Significance
Rheumatoid arthritis (RA) is an autoimmune disorder that affects ∼1% of the population. Macrophage- and fibroblast-like synoviocytes are thought to play central roles in the pathogenesis of RA. They have shared and distinct features defining RA pathology. However, these features have not been systematically explored. Using global gene expression profiling, we identified molecular signature and biological network models underlying the pathologic features of macrophage-like vs. fibroblast-like synoviocytes. Based on the network models, we selected key regulators, including POSTN and TWIST1, responsible for synoviocyte migration/invasion causing joint destruction, and their validity was experimentally confirmed. Our findings provide a comprehensive and systematic basis for mechanisms explaining RA pathogenesis and also for identification of therapeutic targets for RA.
Abstract
Rheumatoid synoviocytes, which consist of fibroblast-like synoviocytes (FLSs) and synovial macrophages (SMs), are crucial for the progression of rheumatoid arthritis (RA). Particularly, FLSs of RA patients (RA-FLSs) exhibit invasive characteristics reminiscent of cancer cells, destroying cartilage and bone. RA-FLSs and SMs originate differently from mesenchymal and myeloid cells, respectively, but share many pathologic functions. However, the molecular signatures and biological networks representing the distinct and shared features of the two cell types are unknown. We performed global transcriptome profiling of FLSs and SMs obtained from RA and osteoarthritis patients. By comparing the transcriptomes, we identified distinct molecular signatures and cellular processes defining invasiveness of RA-FLSs and proinflammatory properties of RA-SMs, respectively. Interestingly, under the interleukin-1β (IL-1β)–stimulated condition, the RA-FLSs newly acquired proinflammatory signature dominant in RA-SMs without losing invasive properties. We next reconstructed a network model that delineates the shared, RA-FLS–dominant (invasive), and RA-SM–dominant (inflammatory) processes. From the network model, we selected 13 genes, including periostin, osteoblast-specific factor (POSTN) and twist basic helix–loop–helix transcription factor 1 (TWIST1), as key regulator candidates responsible for FLS invasiveness. Of note, POSTN and TWIST1 expressions were elevated in independent RA-FLSs and further instigated by IL-1β. Functional assays demonstrated the requirement of POSTN and TWIST1 for migration and invasion of RA-FLSs stimulated with IL-1β. Together, our systems approach to rheumatoid synovitis provides a basis for identifying key regulators responsible for pathological features of RA-FLSs and -SMs, demonstrating how a certain type of cells acquires functional redundancy under chronic inflammatory conditions.
Rheumatoid arthritis (RA) is a common autoimmune disorder that afflicts ∼1% of the population. Despite the advent of anticytokine therapies that ameliorate the inflammatory manifestations of disease, there is no cure, and the pathogenesis of RA is not fully understood. In RA joints, various inflammatory cells, including innate immune cells (e.g., mast cells, macrophages, dendritic cells, and natural killer cells), adaptive immune cells (T and B cells), endothelial cells, and fibroblast-like synoviocytes (FLSs), are activated (1). Identification of the major roles of the participating cells has been a key issue in understanding RA pathogenesis. Evidence is emerging that rheumatoid synoviocytes, consisting of macrophage-like synoviocytes (MLSs) and FLSs, play a central role in the pathogenesis of RA (2). These cells are the major constituents of the synovial lining layer and proactively participate in inflammatory cascades and cartilage/bone destruction (2).
MLSs and FLSs share many pathologic functions for the initiation and perpetuation of RA as innate immune cells, although they originate differently from myeloid and mesenchymal stem cells, respectively (2, 3). MLSs produce proinflammatory cytokines [e.g., tumor necrosis factor-alpha (TNF-α) and interleukin-1-beta (IL-1β)], proteases, and prostaglandins, which lead to inflammatory cascades in the joints. FLSs are similar to MLSs in that they aggressively generate diverse cytokines/chemokines (e.g., IL-6, -8, and monocyte chemotactic protein 1), proteolytic enzymes, and prostaglandins (2). Conversely, MLSs and FLSs also show distinct pathologic features in vitro. MLSs phagocytose cell debris and wastes in the synovial fluid and possess professional antigen presentation capacity (2, 3). FLSs of RA patients (RA-FLSs) can migrate/attach to cartilage and bone and invade the local environment (2, 4). Moreover, RA-FLSs proliferate abnormally and exhibit several oncogenes or tumor suppressor genes, including H-ras and p53, harboring somatic mutations (5, 6). Despite the knowledge provided by these studies (1–6), it is not fully resolved how RA-FLSs exhibit aggressive and invasive phenotypes.
Both MLSs and FLSs are exposed persistently to proinflammatory cytokines, growth factors, and hypoxia in vivo (in the RA joints). A number of innate and adaptive immune cells interact via an array of cytokines and/or cell-to-cell contacts, which also can similarly activate both MLSs and FLSs, leading to the secretion of different and common cytokines/chemokines, growth factors, and other inflammatory mediators (7). Because of the common secretory factors, many biologic functions overlap between MLSs and FLSs (2, 3, 8). This complexity presents challenges in determining the specific roles of MLSs and FLSs and their interplay during the progression of RA. Comparative and unbiased analyses of gene expression profiles in different cell types (9), as well as a computational framework for removing the effects of sample heterogeneity (10), may help to identify distinct and shared molecular signatures involving RA pathogenesis.
The inflammatory role of MLSs in RA, in addition to the potential of FLSs to migrate and perform matrix remodeling, has been reported individually (2, 11). However, an unbiased and systematic study to determine molecular signatures and biological networks of MLSs and FLSs underlying their shared and unique roles in RA pathogenesis has not been explored. Therefore, a comprehensive and integrative understanding of synoviocyte biology in RA has not been well established, and the discovery of key molecules specifically targeting RA-FLSs has been unsuccessful. Here, we performed global transcriptome profiling of RA-FLSs and synovial macrophages (SMs) isolated from RA patients. A comparative analysis of the transcriptomes from RA-FLSs and -SMs revealed molecular signatures defining invasiveness of RA-FLSs and inflammatory potential of RA-SMs, respectively. Interestingly, after the exposure to IL-1β, RA-FLSs underwent a functional transition to acquire inflammatory potential dominant in RA-SMs, while keeping their original invasive potential. We next generated a network model delineating distinct and shared features of RA-FLSs and -SMs. Based on this network model, two potential key regulators, the twist basic helix–loop–helix transcription factor 1 (TWIST1) and the periostin, osteoblast-specific factor (POSTN), responsible for FLS migration and invasion, were selected, and their validity was confirmed in vitro and in vivo. The findings suggest that both molecular signatures and the network model provide a basis for mechanisms highlighting distinct vs. shared pathological features of RA-FLSs and -SMs with different cellular origin and could be used to identify potential therapeutic targets for RA.
Results
Molecular Signatures Defining Invasive Potential of RA-FLSs and Inflammatory Potential of SMs.
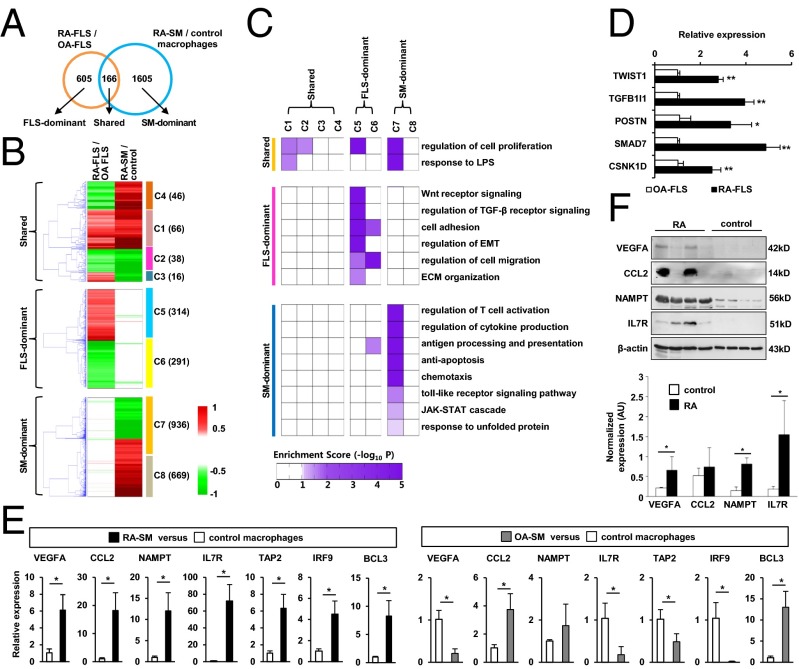
To identify molecular signatures of FLSs and MLSs in RA joints, we isolated FLSs from synovial tissues of RA and osteoarthritis (OA) patients, obtained SMs from synovial fluid of RA patients, and differentiated control macrophages from peripheral blood of healthy subjects. OA is a representative of noninflammatory degenerative arthritis. The primary defect of OA is in cartilage rather than in synovium (12), and thus OA-FLSs have been widely used for comparison with RA-FLSs. In the present study, global gene-expression profiling of those cells was performed by using the Illumina microarray platform. By comparing the gene-expression profiles, we identified 771 differentially expressed genes (DEGs) between RA- and OA-FLSs with P < 0.05 using integrative statistical testing (13), and we also identified 1,771 DEGs between RA-SMs and healthy macrophages (Fig. 1A). Out of the total 2,376 DEGs in RA-FLSs and SMs, a relatively small portion of the DEGs (166 genes; 7%) were shared by both cell types, whereas most of the genes (605/771 DEGs in RA-FLS and 1,605/1,771 DEGs in RA-SM) were predominantly regulated by each cell type (Fig. 1A). These DEGs were grouped into eight clusters (C1–C8) based on their cell-type-dependent differential expression patterns. The eight clusters could be categorized into shared clusters (C1∼C4) dysregulated in both RA-FLSs and -SMs, FLS-dominant clusters (C5 and C6) dysregulated predominantly in FLSs, and SM-dominant clusters (C7 and C8) dysregulated predominantly in RA-SMs (Fig. 1B). These data show that RA-FLSs and SMs have distinct gene expression patterns, indicating that there are unique cellular functions for either RA-FLSs or SMs.
Fig. 1.
Comparison of gene expression profiles of RA-FLSs and SMs. (A) Relationships between two sets of DEGs from RA-FLSs/OA-FLSs and RA-SMs/healthy controls: (i) shared, (ii) FLS-dominant, and (iii) SM-dominant DEGs. (B) Eight clusters of the two sets of DEGs. Red and green denote increase and decrease in mRNA expression, respectively. The dendrogram shows how individual genes in each group are clustered by hierarchical clustering. (C) Cellular processes (rows) enriched by the DEGs in each cluster (columns), which were grouped into three categories: the processes commonly enriched by shared DEGs (shared) and those specifically enriched by FLS-dominant or SM-dominant DEGs. Color gradient represents –log10(P), where P is the enrichment P value from DAVID software. (D and E) Quantitative RT-PCR assays for representative DEGs of FLS-dominant (D) and SM-dominant (E) processes. FLSs (n = 3), RA-SMs (n = 3), and OA-SMs (n = 9) were freshly isolated from synovial tissues of RA and OA patients. Control macrophages (n = 3) were differentiated from peripheral blood monocytes of healthy controls. Data show mean ± SEM. *P < 0.05; **P < 0.01. (F, Upper) Protein expression of proinflammatory genes, including VEGFA, CCL2, NAMPT, and IL7R, in macrophages of RA patients (n = 4) and healthy controls (n = 4), as determined by Western blot analysis. (Lower) A bar graph shows the optical density ratio (proinflammatory genes/β-actin) between RA macrophages and control macrophages. *P < 0.05 vs. normal macrophages.
To investigate the cellular processes specific to each cell type, we performed functional enrichment analysis of the DEGs in the eight clusters using DAVID software (14). The cellular processes enriched by the genes in each cluster were further categorized according to the cell type dominancy: (i) shared, (ii) FLS-dominant, and (iii) SM-dominant processes (Fig. 1C). As expected, key cellular processes associated with RA pathology were enriched mostly by the genes in the up-regulated gene clusters (C1, C5, and C7). Of note, the genes in FLS-dominant up-regulated clusters (C5) were involved uniquely in cell-invasion-related processes, such as Wnt and TGF-β signaling, cell migration and adhesion, epithelial-to-mesenchymal transition (EMT), and extracellular matrix (ECM) organization. In contrast, the genes in SM-dominant up-regulated clusters (C7) were predominantly associated with inflammatory response-related processes, including regulation of T-cell activation, cytokine production, antigen presentation, chemotaxis, Toll-like receptor signaling, and the JAK-STAT cascade. As we reported (15), endoplasmic reticulum stress-related processes, such as antiapoptosis and unfolded protein response, were also functionally enriched by the genes in SM-dominant up-regulated clusters. Conversely, SI Appendix, Fig. S1 shows cellular processes enriched by the genes in the down-regulated gene clusters (C3, C4, C6, and C8). FLS-dominant down-regulated genes (C6) were specifically involved in tissue regeneration and growth. The lipid-associated cellular processes were enriched by the SM-dominant down-regulated cluster (C8).
Because OA is not a systemic inflammatory disease, in contrast to RA, inflammatory gene signatures in peripheral blood mononuclear cells of OA patients are not significantly different from those of healthy individuals (16, 17). We tried to validate differential expression of the selected 12 genes from FLS- and SM-dominant up-regulated clusters (C5 and C7) in independent RA-FLS and -SM samples obtained from different RA and OA patients and healthy controls using quantitative real-time PCR assays. To this end, we freshly isolated tissue macrophages from OA synovium and immediately obtained mRNA from the cells. The selected genes include five genes involved in cell invasion in C5 and seven genes involved in inflammatory response and cell survival in C7. As shown in Fig. 1D, increased mRNA expression levels of TWIST1, transforming growth factor beta 1 induced transcript 1 (TGFB1I1), POSTN, SMAD family member 7 (SMAD7), and CSNK1D genes were confirmed in RA-FLSs (n = 3), compared with OA-FLSs (n = 3). We also observed that independent RA-SMs (n = 3) exhibited higher mRNA expression levels of vascular endothelial growth factor A (VEGFA), chemokine (C-C motif) ligand 2 (CCL2), nicotinamide phosphoribosyltransferase (NAMPT), interleukin 7 receptor (IL7R), TAP2, and IRF9 than independent normal macrophages (n = 3) and OA synovial tissue macrophages (n = 9) (Fig. 1E). This difference was similarly noted between RA (n = 6) and OA (n = 6) tissue macrophages (SI Appendix, Fig. S2), but it was not observed between macrophages differentiated from normal peripheral monocytes and those isolated from OA tissues (Fig. 1E). Higher expression levels of VEGFA, CCL2, and IL7R in RA-SMs (n = 4) than in normal macrophages (n = 4) were confirmed by Western blot analysis performed in independent samples (Fig. 1F).
Inflammatory Signatures in RA-FLSs Additionally Acquired by IL-1β Stimulation.
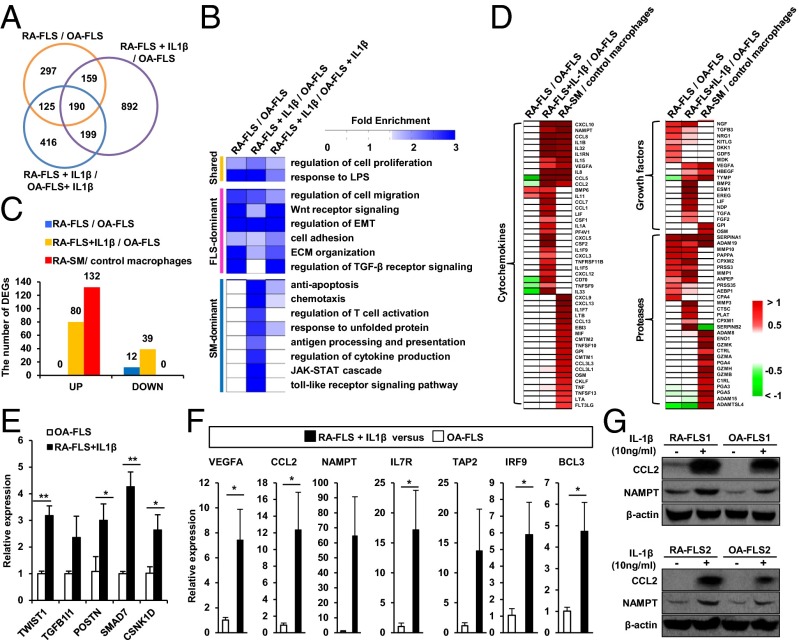
RA-FLSs become quiescent by the third passage after the isolation from RA joints, but their phenotype can be rapidly restored by exposure to proinflammatory cytokines, such as IL-1β and TNF-α (2). However, molecular signatures representing functional transitions of RA-FLSs to acquire the rheumatoid phenotype after cytokine stimulation have not been systemically analyzed. To address this issue, we stimulated FLSs with IL-1β and then analyzed gene-expression profiles of both IL-1β–stimulated RA- and OA-FLSs. Using the gene expression data, we identified 1,440 DEGs in the IL-1β–stimulated RA-FLSs compared with OA-FLSs and 930 DEGs in the IL-1β–stimulated RA-FLSs compared with the IL-1β–stimulated OA-FLSs (Fig. 2A and Dataset S1). The 349 (190 + 159 genes in Fig. 2A) overlapping genes indicate that RA-FLSs retain their original characteristics even after the IL-1β stimulation. Furthermore, 1,091 genes (892 + 199 genes in Fig. 2A and Dataset S1) were additionally changed in RA-FLSs by the IL-1β stimulation. The evidence concerning these 1,091 genes supports the view that RA-FLSs can newly acquire additional functions by IL-1β stimulation.
Fig. 2.
Molecular signatures underlying IL-1β–stimulated RA-FLS. (A) Relationships of the three sets of DEGs from the comparisons of RA-FLS/OA-FLSs, RA-FLS+IL-1β/OA-FLSs, and RA-FLS+IL-1β/OA-FLS+IL-1β. (B) Cellular processes (rows) enriched by the three sets of the DEGs (columns), which are grouped by shared, FLS-, and SM-dominant processes, as in Fig. 1C. The color gradient represents fold enrichment score, which is the enrichment degree of given gene ontology biological processes from the DAVID software. (C) Relative proportions of the DEGs involved in SM-dominant processes in RA-FLSs, IL-1β–stimulated RA-FLSs, and RA-SMs. (D) Up-regulated (red) genes with cytokine, growth factor, and protease activities in RA-FLSs, IL-1β–stimulated RA-FLSs, and RA-SMs. Color gradient, log2-fold-changes. (E and F) Quantitative RT-PCR assays for 12 representative DEGs associated with FLS-dominant (E) and SM-dominant (F) processes in RA-FLSs (n = 3) stimulated with IL-1β (10 ng/mL) for 24 h and OA-FLSs (n = 3). Data show mean ± SEM. *P < 0.05; ** P < 0.01. (G) Expressions of CCL2 and NAMPT protein in RA-FLSs vs. OA-FLSs. FLSs were stimulated without or with IL-1β (10 ng/mL) for 24 h, and CCL2 and NAMPT expression levels were determined by Western blot analysis.
To examine cellular processes that RA-FLSs either keep or newly acquire after the IL-1β stimulation, we performed functional enrichment analysis of the DEGs from the comparison of RA-FLS+IL-1β/OA-FLS and RA-FLS+IL-1β/OA-FLS+IL-1β using DAVID software. Additionally, the resulting processes enriched by these two sets of the DEGs were compared with those enriched by the shared, FLS-dominant, and SM-dominant clusters described in Fig. 1C. The data showed that regardless of IL-1β stimulation, FLS-dominant cellular processes—including cell migration, Wnt receptor signaling, regulation of EMT, adhesion, and ECM organization—were commonly enriched by the DEGs between RA- and OA-FLSs (Fig. 2B), suggesting that RA-FLSs stably keep their intrinsic characteristics even after cytokine stimulation. Of note, the SM-dominant cellular processes (Fig. 1C)—including chemotaxis, regulation of T-cell activation, antigen presentation, cytokine production, and Toll-like receptor pathway—were newly enriched by the DEGs in IL-1β–stimulated RA-FLSs, but not in unstimulated RA-FLSs (Fig. 2B), suggesting that IL-1β stimulation confers SM-dominant proinflammatory features to RA-FLSs. Furthermore, the number of inflammatory response genes involved in SM-dominant cellular processes was significantly increased (from 0 to 80) in RA-FLSs after the IL-1β stimulation (Fig. 2C). Together, these data suggest that RA-FLSs dynamically obtain the proinflammatory potential without losing their invasive properties after the IL-1β stimulation.
The proinflammatory networks in the joints play essential roles in the perpetuation of RA (1). Among the DEGs in IL-1β–stimulated RA-FLSs, 1,091 genes (892 + 199 genes in Fig. 2A) included a variety of proinflammatory cytokines/chemokines, growth factor, and proteases. To understand the quality of proinflammatory DEGs in IL-1β–stimulated RA-FLSs, we compared DEGs newly acquired after IL-1β stimulation with those originally identified in RA-SMs. The result showed that some interleukins (e.g., IL-1B, -1RN, -8, -15, and -32) and chemokines (e.g., CCL2, CCL5, and CXCL10) were commonly up-regulated in both IL-1β–stimulated RA-FLSs and -SMs, whereas other cytokines and chemokines (e.g., CCL7, CXCL12, and IL-33) were specifically increased only in IL-1β–stimulated RA-FLSs (Fig. 2D), but not in RA-SMs. This trend was similarly noted for growth factors and proteases (Fig. 2D). These data indicate that, in addition to the intrinsic invasive properties, RA-FLSs play an additional role in inflammatory cascades by generating a unique profile of cytokines/chemokines, growth factors, and proteases upon IL-1β stimulation.
We then confirmed differential expression of the selected 12 genes in independent RA-FLS samples (n = 3) stimulated by IL-1β using quantitative real-time PCR assays. The 12 genes consist of 5 genes involved in FLS-dominant cellular processes (Fig. 2E) and 7 genes involved in SM-dominant cellular processes (Fig. 2F). Increased expression levels of CCL2 and NAMPT protein were also noted in RA-FLSs stimulated without and/or with IL-1β, as determined by Western blot analysis (Fig. 2G).
Biological Network Representing Molecular Signatures of Invasive and Inflammatory Potentials of FLSs.
Biological networks delineating key cellular processes are essential to understand individual and shared roles of RA-FLSs and SMs in the RA pathogenesis at the molecular level (9). Based on our data (Fig. 1C), RA-FLSs and SMs contribute to the key pathogenic processes related to cell proliferation (shared process), invasion (FLS-dominant process), and inflammation (macrophage-dominant process). To examine the networks defined collectively by RA-FLSs and SMs, we reconstructed a network model capable of delineating the shared, FLS-, and SM-dominant processes by analyzing the up-regulated DEGs in RA-FLSs, -SMs, and IL-1β–stimulated RA-FLSs and by integrating the DEGs with protein–protein interactions (PPIs) obtained from PPI interactome databases (SI Appendix, Fig. S3A; see details in SI Appendix, Table S1). To systematically examine the relative contribution of RA-FLSs and SMs defining the key cellular processes in the network model, we grouped the DEGs into 16 modules representing the shared, FLS-, and SM-dominant processes, based on Gene Ontology (GO) biological processes of the DEGs. We then computed module enrichment scores (see details in SI Appendix, SI Materials and Methods) that represent the contribution of RA-FLSs and SMs to individual modules (SI Appendix, Fig. S3B; see details in SI Appendix, Table S2). The four invasion-related modules—including ECM organization (ECM), regulation of EMT (EMT), WNT signaling (WNT), and cell adhesion and migration (CAM)—were preferentially regulated by RA-FLSs, but not by RA-SMs (SI Appendix, Fig. S3A and the first plot in SI Appendix, Fig. S3B). In contrast, the other 12 inflammation-related modules were preferentially regulated by RA-SMs (SI Appendix, Fig. S3A and the third plot in SI Appendix, Fig. S3B). Upon IL-1β stimulation, RA-FLSs additionally acquired most of the inflammation-related modules, except cytoskeleton organization and leukocyte activation (SI Appendix, Fig. S3A and the second plot in SI Appendix, Fig. S3B).
Selection of Potential Regulators for FLS Migration and Invasiveness.
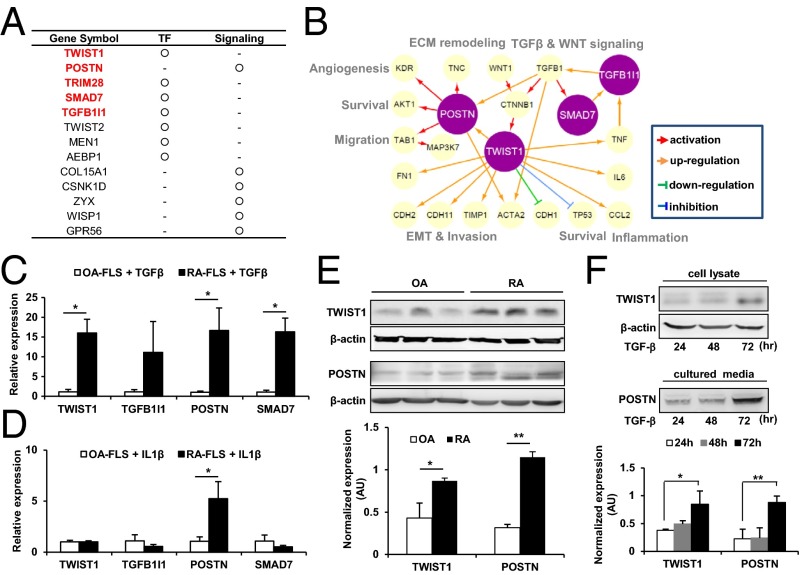
FLSs are the major component of invasive pannus. They migrate and attach to cartilage and bone and then destroy them by secreting proteases within the RA joints (2, 4). Thus, decoding the mechanisms underlying the migration and invasion of RA-FLSs is essential to the understanding of RA pathogenesis and for the identification of therapeutic targets for RA. However, therapeutic agents that emasculate the aggressive phenotype of RA-FLSs have not been tried. To address this issue, we attempted to determine key factors controlling the migration and invasiveness of RA-FLSs. As noted above, the up-regulated genes in RA-FLSs (Fig. 1C) and IL-1β–stimulated RA-FLSs (Fig. 2B) were associated with the migration and invasion of RA-FLSs. Thus, we first focused on the 946 genes up-regulated in either RA-FLSs or IL-1β–stimulated RA-FLSs (SI Appendix, Fig. S4A). Using the network model, we further selected 92 of 946 genes, which belong to the modules related to the invasive potential of RA-FLSs, including ECM organization, regulation of EMT, WNT signaling, and cell adhesion/migration modules (SI Appendix, Fig. S3B). We next examined whether the 92 genes had been implicated in RA using the Genotator database (18). As a result, 56 of 92 genes had no previously reported association with RA pathogenesis. Among the 56 genes, we finally selected 13 genes with regulator activities (e.g., transcriptional regulators and/or signaling molecules) as key regulator candidates responsible for the migration and invasion of RA-FLSs, based on GO terms (Fig. 3A and SI Appendix, Fig. S4B).
Fig. 3.
Selection of potential regulators for FLS invasion. (A) Thirteen potential regulators (transcription factor, TF, or signaling molecule) for migration and invasion of RA-FLS. Five regulators associated with EMT are marked in red. (B) A network model describing regulatory interrelationships of POSTN, TWIST1, SMAD7, and TGFB1I1 and their associated processes. The arrows denote regulator–target gene relationships (see legends at the bottom). (C and D) Comparison of mRNA expression of TWIST1, TGFB1I1, POSTN, and SMAD7 between RA-FLS and OA-FLS using RT-PCR analysis. Independent RA-FLS (n = 3) and OA-FLS (n = 3) were cultured with TGF-β (10 ng/mL) (C) or IL-1β (10 ng/mL) (D) for 24 h. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Data show mean ± SD. (E and F) TWIST1 and POSTN protein levels measured in independent RA-FLSs (n = 3) and OA-FLSs (n = 3) in the absence (E) or presence (F) of TGF-β (10 ng/mL) using Western blot analysis. β-actin was used as an internal control. Data show mean ± SEM. *P < 0.05; **P < 0.01.
Among the four modules related to the invasive potential of RA-FLSs in the above network model, regulation of EMT has been most closely associated with the invasion of cancer, including breast cancer, glioblastoma, and esophageal cancer (19–21). Interestingly, in our network model, 5 of 13 candidates (TWIST1, POSTN, TFGB1I1, TRIM28, and SMAD7) were involved in regulation of EMT (Fig. 3B). However, the role of those five candidates in RA pathogenesis has never been studied. Thus, we tried to define cellular processes to be controlled by five candidates by searching their links to their target genes using the iHOP software (22) and then reconstructed a hypothetical subnetwork model that summarized the relationships between the regulators and their target genes (Fig. 3B and SI Appendix, Table S3). This subnetwork model hypothesizes that POSTN and TWIST1 may regulate the target genes (e.g., ACTA2) involved in invasion-related processes, including migration, EMT, ECM remodeling, and angiogenesis. In addition, it suggests that POSTN, TWIST1, TGFB1I1, and SMAD7 might modulate each other and could be regulated by TGF-β, which promotes EMT (23, 24); TRIM28 was excluded from this network model because of the lack of direct association with the other four candidates, despite its putative role as a transcriptional regulator of EMT (25).
To test our hypothesis on the above subnetwork model, we examined the mRNA expression levels of TWIST1, POSTN, TGFB1I1, and SMAD7 in TGF-β–stimulated FLSs. As shown in Fig. 3C, the mRNA expression levels of TWIST1, POSTN, TGFB1I1, and SMAD7 were significantly increased in RA-FLSs (n = 3) compared with OA-FLSs (n = 3) 24 h after stimulation with 10 ng/mL TGF-β. In contrast, under IL-1β–stimulated conditions, the mRNA expression levels of TWIST1 and POSTN, but not those of TGFB1I1 and SMAD7, were selectively increased in RA-FLSs (Fig. 3D), suggesting that TWIST1 and POSTN may play a more specific role for IL-1β–driven inflammation. We further examined the basal protein expression levels of TWIST1 and POSTN in FLSs by Western blotting. As expected, RA-FLSs (n = 3) showed a higher level of TWIST1 and POSTN protein expressions than OA-FLSs (n = 3) (Fig. 3E). In addition, TGF-β (10 ng/mL) time-dependently increased TWIST1 and POSTN protein expressions in cell lysate and culture supernatants of RA-FLSs, respectively (Fig. 3F). Collectively, our subnetwork model and expression analysis revealed potential functional links of the two regulators, TWIST1 and POSTN, to the invasion-related genes and their associated processes, suggesting their regulatory capability for FLS migration and invasion in RA.
POSTN and TWIST1 Regulate FLS Migration and Invasion.
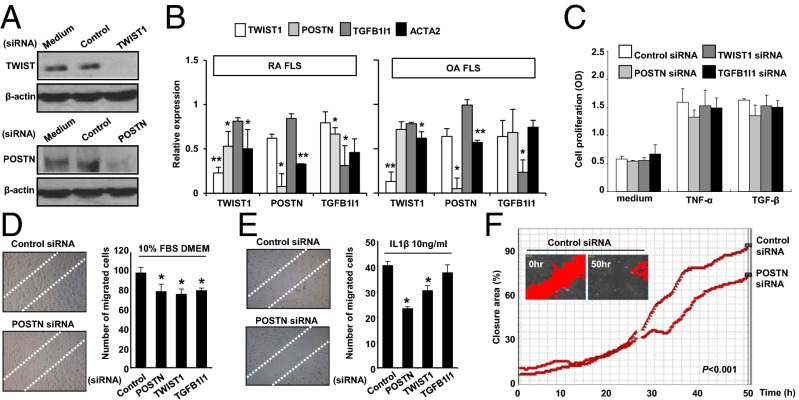
Finally, we wanted to confirm the effect of TWIST1 and POSTN on migration and invasion of IL-1β–stimulated FLS in vitro. To this end, we introduced small interfering RNA (siRNA) for POSTN or TWIST1 into FLSs; siRNA for TGFB1I1 was also used as a control because TGFB1I1 expression was increased by TGF-β, but not by IL-1β (Fig. 3C). The knockdown of transcript for each gene was confirmed by Western blot and real-time PCR analysis (Fig. 4 A and B). Interestingly, knockdown of POSTN, TWIST1, or TGFB1I1 transcript resulted in a marked decrease in mRNA level of ACTA2, which is critical for cell migration (26), in RA-FLSs. In contrast, reduction of ACTA2 mRNA expression was modest in OA-FLSs transfected with POSTN, TWIST1, or TGFB1I1 siRNA (Fig. 4B). Moreover, knockdown of POSTN transcript decreased the TWIST1 mRNA expression in both RA- and OA-FLSs and vice versa (Fig. 4B). However, knockdown TGFB1I1 did not influence gene expression of POSTN and TWIST1. Together, these results support molecular interactions among POSTN, TWIST1, and ACTA2 as suggested in our subnetwork model (Fig. 3B).
Fig. 4.
POSTN and TWIST1 promote migration of RA-FLSs. (A) Confirmation of knockdown of POSTN or TWIST1 at the protein level using Western blot analysis at 48 h after transfecting RA-FLSs with the siRNA. (B) Quantitative RT-PCR assays of mRNA expression of TWIST1, POSTN, and TGFB1I1 in RA-FLSs (Left; n = 3) and OA-FLSs (Right; n = 3) transfected with their siRNAs. Data show mean ± SD of relative mRNA expression in the target gene siRNA compared with the control siRNA. (C) Proliferation of RA-FLSs transfected with siRNA of TWIST1, POSTN, or TGFB1I1 at 72 h after TGF-β (10 ng/mL) and TNF-α (10 ng/mL) stimulations. The proliferation rate was determined by BrdU incorporation assay. Data are means ± SD of three independent experiments performed in triplicate. (D and E) Suppression of wound migration of RA-FLSs by siRNA for POSTN, TGFB1I1, or TWIST1. After 12 h of the transfection with siRNA, FLSs were incubated in DMEM containing 10% FCS for 36 h (D) or 10 ng/mL IL-1β for 24 h (E). Cells migrating beyond the reference line were photographed and counted. Data show mean ± SEM. *P < 0.05. (F) Dynamic quantification of the rate of wound closure. Confluent cultures of RA-FLSs transfected with control or POSTN siRNA were scratched, and wound closure was examined over 50 h. Images were taken with Cell-IQ. A full kinetics profile of cell migration was obtained with the accurate classification of the status of cells present in the population by using the automated Analyzer software. The P value was obtained by linear regression analysis in which the difference in closure area (percent) in the y axis between the two groups was used as a dependent variable and the time in the x axis as an independent variable.
The migration and invasion of FLSs may be accompanied by FLS proliferation in RA joints (27, 28). Thus, we next investigated whether the two regulators, POSTN and TWIST1, mediate TNF-α– or TGF-β–induced synoviocyte proliferation using the BrdU incorporation assays (Fig. 4C). As a result, POSTN, TWIST1, and TGFB1I1 siRNA showed no significant effect on TNF-α– or TGF-β–induced proliferation of RA-FLSs. In contrast, knockdown of POSTN, TWIST, or TGFB1I1 transcript suppressed the wound migration of RA-FLSs stimulated with culture medium containing 10% (vol/vol) FBS (Fig. 4D), which is consistent with earlier reports showing that POSTN and TWIST1 promote migration and invasion of several types of cancer cells (19–21). The IL-1β–induced FLS migration was also mitigated by either POSTN or TWIST1 siRNA, but it was not affected by TGFB1I1 siRNA (Fig. 4E), which paralleled the quantitative PCR data on POSTN and TWIST1 (Fig. 3C). Additionally, we further investigated the real-time migration of FLS over 50 h. There was a significant reduction of wound closure rate in the cells transfected with POSTN siRNA (Fig. 4F and Movie S1), confirming that POSTN regulates FLS migration. Moreover, the IL-1β–stimulated invasion of RA-FLSs in the Matrigel chamber was also markedly impeded by either POSTN or TWIST1 siRNA (SI Appendix, Fig. S5 A and B). Collectively, these data demonstrate that POSTN and TWIST1 are potent regulators promoting the migration and invasion of RA-FLSs stimulated with IL-1β.
Finally, we performed the in vivo experiment to confirm the regulation of FLS migration by POSTN or TWIST1. To this end, we induced skin inflammation by s.c. injecting complete Freund’s adjuvant (CFA) into immune-deficient (SCID) mice. We found that POSTN siRNA or TWIST1 siRNA significantly reduced FLS migration toward the CFA injection site, suggesting that POSTN and TWIST1 siRNA control FLS migration in vivo (SI Appendix, Fig. S5 C–E; for more details, see SI Appendix, SI Results).
Discussion
Diverse types of cells participate in initiation and perpetuation of RA. Identification of novel regulators responsible for the pathologic role of a certain type of cells is essential in better understanding RA pathogenesis and in developing new therapeutic targets. However, because of the complex intercellular interactions, it is challenging to determine the specific roles of individual cells and their interplays during the progression of RA. Among these cells, MLSs and FLSs actively participate in the progression of RA by secreting diverse cytokines/chemokines, proteases, and lipid mediators (2, 11). However, molecular signatures and biological networks underlying the distinct vs. shared features of FLSs and MLSs remain unclear. To systematically tackle these problems, we performed gene expression profiling of the two types of cells with different origin, FLSs and SMs. The comparative analysis of gene expression profiles revealed distinct vs. shared molecular signatures of RA-FLSs and -SMs, explaining how RA-FLSs plays pathologic roles in RA with both invasive and proinflammatory properties. Moreover, network analysis of the signatures provided the model defining the relationships among the signatures and their associated processes. In particular, through an unbiased analysis of gene signatures in FLSs and SMs, as well as dynamic gene profiling of IL-1β–stimulated FLSs, we successfully established a unique molecular network for FLS invasiveness and also identified key regulators for FLS migration and invasion, including POSTN and TWIST1.
By systematically analyzing modules or specific genes in the network, we selected 13 key regulator candidates, including POSTN and TWIST1, associated with FLS invasiveness, which is an essential event in RA progression. The in vitro assays confirmed that POSTN and TWIST1 expressions were elevated in RA-FLSs and were crucial for migration and invasion of FLSs stimulated with IL-1β. Moreover, regulation of FLS migration by POSTN and TWIST1 was confirmed in the in vivo animal model of skin inflammation. Overall, these results suggest that POSTN and TWIST1 could be potential therapeutic targets for RA. Similarly, the effect of the other 11 regulator candidates on FLS invasion can be tested by the in vitro and in vivo functional assays (see SI Appendix, SI Discussion for background information on POSTN and TWSTI).
In the present study, some cytokines and chemokines (e.g., CCL7, CXCL12, and IL-33), growth factors (e.g., NGF, ESM1, and EREG), and proteases [e.g., matrix metalloproteinase 3 (MMP3), CTSC, and SERPINB2] were specifically increased only in IL-1β–stimulated RA-FLSs, but not in RA-SMs (Fig. 2D), indicating that IL-1β–stimulated RA-FLSs newly acquire inflammatory potential in a different way from the pathway embedded in RA-SMs. What are the key factors regulating the inflammatory signature newly imparted to RA-FLSs upon IL-1β stimulation? Using a similar systems approach, we identified an initial set of 15 regulator candidates composed of transcription factors and signaling molecules (SI Appendix, Fig. S6). By searching for the target genes and their associated cellular processes using the iHOP software, we selected (i) TNFAIP8 activating angiogenesis in RA joints; (ii) IFNAP1/2, IRF9, and FYN promoting cytokine production in RA-FLSs; and (iii) AMFR regulating survival and endoplasmic reticulum stress in RA-FLSs. Although the exact role of above candidate genes in inflammatory potential of IL-1β–stimulated RA-FLSs should be confirmed by functional assays, these data may explain how a certain type of cells undergoes transition to acquire a new function under chronic inflammatory conditions, exhibiting functional redundancy. In addition, we believe that the application of the molecular signatures and network model to other pathologic processes can help us to better understand individual roles of RA-FLSs and -SMs and their interplays in RA joints.
The pathologic hallmark of RA is the formation of invasive pannus, the thickened layer of synovial tissue that erodes cartilage and bone. RA-FLSs intrinsically acquire an aggressive phenotype and become the spearhead of inflamed synovium mediating inflammation and destruction of the joint (2, 29). They destroy bone and cartilage indirectly by releasing matrix-degrading enzymes, such as MMPs and cathepsins (30), and also directly invade adjacent structures even without the help of inflammatory cells (4, 31). Such aggressive behavior of RA-FLSs is further potentiated by IL-1β and TNF-α (32). Given that current therapeutic options modulating adaptive immune cells and inflammatory cytokines have met with limited success, RA-FLSs are an attractive alternative as a therapeutic target. For example, treatment targeting cadherin-11, which is selectively expressed on FLSs, prevents arthritis in mouse models (33). Therefore, the future development of new agents could be directed toward targeting RA-FLSs to control their invasiveness and migratory capacity, which could enable more effective control of RA activity without impairment of host defense (2).
RA is a classic model of chronic inflammatory diseases. Therefore, we suggest that our systems approach can serve as a comprehensive basis that sheds insights into fundamental mechanism(s) for the major role of the participating cells in the pathogenesis of complex inflammatory diseases, including RA. Moreover, this approach could be applied to identify potential therapeutic targets for RA, such as anti-POSTN or -TWIST1 inhibitors to block FLS invasiveness.
Materials and Methods
Detailed materials and methods are described in SI Appendix, SI Materials and Methods. Experimental methods include cell culture, primary FLS and macrophages isolation, siRNA transfection, microarray analysis, real-time PCR, Western blot analysis, wound migration, Matrigel invasion assay, and determination of FLS migration in vivo. Bioinformatics analyses include statistical testing, identification of DEGs, and network analysis.
Supplementary Material
Acknowledgments
This work was supported by a Korea Healthcare Technology R&D Project grant; Ministry for Health, Welfare and Family Affairs Grant A092258; the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology Grants 2009-0080087, NRF-M1AXA002-2011-0028392, proteogenomics program, and R31-2008-000-10105-0; POSCO research fund (Project 2013Y008); and Institute for Basic Science Grant CA1308.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.Y. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311239111/-/DCSupplemental.
References
- 1.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 2.Bartok B, Firestein GS. Fibroblast-like synoviocytes: Key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233(1):233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athanasou NA. Synovial macrophages. Ann Rheum Dis. 1995;54(5):392–394. doi: 10.1136/ard.54.5.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefèvre S, et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat Med. 2009;15(12):1414–1420. doi: 10.1038/nm.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roivainen A, et al. H-ras oncogene point mutations in arthritic synovium. Arthritis Rheum. 1997;40(9):1636–1643. doi: 10.1002/art.1780400913. [DOI] [PubMed] [Google Scholar]
- 6.Yamanishi Y, et al. p53 tumor suppressor gene mutations in fibroblast-like synoviocytes from erosion synovium and non-erosion synovium in rheumatoid arthritis. Arthritis Res Ther. 2005;7(1):R12–R18. doi: 10.1186/ar1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7(6):429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 8.Bottini N, Firestein GS. Duality of fibroblast-like synoviocytes in RA: Passive responders and imprinted aggressors. Nat Rev Rheumatol. 2013;9(1):24–33. doi: 10.1038/nrrheum.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lähdesmäki H, Shmulevich L, Dunmire V, Yli-Harja O, Zhang W. In silico microdissection of microarray data from heterogeneous cell populations. BMC Bioinformatics. 2005;6:54. doi: 10.1186/1471-2105-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ospelt C, Neidhart M, Gay RE, Gay S. Synovial activation in rheumatoid arthritis. Front Biosci. 2004;9:2323–2334. doi: 10.2741/1399. [DOI] [PubMed] [Google Scholar]
- 12.Wluka AE, Ding C, Jones G, Cicuttini FM. The clinical correlates of articular cartilage defects in symptomatic knee osteoarthritis: A prospective study. Rheumatology (Oxford) 2005;44(10):1311–1316. doi: 10.1093/rheumatology/kei018. [DOI] [PubMed] [Google Scholar]
- 13.Lee HJ, et al. Direct transfer of alpha-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J Biol Chem. 2010;285(12):9262–9272. doi: 10.1074/jbc.M109.081125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Yoo SA, et al. A novel pathogenic role of the ER chaperone GRP78/BiP in rheumatoid arthritis. J Exp Med. 2012;209(4):871–886. doi: 10.1084/jem.20111783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You S, et al. A systems approach to rheumatoid arthritis. PLoS ONE. 2012;7(12):e51508. doi: 10.1371/journal.pone.0051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos YF, et al. Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann Rheum Dis. 2013 doi: 10.1136/annrheumdis-2013-203405. [DOI] [PubMed] [Google Scholar]
- 18.Wall DP, et al. Genotator: A disease-agnostic tool for genetic annotation of disease. BMC Med Genomics. 2010;3:50. doi: 10.1186/1755-8794-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li QQ, et al. Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin Cancer Res. 2009;15(8):2657–2665. doi: 10.1158/1078-0432.CCR-08-2372. [DOI] [PubMed] [Google Scholar]
- 20.Michaylira CZ, et al. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res. 2010;70(13):5281–5292. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikheeva SA, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36(7):664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 23.Naik PK, et al. COMET Investigators Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303(12):L1046–L1056. doi: 10.1152/ajplung.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkov CD, et al. A proximal activator of transcription in epithelial-mesenchymal transition. J Clin Invest. 2007;117(2):482–491. doi: 10.1172/JCI29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattey DL, Dawes PT, Nixon NB, Slater H. Transforming growth factor beta 1 and interleukin 4 induced alpha smooth muscle actin expression and myofibroblast-like differentiation in human synovial fibroblasts in vitro: Modulation by basic fibroblast growth factor. Ann Rheum Dis. 1997;56(7):426–431. doi: 10.1136/ard.56.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Vicuña R, et al. CC and CXC chemokine receptors mediate migration, proliferation, and matrix metalloproteinase production by fibroblast-like synoviocytes from rheumatoid arthritis patients. Arthritis Rheum. 2004;50(12):3866–3877. doi: 10.1002/art.20615. [DOI] [PubMed] [Google Scholar]
- 28.Mix KS, et al. Orphan nuclear receptor NR4A2 induces synoviocyte proliferation, invasion, and matrix metalloproteinase 13 transcription. Arthritis Rheum. 2012;64(7):2126–2136. doi: 10.1002/art.34399. [DOI] [PubMed] [Google Scholar]
- 29.Pap T, Meinecke I, Müller-Ladner U, Gay S. Are fibroblasts involved in joint destruction? Ann Rheum Dis. 2005;64(Suppl 4):iv52–iv54. doi: 10.1136/ard.2005.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolboom TC, et al. Invasive properties of fibroblast-like synoviocytes: correlation with growth characteristics and expression of MMP-1, MMP-3, and MMP-10. Ann Rheum Dis. 2002;61(11):975–980. doi: 10.1136/ard.61.11.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller-Ladner U, et al. Synovial fibroblasts of patients with rheumatoid arthritis attach to and invade normal human cartilage when engrafted into SCID mice. Am J Pathol. 1996;149(5):1607–1615. [PMC free article] [PubMed] [Google Scholar]
- 32.Wang AZ, Wang JC, Fisher GW, Diamond HS. Interleukin-1beta-stimulated invasion of articular cartilage by rheumatoid synovial fibroblasts is inhibited by antibodies to specific integrin receptors and by collagenase inhibitors. Arthritis Rheum. 1997;40(7):1298–1307. doi: 10.1002/1529-0131(199707)40:7<1298::AID-ART15>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Lee DM, et al. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007;315(5814):1006–1010. doi: 10.1126/science.1137306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.