1d+CD24−CD44+, fourth row). Vα14 rearrangements were amplified by PCR from each population using Vα14- and Cα-specific primers and the Vα14-Cα PCR products were sequenced using the 454 platform. The frequency use of TRAJ genes encoding productive rearrangements with TRAV11 in the indicated sorted population is depicted. The experiment was repeated twice for the DP thymocytes and the individual results are depicted in black and red, respectively. The number of total sequence analyzed for each sample is indicated. Furthermore, the frequency and CDR3 sizes of the Vα14-Jα18 rearrangements for each targeted population are shown. The canonical D94 rearrangement is depicted in blue and the A94 variant sequence is depicted in red. The N94 variant is shown in yellow, the E94 variant in dark purple, and the S94 in pink). Other sequences (i.e., other 94 variants as well as other position variants and CDR3 sizes) are depicted in black.
Analysis of Vα14 rearrangements at the different stages of iNKT cell development in C57BL/6 mice demonstrated, as early as stage 0, a prominent enrichment for Jα18 use composed principally of the canonical D94 sequence, consistent with its positive selection within the iNKT cell repertoire (Fig. 6A). A94 sequences were poorly represented (0.1%) at any of the stages of iNKT cell development in C57BL/6 mice, suggesting that the pairing of this sequence with most of the naturally produced Vβ chains poorly allow for its positive selection within the iNKT cell repertoire.
The situation was entirely different in the 2A3-D mice (Fig. 6B). Although D94 sequences could still be found about 2% of the time in stage 0, this CDR3 sequence was essentially absent from stages 1 and 2/3 (<0.5%), suggesting that clonal deletion of this sequence takes place between the DP and stage 0 of development. Consistent with the sequencing results of peripheral 2A3-D iNKT cells, we found several variant sequences at position 94 of the CDR3 at all stages of development. The A94 variant was the predominant sequence found in stage 0 and stage 1, but this sequence was only found 10% of the time in stage 2/3. Instead, at this stage, the Vα14-Jα18 rearrangements were dominated by N94 (35%) and S94 (17%) sequences, that were also found enriched in 2A3-D liver iNKT-like cells (Fig. S4C). Hence, the forces of positive and negative selection act throughout iNKT cell development to shape the characteristics of the iNKT TCR.
Natural Variants at Position 94 of the Canonical iNKT CDR3α Have Lower Affinity for Self.
The structures of the 2A3-D iNKT TCR in complex with various self and nonself-antigens presented by mouse CD1d revealed that D94 makes multiple contacts with R79 on the α1 helix of CD1d (21, 22). We hypothesized that the 2A3-D TCRs with their variants residues at position 94 would probably have lower affinities for the antigen/CD1d complex. To test this possibility, we first engineered a series of hybridomas in which we expressed, using retroviruses, the 2A3-D TCR-β chain paired with Vα14-Jα18 TCR-α variants that were most frequently detected in our sequencing analysis (D94, A94, N94, S94, and E94). Each hybridoma was sorted for similar levels of TCR expression and subsequently stained with CD1d tetramers composed of CD1d monomers produced in 293 cells and presumably loaded with self-antigens, or with CD1d tetramers loaded with PBS57. The native 2A3-D TCR interacted with both self-antigens and PBS57, as previously reported (21). However, all of the natural variants at position 94 (A94, N94, S94, and E94) that are found in iNKT cells of the peripheral tissues of the 2A3-D Tg mice had lost their capacity to interact with self but could still interact with the PBS57-loaded CD1d tetramer (Fig. S5).
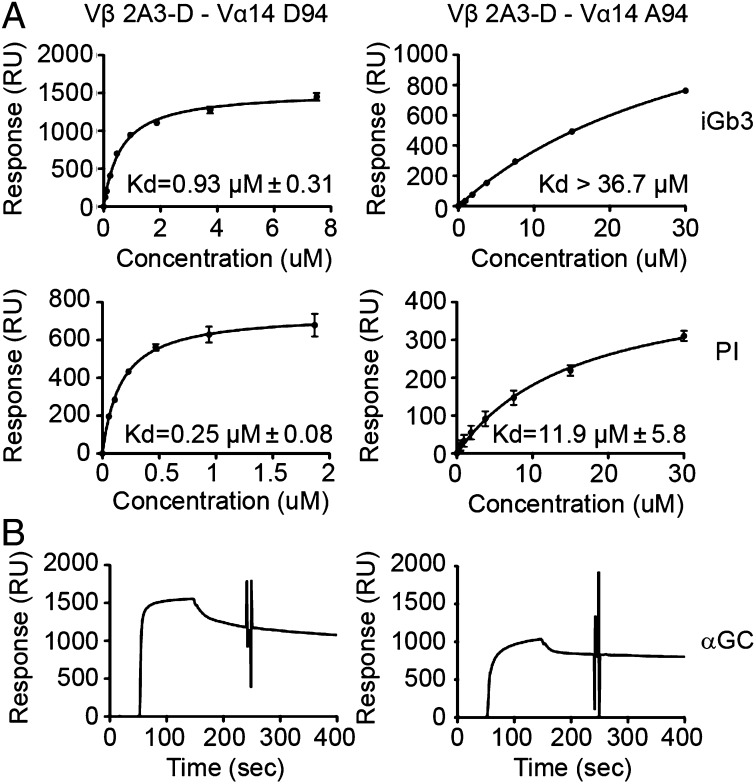
We also created a variant with a sequence that was never detected in any of the 2A3-D-derived samples, replacing the D94 with a bulky tyrosine (Y94). In that case, the created TCR lost binding to both self- and PBS57-loaded CD1d tetramer (Fig. S3). Finally, we produced soluble versions of the native 2A3-D (D94) and the A94 variant TCR and measured by surface plasmon resonance (SPR) the affinity of these two TCRs for the self-antigens iGb3 (29) and phosphatidylinositol (21), as well as the foreign antigen αGC. As seen in Fig. 7A, the A94 variant TCR showed a 40-fold decrease in affinity for self-ligands compared with the native TCR. However, the variant TCR continued to bind with high affinity to the αGC/CD1d complex (Fig. 7B). These results demonstrate that cells bearing the variant TCR escape negative selection in the thymus because the TCR has lower affinity for CD1d plus self-ligands.
Fig. 7.
Natural variants at position 94 of the canonical iNKT CDR3α have a lower affinity for self. (A) Binding of iNKT TCR variants to lipid loaded CD1d as assessed by SPR. Saturation plots demonstrating equilibrium binding of Vβ 2A3-D–Vα14 D94 TCR and the Vβ 2A3-D–Vα14 A94 TCR to immobilized CD1d-iGb3 and CD1d-PI. Affinities (Kd) were calculated from two independent experiments using BIAevaluation software. For visualization, datapoints from duplicates of a single experiment were plotted as the mean ± SEM and curves were fitted to a one-site model using Prism. (B) Sensograms showing the binding of 5 μM Vβ 2A3-D–Vα14 D94 or Vβ 2A3-D–Vα14 A94 to immobilized CD1d-α-GC.
iNKT TCR Affinity Affects the Developmental Program of iNKT Cells.
The above findings showed that in the 2A3-D Tg mice the few iNKT-like cells that have avoided clonal deletion express variant TCR-α chains that confer diminished affinity for “self.” However, this affinity must be sufficient for the cells to be positively selected, because no PBS57-CD1d tetramer-positive cells were detected in 2A3-D Tg × CD1d−/− mice (Fig. S1B). These findings provided us with the unique opportunity to examine, in an unmanipulated situation, the extent to which iNKT TCR affinity might affect agonist selection, and thereby the acquisition of the iNKT program. In agreement with previous results (16), we found high levels of the transcription factor Egr-2 in stage 0 iNKT cells from C57BL/6 mice, consistent with cells having received a strong agonistic signal during selection (Fig. 8 A and B). In contrast in 2A3-D Tg mice, stage 0 iNKT cells expressed only about half the levels of Egr-2 found in wild-type stage 0 cells (Fig. 8 A and B). These levels were nonetheless higher than the Egr-2 levels detected in conventional T cells (Fig. 8B), suggesting that these cells had received a “higher than normal” TCR signal during selection. Subsequently, the levels of Egr-2 dropped and were equivalent between the two strains of mice (Fig. 8 A and B).
Fig. 8.
iNKT TCR affinity affects the developmental program of iNKT cells. (A) Total thymocytes were MACS bead-enriched for PBS57-CD1d+ population and then stained for CD24, CD44, and NK1.1 to characterize the developmental stages of iNKT cells. The stages are defined as follows: stage 0 (CD24highCD44lowNK1.1low), stage 1 (CD24lowCD44lowNK1.1low), stage 2 (CD24lowCD44highNK1.1low), and stage 3 (CD24lowCD44highNK1.1high) (data representative of n = 4). Five thymi were pooled for each staining, enriched for PBS57-CD1d+ cells, and further stained for iNKT cell developmental stages and Egr-2. The histogram show representative Egr-2 gMFI (geometric mean flouorescent intensity) for each stages as defined in A for C57BL/6 and 2A3-D Tg mice (data representative of n = 2). (B) Plot presenting the Egr-2 gMFI results from two independent experiments for a total of four independent stainings. Each dot represents five pooled thymi. (C) Five thymi were pooled for each staining, enriched for PBS57-CD1d+ cells and further stained for iNKT cell developmental stages and PLZF. (D) Plot presenting PLZF gMFI results for each stage from one experiment with three independent samples from C57BL/6 and 2A3-D Tg mice. (E) C57BL/6 and 2A3-D Tg mice received daily intraperitoneal injection of 1 mg BrdU for 3 d. Mice were killed and thymocytes were MACS bead-enriched for PBS57-CD1d+ population. BrdU incorporation was assessed by intracellular staining (data representative of n = 3). (F) Plot presenting the percentage of BrdU incorporation according to stages described in A for C57BL/6 and 2A3-D Tg mice (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
One of the consequences of elevated levels of Egr-2 is transactivation of the Zbtb16 promoter (16, 18). As shown in Fig. 8 C and D, low induction of Egr-2 at stage 0, leads to poor PLZF expression (the product of the Zbtb16 gene) in stage 1 and stage 2 iNKT cells from 2A3-D Tg mice compared with those of C57BL/6 mice. Although PLZF expression peaks in nearly 100% of iNKT cells at stage 1 and 2 in C57BL/6 mice, only a fraction of stage 1 and 2 iNKT cells in 2A3-D Tg mice expressed PLZF (Fig. 8D).
PLZF deficiency prevents the massive intrathymic expansion and memory effector formation that characterizes iNKT cell development (19, 20). Phenotypic characterization (above) showed that the thymus of 2A3-D Tg mice contains few iNKT cells and that these are enriched for immature cells, a result that is consistent with their low PLZF levels. To determine whether intrathymic expansion was also affected, we measured BrdU incorporation at different stages of iNKT cell development (Fig. 8 E and F). Consistent with previous results (19, 30), a large portion of stage 0 and, especially stage 1 iNKT cells in C57BL/6 mice were found to have incorporated BrdU during the 3-d period of the experiment. In contrast, developing iNKT cells in 2A3-D Tg mice failed, for the most part, to undergo this massive expansion (Fig. 8 E and F). This finding was true even though BrdU incorporation in total thymocytes was similar in the two types of mice (Fig. 8F).
Finally, to establish formally a direct relationship between the affinity of the iNKT TCR for self-antigen–CD1d complexes and the induction of Egr-2, we used Ly108− hybridomas expressing the 2A3-D β-chain paired with iNKT TCR-αs expressing either the canonical D or the A variant at CDR3α position 94. Both hybridomas were stimulated for 2 h with A20 cells expressing, or not, mCD1d and with or without the agonist PBS57. We then measured the induction of Egr-2 by intracellular staining. No response was observed when CD1− APCs were used in the assay. Both hybridomas readily induced Egr-2 expression in response to CD1d plus PBS57 antigen. However, only the D94-expressing hybridoma was able to induce Egr-2 in an autoreactive situation with mCD1d-expressing A20 cells (Fig. S6). Taken together, these results demonstrate a direct link between iNKT TCR affinity, the signaling induced by agonist self-ligands during selection, and the triggering and completion of the full iNKT cell program.
Discussion
We provide direct evidence that iNKT cells need particular, defined affinities for the reactions between their TCRs and CD1d ligands for their proper development. We used mice expressing a hybrid TCR-β chain with an unusual CDR3 sequence that allows an iNKT TCR that bears it to react particularly well with CD1d, regardless of the ligand bound. Importantly, the TCR-α locus in these mice was normal; therefore, (iNK) T-cell development occurred with normal kinetics in these animals. Perhaps because of this, the transgenic TCR-β did not lead to any observable change in the development of conventional T cells. However, the population of iNKT cells, tracked using PBS57-loaded CD1d tetramers, was markedly decreased in all examined organs of the TCR-β transgenic mice, except the lymph nodes which had normal numbers. These results are in agreement with the idea that iNKT precursors expressing TCRs of high affinity for self are, like their conventional T-cell counterparts, subject to clonal deletion.
Perhaps unexpectedly, a small percentage of PBS57-CD1d tetramer-positive cells remained in the 2A3-D Tg mice, suggesting that clonal deletion of iNKT cells was either incomplete or that the remaining cells bore a TCR that differed in sequence from the expected canonical Vα14-Jα18 iNKT TCR-α, a phenomenon that would allow the cells to avoid clonal deletion.
The first clue in favor of the latter possibility came from the absence of staining of the remaining iNKT cells in 2A3-D Tg mice with the CD1d-tetramer loaded with what we presume are self-antigens, because we had previously demonstrated that hybridoma expressing this unique combination of the 2A3-D β-chain paired with the canonical Vα14-Jα18 α chain, interacted readily with this reagent (21). This result led us to carefully examine the Vα repertoire of the remaining iNKT cells in the 2A3-D Tg mice. To our surprise, we found that most cells continue to express a Vα14-Jα18 TCR-α chain with a CDR3α size of 15 aa, like that of wild-type iNKT cells. However, the canonical sequence, which encodes for a D residue at position 94, was almost absent from mature 2A3-D iNKT cells. However, the frequency of rearrangements encoding for this particular sequence was comparable in preselection DP thymocytes of 2A3-D Tg and wild-type mice. These results demonstrate that iNKT cell precursors expressing TCRs with high affinity for self are probably clonally deleted. Such deletion must occur very early in iNKT cell development because the canonical CDR3α D94 is lost as early as stage 0. At the moment we do not know which cell type mediates clonal deletion of the D94-expressing iNKT cell precursors. Positive selection of iNKT cells occurs on DP thymocytes themselves rather than the thymic epithelial cells that are used for positive selection of conventional T cells (31). However, previous results using CD1d-overexpressing transgenic mice suggested that dendritic cells in the thymus might be responsible for clonal deletion of high-affinity TCR-expressing iNKT cells (32). Nevertheless, the 2A3-D hybridoma responds as well to CD1d-expressing thymocytes as it does to bone-marrow–derived dendritic cells (21), raising the possibility that thymocytes bearing this TCR might be deleted during reaction with CD1d on the surface of DP cells themselves, as previously proposed (33). The recent description of CD1d floxed mice will be helpful in deciphering whether the two selection events are dissociated or not (34). Furthermore, the nature of the lipid antigens responsible for the positive selection of iNKT cells remains a matter of debate (10). In our view (24), the pattern recognition receptor-like mode of interaction between the iNKT TCR and the CD1d–lipid complex suggests that several different self-antigens might be able to positively select iNKT cells. It will be interesting to determine whether different self-lipids, presumably of higher affinity than the positively selecting ones, are involved in the clonal deletion of iNKT cells.
The few iNKT cells that avoided clonal deletion in 2A3-D Tg mice expressed natural variations of the canonical Vα14-Jα18 rearrangements. Specifically, the D residue at position 94 of the CDR3 was often changed for A, S, E, or N. Importantly, iNKT TCRs containing the 2A3-D β-chain paired with any of these Vα variants lost the capacity to interact with CD1d tetramer loaded with self-antigens, but continued to bind PBS57-CD1d tetramers, suggesting that the affinity of these TCRs might have been sufficiently decreased for them to avoid clonal deletion in vivo. Furthermore, direct affinity measurement by SPR of the 2A3-D A94 TCR variant for two different potential self-antigens, iGb3 and PI, presented by CD1d, showed a loss of affinity of about 40-fold compared with that of the native TCR. However, its affinity for self-antigen–CD1d complexes seemed sufficient to allow for positive selection.
Although the 2A3-D A94 TCR variant has an overall lower affinity for the antigen–CD1d complex compared with the native 2A3-D D94 TCR, it is currently unclear to what extend its affinity for self-Ag-CD1d compares to the affinities of the iNKT TCRs that compose the normal iNKT cell repertoire. Indeed, similarly to the majority of wild-type iNKT cells, cells expressing the 2A3-D A94 variant TCR bound the PBS57-CD1d tetramer but not the self-CD1d tetramer. However, some affinities of the 2A3-D A94 variant TCR could still be measured by SPR for both iGb3 and PI presented by CD1d, albeit with about 40-fold lower affinity than the D94 TCR. Whether iGb3 represents an important self-antigen for iNKT cells remains debatable (10). Nonetheless, we can use the iGb3 SPR data to compare the affinity of the A94 mutant to that of two different mouse iNKT TCRs presumably representative of the “normal” iNKT repertoire (22, 35). In one case, an iNKT TCR composed of the Vα14-Jα18 canonical rearrangement paired with a Vβ8.2 TCR chain isolated by PCR from αGC-CD1d tetramer-positive iNKT thymocytes showed a Kd for the iGb3-CD1d complex of >100 μM (22); however, in the second case, the iNKT TCR isolated from the iNKT cell hybridoma 2C12 (36), which also consist of the Vα14-Jα18 canonical rearrangement paired with a Vβ8.2 TCR chain with a different CDR3β sequence, had a Kd of 0.7 μM for the iGb3-CD1d complex (35). In our experiments, the 2A3-D A94 variant TCR bound the iGb3-CD1d complex with a Kd of 36 μM, further illustrating the importance of the TCR-β chain in modulating the recognition of the antigenic complex. With such a broad range in affinity of the normal iNKT repertoire for at least one representative iNKT cell self-antigen, it is difficult to determine how the affinity of the 2A3-D A94 variant TCR compares to that of typical wild-type iNKT TCRs.
Interestingly, a rather substantial proportion of human iNKT cell clones (4–30%), which also express a conserved Vα24-Jα18 rearrangement, has been found with variations to the canonical sequence (37, 38). Moreover, these TCR-α variations strongly influenced the affinity of human iNKT TCRs for various antigen-CD1d complexes, irrespective of the TCR-β chain with which they were paired (39). However, in contrast with our results where variations were most common at position 94, the variations in human iNKT TCR-α chains occurred most often at position 93 of the CDR3α. A serine residue is found at position 93 in the majority of human iNKT TCR-αs, whereas in the mouse iNKT TCRs this position is a glycine residue. Although this position 93 does not directly contact CD1d, dynamic modeling studies suggested that it can nonetheless affect the conformation of the CDR3α loop (39). The reason for this difference between species is currently unclear. Indeed, although it does not affect the protein sequence, the last nucleotide of codon 93 from the mouse rearrangement is not germ-line–encoded 40% of the time, arguing that position 93 of the mouse rearrangement could potentially also encode variant sequences.
Analysis of Vα14 rearrangements in preselection DP CD69− cells showed a broad range of Jα use, with rearrangements with Jα27, Jα18, and Jα6 occurring about 9% of the time for each. The frequency of rearrangements encoding the canonical sequence among Vα14-Jα18 rearrangements was relatively high, in the range of 10–20%, and the most prevalent nucleotide sequence encoding for this amino acid sequence was germ-line–encoded. Convergent recombination has been proposed as a mechanism that efficiently favors the production of the iNKT TCR-α sequence (40). In silico simulation of Vα14 to Jα18 rearrangement, performed without the introduction of p-additions, predicted that the canonical TCR-α sequence would theoretically represent ∼1% of the total simulated repertoire (40). Experiments are underway to further clarify whether other mechanisms might also be at play in favoring the production of the canonical sequence by the recombination machinery.
All of the iNKT 2A3-D–related TCRs identified in vivo with variant CDR3α sequences had lower affinity for self compared with the native 2A3-D TCR. This result gave us the unique opportunity to observe the development of iNKT cells bearing these TCRs and ask whether it might affect the agonist selection of iNKT cells. The results showed that iNKT cells expressing TCR variants did not properly follow the iNKT cell developmental program. Both in hybridomas and T cells isolated from animals, the cells induced Egr-2 poorly in response to CD1d plus self-antigens. Recent findings have shown that iNKT cell precursors that have received a strong TCR signal induce high levels of Egr-2, which in turn binds the Zbtb16 promoter and induces the expression of PLZF (16). This effect is potentiated by the costimulation through the homophilic signaling lymphocyte activation molecule, Ly108 (18), which is found expressed at high levels on DP thymocytes but is absent from the thymic stroma. We did not find any difference in Ly108 expression on DP thymocytes of 2A3-D Tg vs. wild-type mice, arguing that this signaling pathway is probably intact. Thus, Egr-2 is probably poorly induced by the 2A3-D variants because of their low affinity for CD1d plus self-ligands. These results directly demonstrate that TCR signaling by agonist self-ligands is the trigger of the iNKT cell differentiation pathway. Furthermore, it emphasizes that only a limited range of TCR affinity for self is actually compatible with iNKT cell development versus death by neglect or negative selection.
Several different cell types (i.e., T regulatory cells, iNKT cells, CD8αα, intraepithelial lymphocytes, mucosa-associated invariant T cells), each expressing very different genetic programs, are thought to arise from T-cell precursors that have received an agonistic signal during development. Although the precise nature of the signaling pathways involved in agonist selection will certainly remain an exciting avenue of future research, the current findings directly illustrate the central importance of TCR affinity in inducing different iNKT cell fates.
Materials and Methods
Mice.
The 2A3-D Tg strain with a CD2 driving promoter of the 2A3-D β-chain was produced by the Mouse Genetic Core facility at National Jewish Health. The CD1d1d2−/− mice and Jα18−/− mice have been described previously (41, 42). C57BL/6, TCR-α−/−, and TCR-β−/− were purchased from Jackson Laboratories. All mice were used between 6 and 12 wk and were age-matched for each experiment. All mice were raised in a specific pathogen-free environment at the Biology Resource Center in National Jewish Health. Experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at National Jewish Health.
Cell Lines, TCR-αβ Constructs, and Retroviral Packaging.
Mutant TCR-α chains were constructed by PCR with overlapping primers using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). TCR-α constructs were expressed by retroviral transduction of 5KC C3 derived from the 5KC-78.3.20 TCR-α− and TCR-β− hybridoma that were selected for the loss of CD1d expression. Retroviral plasmids were transfected into Phoenix cells together with the pCLEco accessory plasmid with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s specifications. Retrovirus-containing supernatants were harvested 48 h after transfection, centrifuged, and filtered for removal of debris. Hybridomas were “spin-infected” at 3,300 × g for 90 min at 37 °C in retrovirus-containing supernatants supplemented with polybrene (8 μg/mL). Hybridomas were then sorted on a MoFlo XDP (Beckman Coulter) on the basis of retroviral reporter and TCR-β membrane expression. All cells were maintained in complete RPMI with 10% FCS (vol/vol).
Lymphocyte Isolation.
Cell suspensions were prepared from the thymus, spleen, and peripheral lymph nodes (pool of inguinal, axillary and brachial lymph nodes) by manual disruption using syringe plunger. Liver was perfused with PBS then cut into small pieces, disrupted with a syringe plunger, and separated on a 33% (vol/vol) Percoll gradient (Sigma-Aldrich).
Flow Cytometry.
CD1d-PBS57 and CD1d-self tetramers were obtained from the National Institutes of Health Tetramer Core Facility. CD1d-self tetramers were generated using CD1d monomeric molecules produced by human embryonic kidney 293 cells transduced with a mouse CD1d-encoding lentiviral construct. No external antigen was added during the purification or tetramerization processes. Fluorochrome-labeled monoclonal antibodies against BrdU (3D4), CD4 (GK1.5), CD8α (53-6.7) CD24 (M1/69), CD25 (PC61.5) CD44 (IM7), CD62L (MEL-14), Egr-2 (erongr2), IFN-γ (XMG1.2), IL-4 (11B11), NK1.1 (PK136), PLZF (Mags.21F7), and TCR-β (H57-597) were purchased from eBioscience, BD Biosciences, or BioLegend. For intracellular staining of Egr2 and PLZF, cells were prepared using the Foxp3 fixation/permeabilization kit (eBioscience).
iNKT Cell Enrichment.
For MACS enrichment, pooled thymocytes were labeled with PE-conjugated CD1d-PBS57 tetramer and enriched on an autoMACS cell separator (Miltenyi Biotech), according to the manufacturer’s protocol, with all procedures conducted on ice. Samples were analyzed on an LSRII (Becton Dickinson) or sorted on a MoFlo XDP (Beckman Coulter).
In Vivo Cytokine Response Quantification.
Mice received 2 μg of αGC (Alexis Biochemicals) by intraperitoneal injection. Ninety minutes after injection, blood was collected and serum isolated using Z-gel microtubes (Sarstedt). Cytokines responses were measured by an IFN-γ or IL-4 ELISA in accordance with standard protocols.
BrdU Incorporation.
Mice received an intraperitoneal injection of BrdU (BrdUrd; 1 mg in 200 μL PBS; BD Biosciences) at 0, 24, and 48 h before being killed for analysis of proliferation. To detect the presence of BrdU, the APC BrdU Flow Kit (BD Biosciences) was used according to the manufacturer’s protocol.
High-Throughput Sequencing.
For each sorted population, cells were washed twice in ice-cold PBS and total RNA was extracted using the RNeasy Mini Kit (Qiagen). cDNA was produced using SuperScript III Reverse Transcriptase (Invitrogen). Amplification of the Vα14-Cα region was performed using specific Vα14 (5′-TACAGTGTGACCCCCGACAAC-3′) and Cα (5′-GAGGGTGCTGTCCTGAGACCGAG-3′) labeled with required specific tags. Purified PCR products were sent for high-throughput sequencing with the Roche 454 platform. Sequence analysis was done with in-house software, and gene identity was assigned on the basis of sequence alignment with published sequences (International ImMunoGeneTics Information System).
Protein Production and Purification.
Mouse CD1d was expressed, purified, and biotinylated, as described previously (21). Soluble NKT TCR Vα14 and 2A3-D chains were cloned into PET30 vector upstream of jun and fos leucine zippers, respectively. Protein chains were expressed in Escherichia coli strain BL21 as inclusion bodies and refolded/purified, as described (21). The NKT TCR D94A mutant was generated by site-directed mutagenesis using Pfu polymerase (Promega) according to the manufacturer’s instructions.
SPR.
SPR experiments were performed at 25 °C on a Biacore 3000 using HBS buffer (10 mM Hepes pH 7.4, 150 mM NaCl). CD1d samples loaded with PI, αGC, or iGb3 were coupled to a SA sensor chip (GE Healthcare). To determine equilibrium affinity measurements, NKT TCR were passed over the chip at a flow rate of 5 μL/min for 96 s and the final response was calculated following subtraction of an empty flow cell. All measurements were performed in duplicate and affinities were calculated as the mean of two independent experiments. BIAevaluation Version 3.1 (Biacore AB) was used for all data analysis.
Statistical Analysis.
Unpaired two-tailed Student t test was performed with Prism (Graph Pad Software). *P < 0.05, **P < 0.001, ***P < 0.0001. All error bars indicate the mean ± SEM.
Supplementary Material
Acknowledgments
We thank members of the L.G. laboratory, John Cambier, and Leonard Dragone for thoughtful discussion and critical comments on the manuscript; Derek Sant’Angelo for the antipromyelocytic zinc finger transcription factor mAb used in preliminary experiments; the National Jewish Health flow cytometry facility, the Mucosal and Vaccine Research Colorado Flow Core, and the University of Colorado flow cytometry shared resource facility for assistance with cell sorting; the Center for Genes, Environment, and Health at National Jewish Health for sequencing and the National Institutes of Health core facility for CD1d tetramers. This work was supported by National Institutes of Health Grants AI18785 and AI22295 (to P.M. and J.W.K.), and AI092108 (to L.G.); The Cancer Center Support Grant P30CA046934; the Howard Hughes Medical Institute (P.M. and J.W.K.); the National Health and Medical Research Council (NHMRC) Australia Fellowship (to J.R.); the NHMRC Senior Principal Research Fellowship (to D.I.G.); a NHMRC Peter Doherty fellowship (to Richard Berry); NHMRC Program grants (to D.I.G.); and a Cancer Council of Victoria grant (to J.R. and D.I.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320777110/-/DCSupplemental.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg M. Toward an understanding of NKT cell biology: Progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 3.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20(3):358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uldrich AP, et al. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12(7):616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180(3):1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2(10):971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 8.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: Innate B and T lymphocytes. Nat Rev Immunol. 2001;1(3):177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 9.Gapin L. iNKT cell autoreactivity: What is ‘self’ and how is it recognized? Nat Rev Immunol. 2010;10(4):272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gapin L, Godfrey DI, Rossjohn J. Natural killer T cell obsession with self-antigens. Curr Opin Immunol. 2013;25(2):168–173. doi: 10.1016/j.coi.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202(4):485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellicci DG, et al. NKT cells develop through a thymus-dependent NK1.1−CD4+ CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 14.Berzins SP, McNab FW, Jones CM, Smyth MJ, Godfrey DI. Long-term retention of mature NK1.1+ NKT cells in the thymus. J Immunol. 2006;176(7):4059–4065. doi: 10.4049/jimmunol.176.7.4059. [DOI] [PubMed] [Google Scholar]
- 15.Hu T, Gimferrer I, Simmons A, Wiest D, Alberola-Ila J. The Ras/MAPK pathway is required for generation of iNKT cells. PLoS ONE. 2011;6(5):e19890. doi: 10.1371/journal.pone.0019890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13(3):264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10(3):306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta M, et al. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J Immunol. 2013;190(5):2121–2128. doi: 10.4049/jimmunol.1202145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34(3):315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellicci DG, et al. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12(9):827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185(1):133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 24.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12(12):845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girardi E, Zajonc DM. Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunol Rev. 2012;250(1):167–179. doi: 10.1111/j.1600-065X.2012.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vahl JC, et al. NKT cell-TCR expression activates conventional T cells in vivo, but is largely dispensable for mature NKT cell biology. PLoS Biol. 2013;11(6):e1001589. doi: 10.1371/journal.pbio.1001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallevaey T, et al. T cell receptor CDR2 β and CDR3 β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31(1):60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matulis G, et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3β loop. PLoS Biol. 2010;8(6):e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306(5702):1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 30.Ziętara N, et al. Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc Natl Acad Sci USA. 2013;110(18):7407–7412. doi: 10.1073/pnas.1221984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun T, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003;197(7):907–918. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schümann J, et al. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol. 2005;175(11):7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- 34.Bai L, et al. Distinct APCs explain the cytokine bias of α-galactosylceramide variants in vivo. J Immunol. 2012;188(7):3053–3061. doi: 10.4049/jimmunol.1102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu ED, Girardi E, Wang J, Zajonc DM. Cutting edge: Structural basis for the recognition of β-linked glycolipid antigens by invariant NKT cells. J Immunol. 2011;187(5):2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burdin N, et al. Selective ability of mouse CD1 to present glycolipids: α-Galactosylceramide specifically stimulates V α 14+ NK T lymphocytes. J Immunol. 1998;161(7):3271–3281. [PubMed] [Google Scholar]
- 37.Exley MA, et al. Selective activation, expansion, and monitoring of human iNKT cells with a monoclonal antibody specific for the TCR α-chain CDR3 loop. Eur J Immunol. 2008;38(6):1756–1766. doi: 10.1002/eji.200737389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent SC, Hafler DA, Strominger JL, Wilson SB. Noncanonical Valpha24JalphaQ T cells with conservative α chain CDR3 region amino acid substitutions are restricted by CD1d. Hum Immunol. 1999;60(11):1080–1089. doi: 10.1016/s0198-8859(99)00109-3. [DOI] [PubMed] [Google Scholar]
- 39.Sanderson JP, et al. Natural variations at position 93 of the invariant Vα24-Jα18 α chain of human iNKT-cell TCRs strongly impact on CD1d binding. Eur J Immunol. 2012;42(1):248–255. doi: 10.1002/eji.201141956. [DOI] [PubMed] [Google Scholar]
- 40.Greenaway HY, et al. NKT and MAIT invariant TCRα sequences can be produced efficiently by VJ gene recombination. Immunobiology. 2013;218(2):213–224. doi: 10.1016/j.imbio.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 42.Chen YH, et al. Expression of CD1d2 on thymocytes is not sufficient for the development of NK T cells in CD1d1-deficient mice. J Immunol. 1999;162(8):4560–4566. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.