Significance
Recombination activating gene (RAG)-mediated variable, diversity, and joining [V(D)J] gene recombination is the hallmark of the adaptive immunity that only exists in jawed vertebrates. The evolutionary origin of RAG remains largely unknown although many hypotheses have been proposed. This study indicates that a RAG1-like DNA fragment (bfRAG1L) from the invertebrate amphioxus might encode a functional central domain of vertebrate core RAG1. We show that bfRAG1L (if translated) is a virus-related protein that can degrade both DNA and RNA and is able to introduce V(D)J gene recombination in RAG1-deficient mice after the reconstitution of bfRAG1L into a core RAG1-like protein. In this paper, we propose a model for the evolutionary process of RAG1 in the combination of previous hypotheses, and it provides unique insights into the origin and evolution of RAG1.
Abstract
The highly diversified repertoire of antigen receptors in the vertebrate immune system is generated via proteins encoded by the recombination activating genes (RAGs) RAG1 and RAG2 by a process known as variable, diversity, and joining [V(D)J] gene recombination. Based on the study of vertebrate RAG proteins, many hypotheses have been proposed regarding the origin and evolution of RAG. This issue remains unresolved, leaving a significant gap in our understanding of the evolution of adaptive immunity. Here, we show that the amphioxus genome contains an ancient RAG1-like DNA fragment (bfRAG1L) that encodes a virus-related protein that is much shorter than vertebrate RAG1 and harbors a region homologous to the central domain of core RAG1 (cRAG1). bfRAG1L also contains an unexpected retroviral type II nuclease active site motif, DXN(D/E)XK, and is capable of degrading both DNA and RNA. Moreover, bfRAG1L shares important functional properties with the central domain of cRAG1, including interaction with RAG2 and localization to the nucleus. Remarkably, the reconstitution of bfRAG1L into a cRAG1-like protein yielded an enzyme capable of recognizing recombination signal sequences and performing V(D)J recombination in the presence of mouse RAG2. Moreover, this reconstituted cRAG1-like protein could mediate the assembly of antigen receptor genes in RAG1-deficient mice. Together, our results demonstrate that amphioxus bfRAG1L encodes a protein that is functionally equivalent to the central domain of cRAG1 and is well prepared for further evolution to mediate V(D)J recombination. Thus, our findings provide unique insights into the evolutionary origin of RAG1.
The adaptive immune system, with the high degree of antigen receptor diversity, gives the host the potential to recognize and evade any invader. Antigen receptor genes are assembled from variable (V), diversity (D), and joining (J) gene segments in a somatic DNA rearrangement reaction termed V(D)J recombination (1, 2), a process mediated by the recombination signal sequences (RSSs) and the proteins encoded by the recombination activating genes (RAGs) RAG1 and RAG2. RSSs that flank the V, D, and J gene segments consist of well conserved heptamer and nonamer sequences that are separated by relatively nonconserved spacer region of 12 or 23 base pairs (12-RSS or 23-RSS, respectively) (3), whereas RAG1 and RAG2 (thereafter referred to as RAG) proteins are responsible for sequence-specific DNA recognition and DNA cleavage. During V(D)J recombination, RAG1 binds to nonamer through its nonamer binding domain (NBD) and RAG2 acts as a regulator for the formation of RSS/RAG complex. Double-strand breaks are then introduced at the heptamer-coding flank border by the RAG1 enzymatic activity, generating covalently sealed hairpin coding ends and blunt 5′-phosphorylated signal ends. Finally, the signal and coding ends are repaired by nonhomologous end joining (NHEJ) machinery (4, 5).
RAG-mediated V(D)J recombination is the main mechanism for generating antigen receptor diversity and is the hallmark of jawed vertebrate-specific adaptive immunity (6). Although SpRAG1L and SpRAG2L, a pair of RAG-like genes from the invertebrate purple sea urchin (Strongylocentrotus purpuratus), have been shown to represent ancient homologs of vertebrate RAG1 and RAG2 (7), the earliest known RAG-mediated adaptive immune system was demonstrated in cartilaginous fish, e.g., the horned shark (8). Thus far, no evidence has been provided that supports the existence of functional RAG in either invertebrates or jawless vertebrates.
It has been reported that the hypothetical gene bfRAG1L, from the invertebrate amphioxus (Branchiostoma floridae), is a vertebrate RAG1-like DNA fragment (9). Here, we combined bioinformatic and experimental approaches to explore the relationship between bfRAG1L and vertebrate mouse RAG1 from basic structure to recombination function. We showed that bfRAG1L contains a retroviral type II nuclease active site motif, DXN(D/E)XK, and is capable of degrading both DNA and RNA. Moreover, bfRAG1L shares important functional properties with the central domain of cRAG1, and the reconstitution of bfRAG1L into a cRAG1-like protein yielded an enzyme capable of recognizing RSSs and performing V(D)J recombination together with mouse RAG2. Moreover, this reconstituted cRAG1-like protein could mediate antigen receptor gene assembly in RAG1-deficient mice. Our findings suggest that the bfRAG1L DNA fragment is likely an ancient predecessor of vertebrate RAG1 and, thus, provide unique insights into the evolutionary origin of RAG1.
Results
bfRAG1L Product Is a Virus-Related Protein That Can Degrade both DNA and RNA.
No sense cDNA of bfRAG1L was found after searching the cDNA database of Florida amphioxus B. floridae released by Institute of Cellular and Organism Biology EST project (http://amphioxus.icob.sinica.edu.tw). However, the product of bfRAG1L if translated shares ∼35% similarity with that of cRAG1 (over the amino acid region 508–820) (9). Notably, the region where bfRAG1L is homologous to RAG1 contains the entire central domain of cRAG1 (residues 528–760) (10), including the two key aspartic acid residues (D600 and D708) of the catalytic DDE triad (D600, D708, and E962) (Fig. 1A). With the PSI-BLAST search engine for GenBank proteins, excluding proteins from jawed vertebrates, we found that bfRAG1L bears the closest homology to SpRAG1L and the next closest homology to viral RNA polymerases with 19% identity (SI Appendix, Fig. S1 and Table S1), although the identity doesn’t include the conserved acidic residues (D600, D708) of vertebrate RAG1.
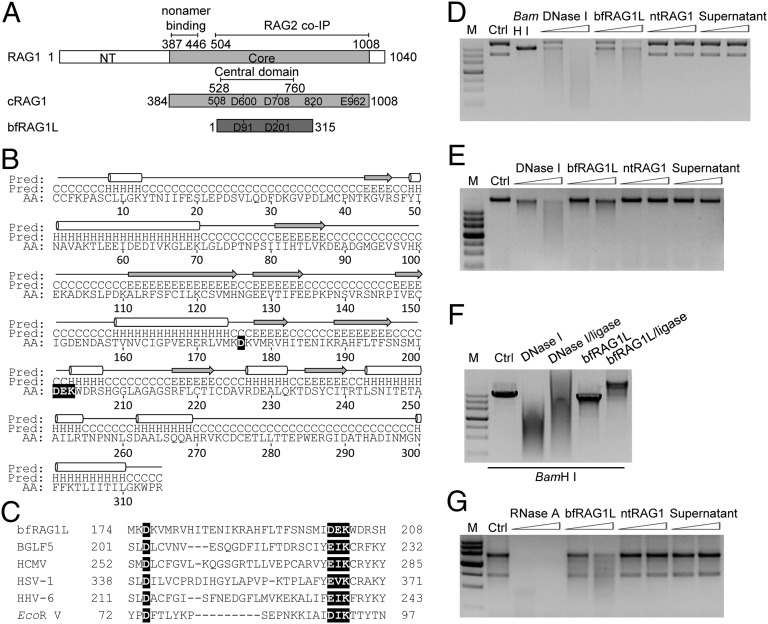
Fig. 1.
The structural and functional analyses of bfRAG1L. (A) Alignment between RAG1 and bfRAG1L. The conserved DDE motif residues are indicated in the box. Numbers represent positions of amino acids. The nonamer-binding domain, RAG2 binding domain, and the central domain are marked with straight lines. NT, N-terminal. Core, the core region of mouse RAG1. (B) The predicted secondary structure is labeled with cylinders (α-helices) and arrows (β-strands). (C) Sequences alignment of the conserved nuclease active motif (highlighted in black) in bfRAG1L, several herpes virus nucleases, and EcoR V. Nuclease activity assays were performed (SI Appendix). The nuclease activities of bfRAG1L (0.1 μg or 0.2 μg) on the substrates plasmid pBR322 (D), genomic DNA (E), or RNA from HEK-293T cells (G) are shown. The incubations with substrates DNA are executed for 8 h, or with substrate RNA for 3 h. Ctrl, represents respective untreated substrate; M, DNA marker. (F) The religation products of BamH I-digested linear plasmid pBR322 degraded by 0.1 μg of bfRAG1L or DNase I. Ctrl, untreated BamH I-digested linear plasmid pBR322. Supernatant, purified bfRAG1L protein immunodepleted with anti-bfRAG1L serum. The results are representative of three independent experiments.
Before examining the potential function of bfRAG1L, we predicted the secondary structure of bfRAG1L by using the publically available PSI-PRED server (11). We found that bfRAG1L shares certain structural characteristics with retroviral type II nucleases, such as the nuclease BGLF5 (from the herpes virus family EBV) (12). Like BGLF5, bfRAG1L contains a highly conserved DXN(D/E)XK active site motif (D176XND201K203) that localizes to several planar β-sheets, which are surrounded by α-helices (Fig. 1 B and C), suggesting that bfRAG1L might be able to function as a nuclease.
Consistent with the above prediction, purified bfRAG1L, but not the N-terminal domain of mouse RAG1 (ntRAG1), could hydrolyze double-strand DNA (e.g., covalently closed circular plasmid DNA or genomic DNA) in a non-DNA sequence-specific manner (Fig. 1 D and E and SI Appendix, Fig. S2 and Table S2). In contrast, this hydrolytic activity was not detected when bfRAG1L was immunodepleted with anti-bfRAG1L serum, whereas IgG-depleted samples retained their activity (Fig. 1 D and E and SI Appendix, Fig. S3). Notably, the partially digested linear DNA fragments could be religated by T4 DNA ligase, indicating that bfRAG1L could generate 5′-phosphorylated and 3′-OH end groups (Fig. 1F). Given that a putative RNA-binding motif was also identified in bfRAG1L by performing a BindN search (http://bioinfo.ggc.org/bindn), its RNase activity was also examined. Interestingly, bfRAG1L could degrade RNA, whereas neither the anti-bfRAG1L serum-depleted bfRAG1L sample, nor the ntRAG1 control sample, showed detectable RNase activity (Fig. 1G).
The DXN(D/E)XK Motif Is Critical for the Nuclease Activity of bfRAG1L.
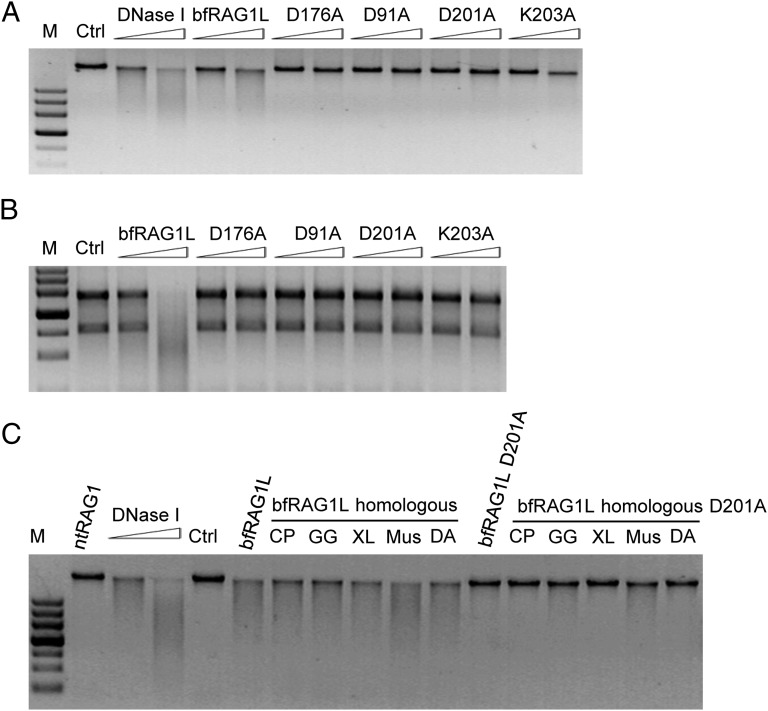
After verifying that bfRAG1L was a metal ion-dependent nuclease (SI Appendix, Fig. S4), we examined whether the key residues of the putative D176XND201K203 active site motif and the catalytic DDE triad (D91 and D201) in bfRAG1L are essential for its nuclease activity. Mutation of D176, D201, or K203 significantly attenuated the DNA and RNA degradation activity compared with the wild-type control (Fig. 2 A and B and SI Appendix, Fig. S2), indicating that the DXN(D/E)XK motif is critical for the nuclease activity of bfRAG1L. Notably, D91 also plays a role in the nuclease activity of bfRAG1L (Fig. 2 A and B and SI Appendix, Fig. S2). Considering that the central domain of cRAG1 is similar to bfRAG1L and is essential for the cleavage activity of RAG1 (10), we wondered whether the bfRAG1L-like region from vertebrate RAG1 could degrade DNA. Intriguingly, like bfRAG1L, these vertebrate bfRAG1L-like regions showed similar non-DNA sequence-specific nuclease activities (Fig. 2C and SI Appendix, Fig. S2), which is consistent with previous observation that the central domain of mouse cRAG1 contains an inherent nuclease activity (13).
Fig. 2.
The effects of mutations in bfRAG1L on nuclease activity. The nuclease activities of bfRAG1L mutants on genomic DNA (A) and RNA substrates from 293T cell lines (B). (C) The nuclease activities of bfRAG1L-homologous or bfRAG1L-homologous D201A mutants of the indicated vertebrates RAG1 on genomic DNA substrate (CP, bull shark; DA, zebrafish; GG, chicken; Mus, mouse; nt, N-terminal; XL, frog). Ctrl, untreated genomic DNA substrate from 293T cell lines. The results are representative of three (A and B) or two (C) independent experiments.
bfRAG1L Is a Nuclear Protein That Binds Vertebrate RAG2.
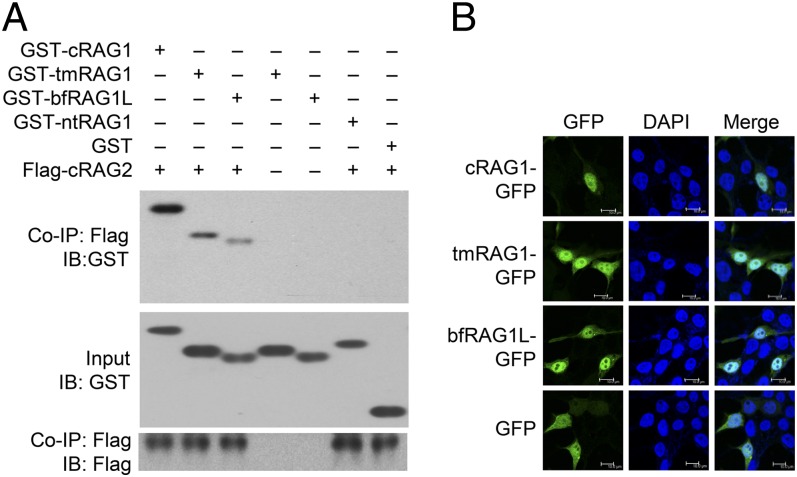
The central domain of cRAG1 is important for both the DNA cleavage activity of RAG1 and its association with RAG2 (14). We therefore asked whether bfRAG1L could also interact with RAG2. Although no RAG2-like gene has been identified in the Florida amphioxus genome (9), bfRAG1L could specifically interact with mouse core RAG2 (cRAG2), albeit with lower efficiency than tmRAG1, a bfRAG1L-like truncated form of mouse cRAG1 (comprising amino acid residues 508–820 of RAG1) (Fig. 3A). Moreover, bfRAG1L preferentially localized to the nucleus (in a similar manner to cRAG1 and tmRAG1) upon overexpression in HEK-293T cells (Fig. 3B). However, bfRAG1L failed to perform RSS-mediated recombination in the presence of mouse cRAG2 through cell-based bacterial colony assay as described (15), although nonsequence-specific DNA breaks were generated in the plasmid substrates (SI Appendix, Figs. S5 and S6).
Fig. 3.
bfRAG1L binds RAG2 and localizes to the nucleus. (A) Coimmunoprecipitation (Co-IP) assay to examine the interaction between bfRAG1L and cRAG2 in HEK-293T cells. IB, immunoblot; nt, N-terminal; tmRAG1, bfRAG1L-like truncation of mouse RAG1. (B) Subcellular localization of indicated GFP-tagged constructs in HEK-293T cells. The results are representative of three (A) or two (B) independent experiments.
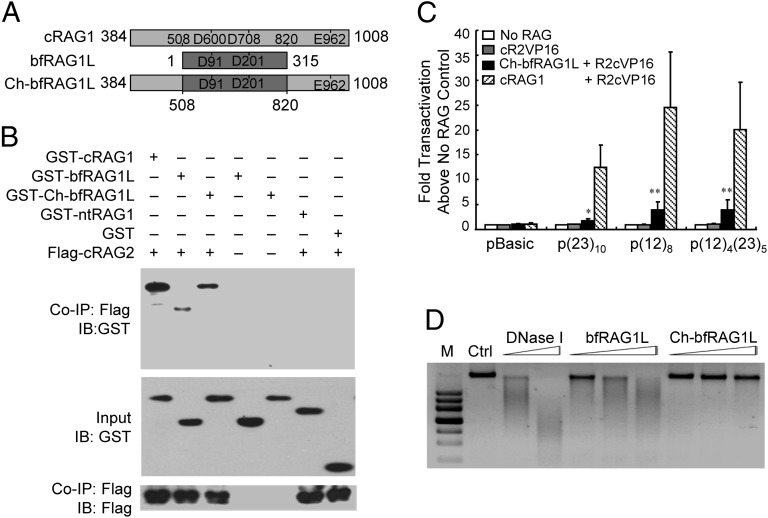
bfRAG1L Specially Recognizes RSS After Reconstitution.
As noted above, compared with cRAG1, bfRAG1L lacks the NBD and the C-terminal conserved region (Fig. 4A), which are responsible for RSS binding and dimerization (10), respectively. In addition, the C-terminal conserved region contains the third residue E of the triad DDE motif that is essential for RAG1 catalytic activity. We therefore wondered whether bfRAG1L could recognize RSSs and introduce RSS-specific DNA breaks when reconstituted with the two parts mentioned above to be a cRAG1-like protein, Ch-bfRAG1L (Fig. 4A). Upon reconstitution, the RAG2-binding activity of Ch-bfRAG1L was slightly increased, as demonstrated by co-IP (Fig. 4B). More importantly, Ch-bfRAG1L could recruit RAG2 to arrays of 12-RSSs and/or 23-RSSs in vivo, as assessed by using a one-hybrid assay, although its activity was lower than that of mouse cRAG1 (Fig. 4C and SI Appendix, Fig. S7). Notably, the non-DNA sequence-specific nuclease activity of Ch-bfRAG1L was significantly reduced compared with bfRAG1L, implying that either the N- or C-terminal regions of cRAG1 are required not only for RSS recognition but also for the restriction of the non-DNA sequence-specific DNA cleavage activity of cRAG1 (Fig. 4D).
Fig. 4.
bfRAG1L can recognize RSS after reconstitution. (A) Scheme of the constitution of Ch-bfRAG1L. (B) Co-IP assay to examine the interaction between Ch-bfRAG1L and cRAG2 in HEK-293T cells. IB, immunoblot; nt, N-terminal. (C) The recognition of RSS by Ch-bfRAG1L. The “fold transactivation” of luciferase activity was normalized against ”no RAG control” with the indicated plasmids (the means ± SD is calculated from triplicate experiments). Error bars reflect the SD. Asterisks indicate the P value by using the Student t test (*P < 0.05; **P < 0.01). (D) The nuclease activities of bfRAG1L and Ch-bfRAG1L on a genomic DNA substrate. Ctrl, represent untreated genomic DNA from 293T cell lines. The results are representative of two (B) or four (D) independent experiments.
Ch-bfRAG1L Introduces Gene Recombination in the Presence of RAG2.
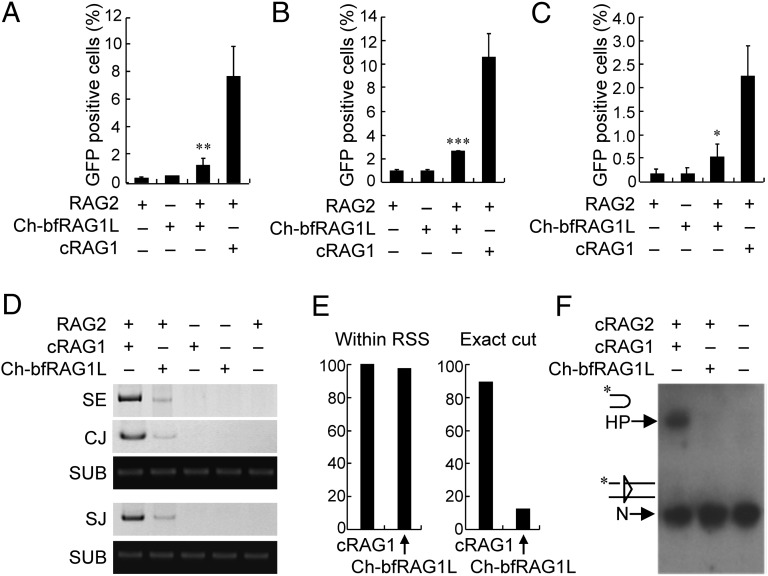
We next asked whether Ch-bfRAG1L could perform RSS-mediated gene recombination. Reporter substrates pCJGFP and pSJGFP were used to measure coding joint and signal joint formation activity, respectively, as described (16) (SI Appendix, Fig. S8 A and B). Together with RAG2, Ch-bfRAG1L was able to induce GFP-positive cells in HEK-293T cells, whereas neither Ch-bfRAG1L alone nor RAG2 alone could do so (Fig. 5 A and B). Furthermore, Ch-bfRAG1L was also able to mediate chromosomal DNA recombination, as demonstrated with the chromosomally integrated substrate pMX-INV in pre–B-cell lines (Fig. 5C and SI Appendix, Fig. S8C). Mutation of D217, D327, or E585 significantly attenuated the DNA recombination activity compared with the wild-type Ch-bfRAG1L (SI Appendix, Fig. S9), suggesting that the three key residues in DDE motif of Ch-bfRAG1L were critical for the optimal recombination activity. We subsequently characterized DNA cleavage by Ch-bfRAG1L using a cell-based PCR assay. These experiments demonstrated that in the presence of mouse RAG2, Ch-bfRAG1L could generate signal ends, signal joints, and coding joints when the plasmid substrates pJH290 or pJH289 were used, although at lower efficiency than mouse cRAG1 (Fig. 5D and SI Appendix, Fig. S8D). Further sequence analysis revealed that although only 13% of double-strand breaks were introduced at the expected site adjacent to the heptamer, more than 90% of the double-strand breaks were within the RSS when Ch-bfRAG1Lwas used (Fig. 5E); in addition, excessive deletion or insertion occurred during Ch-bfRAG1L–mediated coding joint formation in HEK-293T cells (SI Appendix, Table S3). Notably, the hairpin formation by Ch-bfRAG1L was barely detectable (Fig. 5F), which may be due to the low activity of Ch-bfRAG1L in this test.
Fig. 5.
Ch-bfRAG1L introduces RSS-mediated gene recombination. HEK-293T (A and B) or Raji (C) cells were transfected with the indicated plasmids, and the GFP level was subsequently measured by flow cytometry. The percentages of GFP+ cells are shown (the means ± SD is calculated from triplicate experiments). Error bars indicate the SDs. The asterisks reflect the values significantly different from RAG2 only (*P < 0.05; **P < 0.01; ***P < 0.001, by the Student t test). (D) PCR analysis of signal end (SE), coding joint (CJ), and signal joint (SJ) from Ch-bfRAG1L–mediated recombination on substrates pJH290 and pJH289. (E) Location analysis of cleavage by Ch-bfRAG1L and cRAG1. (F) In vitro cleavage assay. Nicked 12-RSS and 23-RSS substrates were incubated with purified RAG complex as indicated. The nicked substrate (N) and hairpin (HP) were shown on the left. The results are representative of three (D) or two (F) independent experiments.
Ch-bfRAG1L Mediates Antigen Receptor Gene Assembly in RAG1-Deficient Mice.
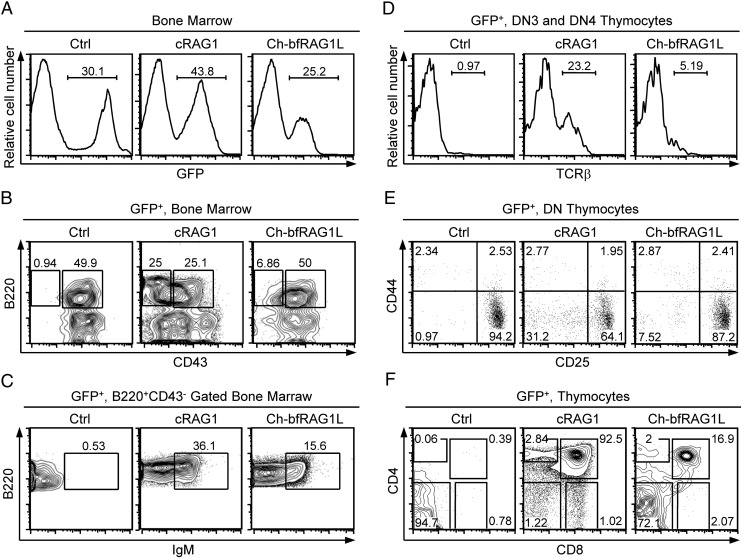
To assess the capacity of Ch-bfRAG1L to perform V(D)J gene recombination in vivo, bone marrow transplantation following retroviral infection of RAG1-deficient bone marrow cells was performed (17). RAG1-deficient mice are unable to rearrange Ig genes, and B-cell development is arrested at the B220+CD43+ pro–B-cell stage (18). RAG1-deficient pro-B cells infected with a cRAG1-expressing retrovirus were able to progress from the pro–B-cell stage to the pre-B stage (B220+CD43−) and beyond (B220+IgM+), which was also the case for cells transfected with Ch-bfRAG1L (Fig. 6 A–C), suggesting that functional heavy and light chains have been expressed.
Fig. 6.
Flow cytometric analysis of B-cell and T-cell development in Ch-bfRAG1L–transfected RAG1-deficient mice. (A) The percentages of GFP-positive bone marrow cells are shown. (B) GFP-positive bone marrow cells are analyzed for the surface markers B220 and CD43. B220+CD43+ pro-B and B220+CD43− pre–B-cell populations are shown. (C) Surface expression of IgM on GFP-positive B220+CD43− bone marrow cells. (D) Surface expression of TCRβ on DN3 and DN4 thymocytes. (E) Transduced DN subsets are presented by CD44 versus CD25 profiles. Numbers indicate the percentage of DN subsets from DN1 to DN4. (F) CD4 and CD8 expression on GFP-positive thymocytes; number represents the percentage of cells in that gate. Ctrl, bone marrow cells only expressing GFP protein. The results are representative of two (A–C) or three (D–F) independent experiments.
Thymocytes fail to undergo TCRβ and TCRα recombination and are arrested at the double negative 3 (DN3) stage in the absence of RAG1. Interestingly, TCRβ-positive cells were present in Ch-bfRAG1L-transfected and cRAG1-transfected DN3 and DN4 thymocytes, but they were not present in control cells (Fig. 6D and SI Appendix, Fig. S10A). Moreover, RAG1-deficient thymocytes were able to differentiate into DN4, DP, and later stages after transfected with Ch-bfRAG1L (Fig. 6 E and F). Thus, Ch-bfRAG1L might function in an analogous manner to cRAG1 by introducing V(D)J gene recombination in vivo, albeit at a lower efficiency. Notably, sequence analyses of DJ or VD coding joints showed that excessive deletion or insertion also occurred in Ch-bfRAG1L–mediated TCRβ gene recombination (SI Appendix, Fig. S10B and Tables S4 and S5).
Discussion
The study on the evolution of the adaptive immune system has been largely concentrated on the identification of ancestral RAG homologs (7, 9, 19, 20). Our findings indicate that the ectopic expressed product of the RAG1-like bfRAG1L from the invertebrate amphioxus exhibits nuclease activity. Importantly, bfRAG1L is functionally equivalent to the central domain of cRAG1 and is able to perform V(D)J recombination after reconstituting it into a cRAG1-like protein, Ch-bfRAG1L. However, it is noteworthy that both the efficiency and the accuracy of Ch-bfRAG1L–mediated recombination are lower than those observed for cRAG1, which could be due to many reasons, such as Ch-bfRAG1L fails to induce a precise conformational change near the active site along with mouse RAG2 and RSS and/or is deficient in hairpin formation during DNA cleavage (21).
SpRAG1L and SpRAG2L from the invertebrate sea urchin are similar in both sequence and genomic organization to the vertebrate RAG1 and RAG2 genes (7). However, SpRAG1L failed to recognize RSS and introduce gene recombination. Importantly, the reconstruction of SpRAG1L into mouse cRAG1-like protein by replacement of NBD failed to improve its ability to perform V(D)J recombination in the presence of mouse RAG2 or SpRAG2L (SI Appendix, Fig. S11), which might be due to the fact that SpRAG1L could not interact with mouse cRAG2 as reported (7). Although no RAG2-like gene could be identified in Florida amphioxus genome near the bfRAG1L DNA fragment because of the incompleted DNA sequence information (9), we observed that bfRAG1L bears closer homology to mouse RAG1 (35%) than SpRAG1L (26%), although bfRAG1L is ∼48% identical to SpRAG1L in terms of amino acid sequence homology (SI Appendix, Fig. S12). SpRAG1L and bfRAG1L might be distinct ancestors of jawed vertebrates RAG1 existing in invertebrates. Although bfRAG1L does not have a complete structure of RAG1 as SpRAG1L from sea urchin, it is more closely related to RAG1 with respect to the function of recombination activity in phylogeny of evolution.
Intriguingly, other data indicate that nuclease activity is a prerequisite, but is insufficient, for the experimental evolution of a functional RAG1. The retroviral type II nuclease BGLF5 was unable to introduce gene recombination after a cRAG1-like reconstruction (SI Appendix, Fig. S13). Amphioxus shares a common ancestor with modern-day vertebrates (22, 23), and our functional characterization of bfRAG1L is consistent with that observation.
Notably, the residue D176 in the DXN(D/E)XK motif is critical for the nuclease activity of bfRAG1L, whereas it is not conserved among vertebrate RAG1s. This phenomenon might be due to the fact that some other residues (even D91) substitute for the function of this D, or that this D is eliminated in the process of evolution, which, in turn, improves the accuracy of RAG1-mediated DNA recombination in vertebrates. Considering that the DXN(D/E)XK motif and the DDE motif in bfRAG1L share one key residue D201, it would be interesting to investigate whether the DXN(D/E)XK motif for nuclease activity evolves into a DDE motif important for V(D)J recombinase in the future.
With regards to the origin of RAG, the “RAG transposon” hypothesis states that RAG derived from a transposon by horizontal transfer into the genome of vertebrates, an idea supported by the observation that RAG1 and RAG2 can mediate a complete transposition reaction (24, 25). In comparison, the “viral recombination” hypothesis predicts that RAG is more likely to have evolved from a viral recombinase, including retroviral integrases, RNase H, and RNA-induced silencing complex (19, 20). Given that bfRAG1L bears homology to some viral RNA polymerases and contains the DXN(D/E)XK active site motif characteristic of many viral nucleases, we propose that bfRAG1L is a virus-related nuclease that resulted from the integration of the virus gene into the genome of amphioxus. However, bfRAG1L cannot recognize the RSS, implying that further evolution would have been needed for it to gain recombination activity. It was reported that the DNA binding domain of Tc1/mariner transposases (26, 27) or the N-terminal region of Transib TPases has sequence similarities to the RAG1 NBD (9), and recent observations suggest that RAG1 represents a functional fusion of separate proteins (9). Presumably, the NBD of RAG1 with specific RSS recognition may well result from the fusion of the DNA binding region of Transib Tpases into the upstream of bfRAG1L DNA fragment thereafter, given that bfRAG1L is located in a transcriptionally active region as evidenced by the expression of an antisense cDNA (information from the cDNA database of Florida amphioxus B. floridae). Together, our findings provide unique insights into the origin and evolution of RAG1.
Materials and Methods
Full methods are provided in the SI Appendix. Briefly, the different forms of bfRAG1L fragment were cloned into vector pET28A and expressed in Escherichia coli BL21 (DE3). The recombinant proteins were purified by Ni-affinity chromatography (GE Healthcare) for detecting nuclease activities. HEK-293T cells were transfected with the different sets of RAG expression vectors for testing protein–protein interaction, localization, and recombination activities. To examine the recombination activity of Ch-bfRAG1L in vivo, retroviral-mediated bone marrow gene transfer was introduced.
Supplementary Material
Acknowledgments
We thank Dr. Linda Holland for the amphioxus genomic DNA; Drs. David G. Schatz, David Roth, Stephen Desiderio, Barry P. Sleckman, and Sebastian D. Fugmann for providing recombination plasmids; Dr. Barry P. Sleckman for Raji cells; and Mi Liu for bioinformation support. This work was supported by National Basic Research Program of China Grants 2013CB835300 and 2012CB518700; National Natural Science Foundation of China Grants 30925031, 81261120380, and 30901305; and Shanghai Municipal Government Grant 12XD1405800.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318843111/-/DCSupplemental.
References
- 1.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3(11):890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 2.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 3.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22(10):1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBlane JF, et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83(3):387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 5.Lewis SM. The mechanism of V(D)J joining: Lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 6.Sato A, et al. Class I mhc genes of cichlid fishes: Identification, expression, and polymorphism. Immunogenetics. 1997;46(1):63–72. doi: 10.1007/s002510050243. [DOI] [PubMed] [Google Scholar]
- 7.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103(10):3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: New perspectives. Nat Rev Immunol. 2005;5(11):866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arbuckle JL, Fauss LA, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276(40):37093–37101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- 11.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 12.Buisson M, et al. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J Mol Biol. 2009;391(4):717–728. doi: 10.1016/j.jmb.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 13.De P, Peak MM, Rodgers KK. DNA cleavage activity of the V(D)J recombination protein RAG1 is autoregulated. Mol Cell Biol. 2004;24(15):6850–6860. doi: 10.1128/MCB.24.15.6850-6860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De P, Rodgers KK. Putting the pieces together: Identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 15.Kienker LJ, Shin EK, Meek K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000;28(14):2752–2761. doi: 10.1093/nar/28.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449(7161):483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 17.Persons DA, et al. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90(5):1777–1786. [PubMed] [Google Scholar]
- 18.Spanopoulou E, et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8(9):1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfus DH. The DDE recombinases: Diverse roles in acquired and innate immunity. Ann Allergy Asthma Immunol. 2006;97(5):567–576, quiz 576–578, 602. doi: 10.1016/S1081-1206(10)61083-6. [DOI] [PubMed] [Google Scholar]
- 20.Dreyfus DH. Immune system: Success owed to a virus? Science. 2009;325(5939):392–393. doi: 10.1126/science.325_392c. [DOI] [PubMed] [Google Scholar]
- 21.Kale SB, Landree MA, Roth DB. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol Cell Biol. 2001;21(2):459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124(4):815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Wada H, Satoh N. Details of the evolutionary history from invertebrates to vertebrates, as deduced from the sequences of 18S rDNA. Proc Natl Acad Sci USA. 1994;91(5):1801–1804. doi: 10.1073/pnas.91.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394(6695):744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 25.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94(4):463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 26.Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: The Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15(8):326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 27.Dreyfus DH. Evidence suggesting an evolutionary relationship between transposable elements and immune system recombination sequences. Mol Immunol. 1992;29(6):807–810. doi: 10.1016/0161-5890(92)90191-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.