Significance
Biological dispersal is a key driver of several fundamental processes in nature, crucially controlling the distribution of species and affecting their coexistence. Despite its relevance for important ecological processes, however, the subject suffers an acknowledged lack of experimentation, and current assessments point at inherent limitation to predictability even in the simplest ecological settings. We show, by combining replicated experimentation on the spread of the ciliate Tetrahymena sp. with a theoretical approach based on stochastic differential equations, that information on local unconstrained movement and reproduction of organisms (including demographic stochasticity) allows reliable prediction of both the propagation speed and range of variability of invasion fronts over multiple generations.
Keywords: Fisher wave, microcosm, colonization, spatial, frontiers
Abstract
Biological dispersal shapes species’ distribution and affects their coexistence. The spread of organisms governs the dynamics of invasive species, the spread of pathogens, and the shifts in species ranges due to climate or environmental change. Despite its relevance for fundamental ecological processes, however, replicated experimentation on biological dispersal is lacking, and current assessments point at inherent limitations to predictability, even in the simplest ecological settings. In contrast, we show, by replicated experimentation on the spread of the ciliate Tetrahymena sp. in linear landscapes, that information on local unconstrained movement and reproduction allows us to predict reliably the existence and speed of traveling waves of invasion at the macroscopic scale. Furthermore, a theoretical approach introducing demographic stochasticity in the Fisher–Kolmogorov framework of reaction–diffusion processes captures the observed fluctuations in range expansions. Therefore, predictability of the key features of biological dispersal overcomes the inherent biological stochasticity. Our results establish a causal link from the short-term individual level to the long-term, broad-scale population patterns and may be generalized, possibly providing a general predictive framework for biological invasions in natural environments.
What is the source of variance in the spread rates of biological invasions? The search for processes that affect biological dispersal and sources of variability observed in ecological range expansions is fundamental to the study of invasive species dynamics (1–10), shifts in species ranges due to climate or environmental change (11–13), and, in general, the spatial distribution of species (3, 14–16). Dispersal is the key agent that brings favorable genotypes or highly competitive species into new ranges much faster than any other ecological or evolutionary process (1, 17). Understanding the potential and realized dispersal is thus key to ecology in general (18). When organisms’ spread occurs on the timescale of multiple generations, it is the byproduct of processes that take place at finer spatial and temporal scales that are the local movement and reproduction of individuals (5, 10). The main difficulty in causally understanding dispersal is thus to upscale processes that happen at the short-term individual level to long-term and broad-scale population patterns (5, 18–20). Furthermore, the large fluctuations observed in range expansions have been claimed to reflect an intrinsic lack of predictability of the phenomenon (21). Whether the variability observed in nature or in experimental ensembles might be accounted for by systematic differences between landscapes or by demographic stochasticity affecting basic vital rates of the organisms involved is an open research question (10, 18, 21, 22).
Modeling of biological dispersal established the theoretical framework of reaction–diffusion processes (1–3, 23–25), which now finds common application in dispersal ecology (5, 14, 22, 26–30) and in other fields (17, 23, 25, 31–36). Reaction–diffusion models have also been applied to model human colonization processes (31), such as the Neolithic transition in Europe (25, 37, 38). The classical prediction of reaction–diffusion models (1, 2, 24, 25) is the propagation of an invading wavefront traveling undeformed at a constant speed (Fig. 1E). Such models have been widely adopted by ecologists to describe the spread of organisms in a variety of comparative studies (5, 10, 26) and to control the dynamics of invasive species (3, 4, 6). The extensive use of these models and the good fit to observational data favored their common endorsement as a paradigm for biological dispersal (6). However, current assessments (21) point at inherent limitations to the predictability of the phenomenon, due to its intrinsic stochasticity. Therefore, single realizations of a dispersal event (as those addressed in comparative studies) might deviate significantly from the mean of the process, making replicated experimentation necessary to allow hypothesis testing, identification of causal relationships, and to potentially falsify the models’ assumptions (39).
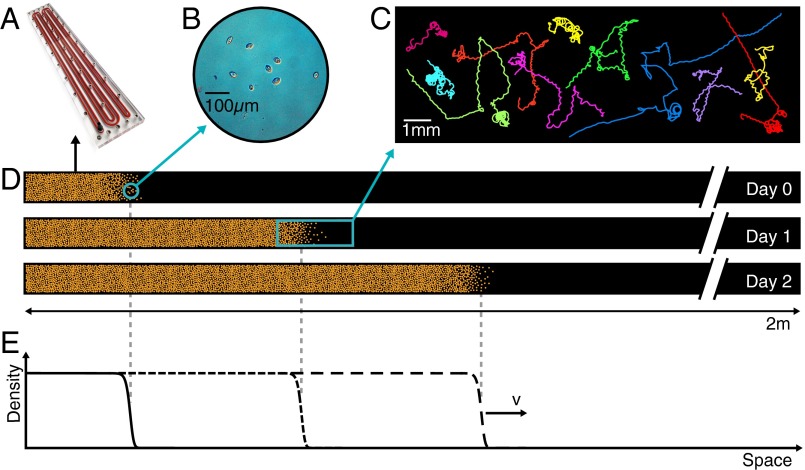
Fig. 1.
Schematic representation of the experiment. (A) Linear landscape. (B) Individuals of the ciliate Tetrahymena sp. move and reproduce within the landscape. (C) Examples of reconstructed trajectories of individuals (Movie S1). (D) Individuals are introduced at one end of a linear landscape and are observed to reproduce and disperse within the landscape (not to scale). (E) Illustrative representation of density profiles along the landscape at subsequent times. A wavefront is argued to propagate undeformed at a constant speed v according to the Fisher–Kolmogorov equation.
Here, we provide replicated and controlled experimental support to the theory of reaction–diffusion processes for modeling biological dispersal (23–25) in a generalized context that reproduces the observed fluctuations. Firstly, we experimentally substantiate the Fisher–Kolmogorov prediction (1, 2) on the existence and the mean speed of traveling wavefronts by measuring the individual components of the process. Secondly, we manipulate the inclusion of demographic stochasticity in the model to reproduce the observed variability in range expansions. We move from the Fisher–Kolmogorov equation (Materials and Methods) to describe the spread of organisms in a linear landscape (1, 2, 24, 25). The equation couples a logistic term describing the reproduction of individuals with growth rate r
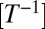 and carrying capacity K
and carrying capacity K
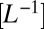 and a diffusion term accounting for local movement, epitomized by the diffusion coefficient D
and a diffusion term accounting for local movement, epitomized by the diffusion coefficient D
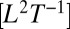 . These species’ traits define the characteristic scales of the dispersal process. In this framework, a population initially located at one end of a linear landscape is predicted to form a wavefront of colonization invading empty space at a constant speed
. These species’ traits define the characteristic scales of the dispersal process. In this framework, a population initially located at one end of a linear landscape is predicted to form a wavefront of colonization invading empty space at a constant speed 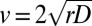 (1, 2, 24, 25), which we measured in our dispersal experiment (Fig. 1D and SI Text).
(1, 2, 24, 25), which we measured in our dispersal experiment (Fig. 1D and SI Text).
Results
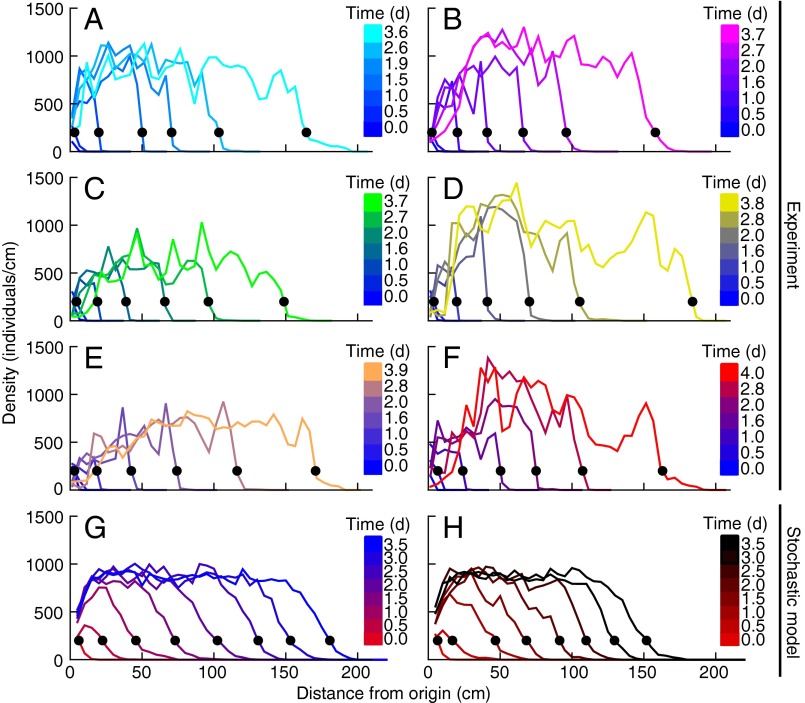
In the experiments, we used the freshwater ciliate Tetrahymena sp. (Materials and Methods) because of its short generation time (16) and its history as a model system in ecology (16, 40, 41). The experimental setup consisted of linear landscapes, filled with a nutrient medium, kept in constant environmental conditions and of suitable size to meet the assumptions about the relevant dispersal timescales (Materials and Methods). Replicated dispersal events were conducted by introducing an ensemble of individuals at one end of the landscape and measuring density profiles throughout the system at different times, through image analysis (Materials and Methods). Density profiles are shown in Fig. 2, in six replicated dispersal events (Fig. 2 A–F). Organisms introduced at one end of the landscape rapidly formed an advancing front that propagated at a remarkably constant speed (Fig. 3 and Table S1). The front position at each time was calculated as the first occurrence, starting from the end of the landscape, of a fixed value of the density (Fig. 2). As for traveling waves predicted by the Fisher–Kolmogorov equation, the mean front speed in our experiment is notably constant for different choices of the reference density value (Fig. 3C).
Fig. 2.
Density profiles in the dispersal experiment and in the stochastic model. (A–F) Density profiles of six replicated experimentally measured dispersal events, at different times. Legends link each color to the corresponding measuring time. Black dots are the estimates of the front position at each time point. Organisms were introduced at the origin and subsequently colonized the whole landscape in 4 d (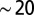 generations). (G and H) Two dispersal events simulated according to the generalized model equation, with initial conditions as at the second experimental time point. Data are binned in 5-cm intervals, typical length scale of the process.
generations). (G and H) Two dispersal events simulated according to the generalized model equation, with initial conditions as at the second experimental time point. Data are binned in 5-cm intervals, typical length scale of the process.
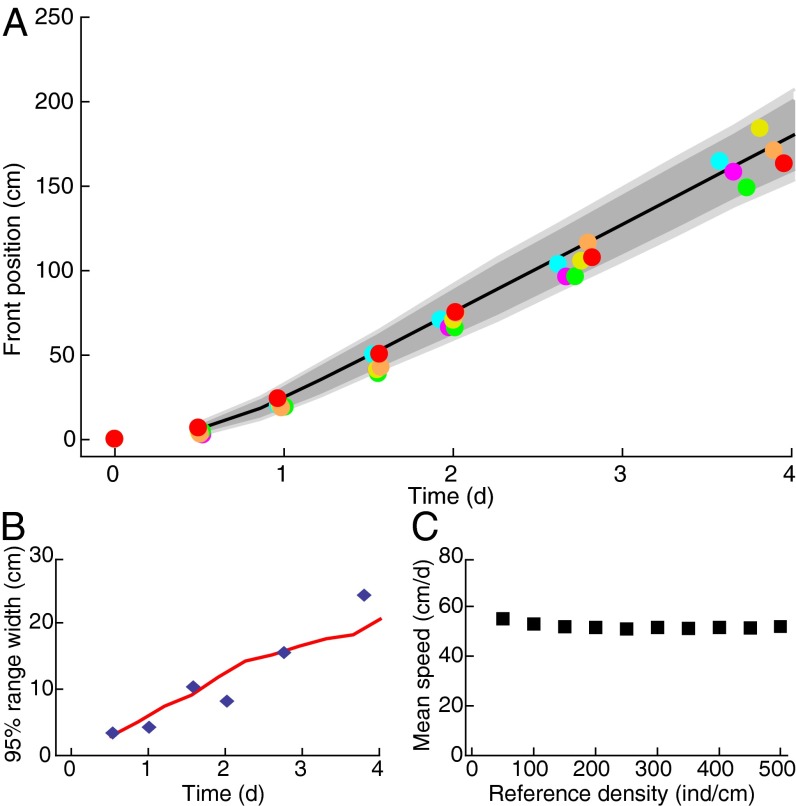
Fig. 3.
Range expansion in the dispersal experiment and in the stochastic model. (A) Front position of the expanding population in six replicated dispersal events; colors identify replicas as in Fig. 2. The dark and light gray shadings are, respectively, the  and
and 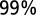 confidence intervals computed by numerically integrating the generalized model equation, with initial conditions as at the second experimental time point, in 1,020 iterations. The black curve is the mean front position in the stochastic integrations. (B) The increase in range variability between replicates in the dispersal experiment (blue diamonds) is well described by the stochastic model (red line). (C) Mean front speed for different choices of the reference density value at which we estimated the front position in the experiment; error bars are smaller than symbols.
confidence intervals computed by numerically integrating the generalized model equation, with initial conditions as at the second experimental time point, in 1,020 iterations. The black curve is the mean front position in the stochastic integrations. (B) The increase in range variability between replicates in the dispersal experiment (blue diamonds) is well described by the stochastic model (red line). (C) Mean front speed for different choices of the reference density value at which we estimated the front position in the experiment; error bars are smaller than symbols.
The species’ traits r, K, and D were measured in independent experiments (Table 1). In the local-growth experiment, a low-density population of Tetrahymena sp. was introduced evenly across the landscape, and its density was measured locally at different times. Recorded density measurements were fitted to the logistic growth model, which gave the estimates for r and K (Table 1 and Table S2). In the local unimpeded movement experiment, we computed the mean-square displacement (SI Text and Fig. S1) of individuals’ trajectories (42–44) to estimate the diffusion coefficient D in density-independent conditions (Table 1 and Materials and Methods). The growth and movement measurements were performed in the same linear landscape settings as in the dispersal experiment and therefore are assumed to accurately describe the dynamics at the front of the traveling wave in the dispersal events.
Table 1.
Experimentally measured species’ traits (mean ± SE)
| Demographic traits |
Movement traits | |
| Deterministic model | Stochastic model | |
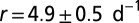 |
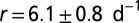 |
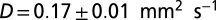 |
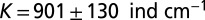 |
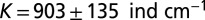 |
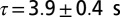 |
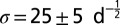 |
||
Demographic traits were estimated both in the framework of the deterministic logistic equation and in the framework of the stochastic logistic Eq. 3. Demographic stochasticity strongly affects the dynamics at low densities; thus, a different value for the growth rate r is obtained in the stochastic model, compared with the deterministic one. ind, individual.
The comparison of the predicted front speed 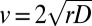 to the wavefront speed measured in the dispersal experiment,
to the wavefront speed measured in the dispersal experiment, 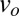 , yields a compelling agreement. The observed speed in the dispersal experiment was
, yields a compelling agreement. The observed speed in the dispersal experiment was 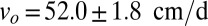 (mean ± SE) (Table S1), which we compare with the predicted one
(mean ± SE) (Table S1), which we compare with the predicted one 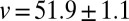 cm/d (mean ± SE). The two velocities are compatible within one SE. A t test between the replicated observed speeds and bootstrap estimates of
cm/d (mean ± SE). The two velocities are compatible within one SE. A t test between the replicated observed speeds and bootstrap estimates of 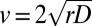 gives a P value of
gives a P value of 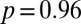 (
(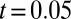 ,
, 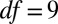 ). Thus, the null hypothesis that the mean difference is 0 is not rejected at the 5% level, and there is no indication that the two means are different. As the measurements of r and D were performed in independent experiments, at scales that were orders of magnitude smaller than in the dispersal events, the agreement between the two estimates of the front velocity is deemed remarkable.
). Thus, the null hypothesis that the mean difference is 0 is not rejected at the 5% level, and there is no indication that the two means are different. As the measurements of r and D were performed in independent experiments, at scales that were orders of magnitude smaller than in the dispersal events, the agreement between the two estimates of the front velocity is deemed remarkable.
Although the Fisher–Kolmogorov equation correctly predicts the mean speed of the experimentally observed invading wavefront, its deterministic formulation prevents it from reproducing the variability that is inherent to biological dispersal (21). In particular, it cannot reproduce the fluctuations in range expansion between different replicates of our dispersal experiment (Fig. 3A). We propose a generalization of the Fisher–Kolmogorov equation (Materials and Methods) accounting for demographic stochasticity that is able to capture the observed variability. The strength of demographic stochasticity is embedded in an additional species’ trait σ
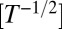 . In this stochastic framework, the demographic parameters r, K, and σ were estimated from the local growth experiment with a maximum-likelihood approach (Table 1 and Materials and Methods) whereas the estimate of the diffusion coefficient D was left unchanged (SI Text). We then used these local independent estimates to numerically integrate the generalized model equation (45, 46), with initial conditions as in the dispersal experiment, and found that the measured front positions are in accordance with simulations (Fig. 3A and Fig. S2). In particular, most experimental data are within the 95% confidence interval for the simulated front position, and the observed range variability is well-captured by our stochastic model (Fig. 3B). Accordingly, the estimate for the front speed and its variability in the experiment are in good agreement with simulations (SI Text). Demographic stochasticity can therefore explain the observed variability in range expansions.
. In this stochastic framework, the demographic parameters r, K, and σ were estimated from the local growth experiment with a maximum-likelihood approach (Table 1 and Materials and Methods) whereas the estimate of the diffusion coefficient D was left unchanged (SI Text). We then used these local independent estimates to numerically integrate the generalized model equation (45, 46), with initial conditions as in the dispersal experiment, and found that the measured front positions are in accordance with simulations (Fig. 3A and Fig. S2). In particular, most experimental data are within the 95% confidence interval for the simulated front position, and the observed range variability is well-captured by our stochastic model (Fig. 3B). Accordingly, the estimate for the front speed and its variability in the experiment are in good agreement with simulations (SI Text). Demographic stochasticity can therefore explain the observed variability in range expansions.
Discussion
To summarize, we suggest that measuring and suitably interpreting local processes allows us to accurately predict the main features of global invasions. The deterministic Fisher–Kolmogorov equation is shown to correctly predict the mean speed of invasion but cannot capture the observed variability. Instead, characterizing the inherent stochasticity of the biological processes involved allows us to predict both the mean and the variability of range expansions, which is of interest for practical purposes, such as the delineation of worst-case scenarios for the spread of invasive species. Our phenomenological approach allows us to make predictions on the spread of organisms without the need to introduce all details on the movement behavior, biology, or any other information. Such details are synthesized in three parameters describing the density-independent yet stochastic behavior of individuals riding the invasion wave. The parsimony of the model allows generalization to organisms with different biology (e.g., growth rates and diffusion coefficients are available for several species in the literature) (6) and supports the view that our protocol may possibly provide a general predictive framework for biological invasions in natural environments.
In conclusion, we have shown that, at least in the simple ecological settings investigated here, predictability remains, notwithstanding biological fluctuations, owing to the stochastic treatment devised. We confirm that deterministic models can be applied to describe ecological processes and show that additional information on the stochasticity acting at the mesoscopic scale allows us to estimate fluctuations at the macroscopic scale. We believe that our results might have implications for the dynamics of phenomena other than species’ invasions, such as morphogenesis (23, 47), tumor growth (23, 25, 36), and the spreading of epidemics (23, 30, 34, 35), which have been traditionally modeled with reaction–diffusion equations.
Materials and Methods
Study Species.
The species used in this study is Tetrahymena sp. (Fig. 1B), a freshwater ciliate, purchased from Carolina Biological Supply. Individuals of Tetrahymena sp. have typical linear size (equivalent diameter) of 14 μm (41). Freshwater bacteria of the species Serratia fonticola, Breviacillus brevis, and Bacillus subtilis were used as a food resource for ciliates, which were kept in a medium made of sterilized spring water and protozoan pellets (Carolina Biological Supply) at a density of 0.45 gL−1. The experimental units were kept under constant fluorescent light for the whole duration of the study, at a constant temperature of 22 °C. Overall, experimental protocols are well-established (16, 41, 48–50), and the contribution of laboratory experiments on protists to the understanding of population and metapopulation dynamics proved noteworthy (48).
Experimental Setup.
Experiments were performed in linear landscapes (Fig. 1A) filled with a nutrient medium and bacteria of the three species above mentioned. The linear landscapes were 2 m long, 5 mm wide, and 3 mm deep, respectively, and 105, 350, and 200 times the size of the study organism (41). Landscapes consisted of channels drilled on a Plexiglas sheet. A second sheet was used as lid, and a gasket was introduced to avoid water spillage (Fig. 1A). At one end of the landscapes, an opening was placed for the introduction of ciliates. The Plexiglas sheets were sterilized with a 70% (vol/vol) alcohol solution, and gaskets were autoclaved at 120 °C before filling the landscape with medium. As Plexiglas is transparent, the experimental units could be placed under the objective of a stereomicroscope, to record pictures (for counting of individuals) or videos (to track ciliates). Individuals were observed to distribute mainly at the bottom of the landscape, whose length was three orders of magnitude larger than its width (w) and depth and two orders of magnitude larger than the typical length scale of the process 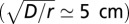 . The population was thus assumed to be confidently well-mixed within the cross section after a time
. The population was thus assumed to be confidently well-mixed within the cross section after a time 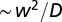 , which in our case is of the order of a minute after introduction of the ciliates in the landscape.
, which in our case is of the order of a minute after introduction of the ciliates in the landscape.
Experimental Protocol.
We performed three independent and complementing experiments, specifically: (i) a dispersal experiment was carried out to study the possible existence and the propagation of traveling invasion wavefronts in replicated dispersal events; (ii) a growth experiment was run to obtain estimates of the demographic species’ traits, which are r and K in the deterministic framework of Eq. 1 and r, K, and σ in the stochastic framework of Eq. 2; (iii) a local movement experiment was performed to study the local unimpeded movement of Tetrahymena sp. over a short timescale (in a time window 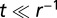 ), to estimate the diffusion coefficient D for our study species, independently from the dispersal and growth experiments.
), to estimate the diffusion coefficient D for our study species, independently from the dispersal and growth experiments.
Dispersal experiment.
We performed six replicated dispersal events in the linear landscapes. After filling the landscapes with medium and bacteria, a small ensemble of Tetrahymena sp. was introduced at the origin. Subsequently, the density of Tetrahymena sp. was measured at 1-cm intervals, five times in the first 48 h and twice in the last 48 h. The whole experiment lasted for about 20 generations of the study species.
Local growth experiment.
We performed five replicated growth measurements in the linear landscapes, to measure the demographic species’ traits, in the same environmental conditions as in the dispersal experiment, but independently from it. A low-density culture of Tetrahymena sp. was introduced in the whole landscape, and its density was measured by taking pictures and counting individuals, covering a region of 7 cm along the landscape. Density measurements were performed at several time points for each of the five replicates, in a time window of 3 d.
Local movement experiment.
We performed four additional, replicated dispersal events in the linear landscapes, initialized in the same way as in the dispersal experiment, to measure the diffusion coefficient of Tetrahymena sp. The diffusion coefficient D is the proportionality constant that links the mean square displacement of organisms’ trajectories to time (42, 44) (SI Text). Macroscopically, it relates the local flux to the density of individuals, under the assumption of steady state (44). To estimate the diffusion coefficient, we recorded several videos of individuals moving at the front of the traveling wave (at low density), reconstructed their trajectories (42, 43), and computed their mean square displacement 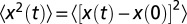 .
.
Video recording.
We recorded videos of Tetrahymena sp. at the front of the traveling wave in four replicated dispersal events, at various times over 4 d. The area covered in each video was of 24 mm in the direction of the landscape and 5 mm orthogonal to it. Each video lasted for 12 min.
Trajectories reconstruction.
For each recorded video, we extracted individuals’ spatial coordinates in each frame and used the MOSAIC plugin for the software ImageJ to reconstruct trajectories (43). The goodness of the tracking was checked on several trajectories by direct comparison with the videos. Examples of reconstructed trajectories can be seen in Fig. 1C or in Movie S1.
Diffusion coefficient estimate.
For each video, the square displacement of each trajectory in the direction parallel to the landscape was computed at all time points and then averaged across trajectories. Precisely, for each trajectory i we computed the quantity  , where
, where 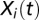 is the 1-dimensional coordinate of organism i at time t in the direction parallel to the landscape and
is the 1-dimensional coordinate of organism i at time t in the direction parallel to the landscape and 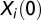 is its initial position. The mean square displacement in a video was then computed as the mean of
is its initial position. The mean square displacement in a video was then computed as the mean of 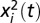 across all trajectories, that is,
across all trajectories, that is, 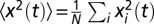 (where N is the total number of trajectories). A typical measurement of
(where N is the total number of trajectories). A typical measurement of 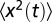 is shown in Fig. S1. As shown in the figure, there exists an initial correlated phase, which we discuss in SI Text. To estimate the diffusion coefficient from the mean square displacement, we fitted the measured
is shown in Fig. S1. As shown in the figure, there exists an initial correlated phase, which we discuss in SI Text. To estimate the diffusion coefficient from the mean square displacement, we fitted the measured 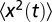 to the function
to the function 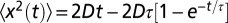 (SI Text) with the two parameters D (diffusion coefficient) and τ (correlation time). The total number of recorded videos was 28, that is, 7 for each replica.
(SI Text) with the two parameters D (diffusion coefficient) and τ (correlation time). The total number of recorded videos was 28, that is, 7 for each replica.
Mathematical Models.
Deterministic framework.
The Fisher–Kolmogorov equation (1, 2). reads:
where 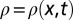 is the density of organisms, r the species’ growth rate, D the diffusion coefficient, and K the carrying capacity. Eq. 1 is known to foster the development of undeformed traveling waves of the density profile. Mathematically, the existence of traveling wave solutions implies that
is the density of organisms, r the species’ growth rate, D the diffusion coefficient, and K the carrying capacity. Eq. 1 is known to foster the development of undeformed traveling waves of the density profile. Mathematically, the existence of traveling wave solutions implies that 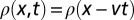 , where v is the speed of the advancing wave. Fisher (1) proved that traveling wave solutions can only exist with speed
, where v is the speed of the advancing wave. Fisher (1) proved that traveling wave solutions can only exist with speed 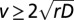 , and Kolmogorov (2) demonstrated that, with suitable initial conditions, the speed of the wavefront is the lower bound.
, and Kolmogorov (2) demonstrated that, with suitable initial conditions, the speed of the wavefront is the lower bound.
The microscopic movement underlying the Fisher–Kolmogorov Eq. 1 is brownian motion (25, 51). Investigation of the movement behavior of Tetrahymena sp., instead, shows that individuals’ trajectories are consistent with a persistent random walk with an autocorrelation time 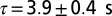 . The corresponding macroscopic equation for the persistent random walk should thus be the reaction–telegraph equation (25) (SI Text). Nonetheless, as the autocorrelation time for our study species is much smaller than the growth rate r
. The corresponding macroscopic equation for the persistent random walk should thus be the reaction–telegraph equation (25) (SI Text). Nonetheless, as the autocorrelation time for our study species is much smaller than the growth rate r
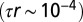 , Eq. 1 provides an excellent approximation to the reaction–telegraph equation. See SI Text for a detailed discussion.
, Eq. 1 provides an excellent approximation to the reaction–telegraph equation. See SI Text for a detailed discussion.
Stochastic framework.
The stochastic model equation reads:
where 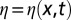 is a Gaussian, zero-mean white noise (i.e., with correlations
is a Gaussian, zero-mean white noise (i.e., with correlations 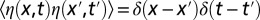 , where δ is the Dirac’s delta distribution) and
, where δ is the Dirac’s delta distribution) and 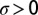 is constant. We adopt the Itô’s stochastic calculus (51), as appropriate in this case. Note, in fact, that the choice of the Stratonovich framework would make no sense here, as the noise term would have a constant nonzero mean (22, 51), which would allow an extinct population to possibly escape the zero-density absorbing state. The square-root multiplicative noise term in Eq. 2 is commonly interpreted as describing demographic stochasticity in a population (46) and needs extra care in simulations (45, 52). In particular, standard stochastic integration schemes fail to preserve the positivity of ρ. We adopted a recently developed split-step method (45) to numerically integrate Eq. 2. This method allows us to perform the integration with relatively large spatial and temporal steps maintaining numerical accuracy.
is constant. We adopt the Itô’s stochastic calculus (51), as appropriate in this case. Note, in fact, that the choice of the Stratonovich framework would make no sense here, as the noise term would have a constant nonzero mean (22, 51), which would allow an extinct population to possibly escape the zero-density absorbing state. The square-root multiplicative noise term in Eq. 2 is commonly interpreted as describing demographic stochasticity in a population (46) and needs extra care in simulations (45, 52). In particular, standard stochastic integration schemes fail to preserve the positivity of ρ. We adopted a recently developed split-step method (45) to numerically integrate Eq. 2. This method allows us to perform the integration with relatively large spatial and temporal steps maintaining numerical accuracy.
Data from the growth experiment were fitted to the equation:
where 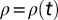 is the local density,
is the local density, 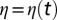 is a Gaussian, zero-mean white noise (i.e., with correlations
is a Gaussian, zero-mean white noise (i.e., with correlations 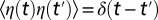 ),
), 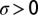 is constant, and l is the size of the region over which densities were measured (SI Text). Eq. 3 describes the time-evolution of the density in a well-mixed patch of length l (SI Text). The likelihood function for Eq. 3 can be written as:
is constant, and l is the size of the region over which densities were measured (SI Text). Eq. 3 describes the time-evolution of the density in a well-mixed patch of length l (SI Text). The likelihood function for Eq. 3 can be written as:
where n is the total number of observations in the growth time series, 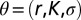 is the vector of demographic parameters, and
is the vector of demographic parameters, and 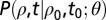 is the transitional probability density of having a density of individuals ρ at time t, given that the density at time
is the transitional probability density of having a density of individuals ρ at time t, given that the density at time 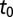 was
was 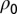 (for a given θ). The transitional probability density
(for a given θ). The transitional probability density 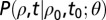 satisfies the Fokker–Planck equation associated to Eq. 3 (SI Text), which was solved numerically for all observed transitions and choices of parameters (SI Text), adopting the implicit Crank–Nicholson scheme. The best fit parameters were those that maximized the likelihood function Eq. 4 (Table 1 and SI Text).
satisfies the Fokker–Planck equation associated to Eq. 3 (SI Text), which was solved numerically for all observed transitions and choices of parameters (SI Text), adopting the implicit Crank–Nicholson scheme. The best fit parameters were those that maximized the likelihood function Eq. 4 (Table 1 and SI Text).
Supplementary Material
Acknowledgments
We thank Amos Maritan and Enrico Bertuzzo for insightful discussions, Janick Cardinale for support with the MOSAIC software, and Regula Illi for the ciliates’ picture. We acknowledge the support provided by the discretionary funds of Eawag: Swiss Federal Institute of Aquatic Science and Technology; by the European Research Council advanced grant program through the project “River networks as ecological corridors for biodiversity, populations and waterborne disease” (RINEC-227612); and by Swiss National Science Foundation Projects 200021_124930/1 and 31003A_135622.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321167110/-/DCSupplemental.
References
- 1.Fisher RA. The wave of advance of advantageous genes. Ann Eugen. 1937;7:355–369. [Google Scholar]
- 2.Kolmogorov AN, Petrovskii IG, Piskunov NS. A study of the diffusion equation with increase in the amount of substance, and its application to a biological problem. Bull. Moscow Univ. Math. Mech. 1937;1(1):1–25. [Google Scholar]
- 3.Skellam JG. Random dispersal in theoretical populations. Biometrika. 1951;38(1-2):196–218. [PubMed] [Google Scholar]
- 4.Elton CS. The Ecology of Invasions by Animals and Plants. London: Methuen; 1958. [Google Scholar]
- 5.Andow DA, Kareiva PM, Levin SA, Okubo A. Spread of invading organisms. Landscape Ecol. 1990;4(2/3):177–188. [Google Scholar]
- 6.Grosholz ED. Contrasting rates of spread for introduced species in terrestrial and marine systems. Ecology. 1996;77(6):1680–1686. [Google Scholar]
- 7.Shigesada N, Kawasaki K. Biological Invasions: Theory and Practice. Oxford: Oxford University Press; 1997. [Google Scholar]
- 8.Clobert J, Danchin E, Dhondt AA, Nichols JD, editors. Dispersal. Oxford: Oxford University Press; 2001. [Google Scholar]
- 9.Okubo A, Levin SA. Diffusion and Ecological Problems: Modern Perspectives. 2nd Ed. Berlin: Springer; 2002. [Google Scholar]
- 10.Hastings A, et al. The spatial spread of invasions: New developments in theory and evidence. Ecol Lett. 2005;8(1):91–101. [Google Scholar]
- 11.Parmesan C, et al. Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature. 1999;399(6736):579–583. [Google Scholar]
- 12.Bell G, Gonzalez A. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science. 2011;332(6035):1327–1330. doi: 10.1126/science.1203105. [DOI] [PubMed] [Google Scholar]
- 13.Leroux SJ, et al. Mechanistic models for the spatial spread of species under climate change. Ecol Appl. 2013;23(4):815–828. doi: 10.1890/12-1407.1. [DOI] [PubMed] [Google Scholar]
- 14.Bertuzzo E, Maritan A, Gatto M, Rodriguez-Iturbe I, Rinaldo A. River networks and ecological corridors: Reactive transport on fractals, migration fronts, hydrochory. Water Resour Res. 2007;43(4):W04419. [Google Scholar]
- 15.Muneepeerakul R, et al. Neutral metacommunity models predict fish diversity patterns in Mississippi-Missouri basin. Nature. 2008;453(7192):220–222. doi: 10.1038/nature06813. [DOI] [PubMed] [Google Scholar]
- 16.Carrara F, Altermatt F, Rodriguez-Iturbe I, Rinaldo A. Dendritic connectivity controls biodiversity patterns in experimental metacommunities. Proc Natl Acad Sci USA. 2012;109(15):5761–5766. doi: 10.1073/pnas.1119651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallatschek O, Nelson DR. Gene surfing in expanding populations. Theor Popul Biol. 2008;73(1):158–170. doi: 10.1016/j.tpb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Sutherland WJ, et al. Identification of 100 fundamental ecological questions. J Ecol. 2013;101(1):58–67. [Google Scholar]
- 19.Levin SA. The problem of pattern and scale in ecology. Ecology. 1992;73(6):1943–1967. [Google Scholar]
- 20.Bascompte J, Solé RV. Rethinking complexity: Modelling spatiotemporal dynamics in ecology. Trends Ecol Evol. 1995;10(9):361–366. doi: 10.1016/s0169-5347(00)89134-x. [DOI] [PubMed] [Google Scholar]
- 21.Melbourne BA, Hastings A. Highly variable spread rates in replicated biological invasions: Fundamental limits to predictability. Science. 2009;325(5947):1536–1539. doi: 10.1126/science.1176138. [DOI] [PubMed] [Google Scholar]
- 22.Méndez V, Llopis I, Campos D, Horsthemke W. Effect of environmental fluctuations on invasion fronts. J Theor Biol. 2011;281(1):31–38. doi: 10.1016/j.jtbi.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Murray JD. Mathematical Biology I: An Introduction. 3rd Ed. Berlin: Springer; 2004. [Google Scholar]
- 24.Volpert V, Petrovskii S. Reaction-diffusion waves in biology. Phys Life Rev. 2009;6(4):267–310. doi: 10.1016/j.plrev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Méndez V, Fedotov S, Horsthemke W. Reaction-Transport Systems. Berlin: Springer; 2010. [Google Scholar]
- 26.Lubina JA, Levin SA. The spread of a reinvading species: Range expansion in the California sea otter. Am Nat. 1988;131(4):526–543. [Google Scholar]
- 27.Holmes EE. Are diffusion models too simple? A comparison with telegraph models of invasion. Am Nat. 1993;142(5):779–795. doi: 10.1086/285572. [DOI] [PubMed] [Google Scholar]
- 28.Solé RV, Bascompte J. Self-Organization in Complex Ecosystems. Princeton: Princeton Univ Press; 2006. [Google Scholar]
- 29.Roques L, Garnier J, Hamel F, Klein EK. Allee effect promotes diversity in traveling waves of colonization. Proc Natl Acad Sci USA. 2012;109(23):8828–8833. doi: 10.1073/pnas.1201695109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pybus OG, et al. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci USA. 2012;109(37):15066–15071. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos D, Fort J, Méndez V. Transport on fractal river networks: application to migration fronts. Theor Popul Biol. 2006;69(1):88–93. doi: 10.1016/j.tpb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104(50):19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fort J, Pujol T. Progress in front propagation research. Rep Prog Phys. 2008;71(8):086001. [Google Scholar]
- 34.Bertuzzo E, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A. On spatially explicit models of cholera epidemics. J R Soc Interface. 2010;7(43):321–333. doi: 10.1098/rsif.2009.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Righetto L, et al. Modelling human movement in cholera spreading along fluvial systems. Ecohydrology. 2010;4(1):49–55. [Google Scholar]
- 36.Fort J, Solé RV. Accelerated tumor invasion under non-isotropic cell dispersal in glioblastomas. New J Phys. 2013;15(5):055001. [Google Scholar]
- 37.Ammerman AJ, Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton: Princeton Univ Press; 1984. [Google Scholar]
- 38.Fort J. Synthesis between demic and cultural diffusion in the Neolithic transition in Europe. Proc Natl Acad Sci USA. 2012;109(46):18669–18673. doi: 10.1073/pnas.1200662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hekstra DR, Leibler S. Contingency and statistical laws in replicate microbial closed ecosystems. Cell. 2012;149(5):1164–1173. doi: 10.1016/j.cell.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 40.Schtickzelle N, Fjerdingstad EJ, Chaine A, Clobert J. Cooperative social clusters are not destroyed by dispersal in a ciliate. BMC Evol Biol. 2009;9(251):251. doi: 10.1186/1471-2148-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giometto A, Altermatt F, Carrara F, Maritan A, Rinaldo A. Scaling body size fluctuations. Proc Natl Acad Sci USA. 2013;110(12):4646–4650. doi: 10.1073/pnas.1301552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg HC. Random Walks in Biology. Princeton: Princeton Univ Press; 1993. [Google Scholar]
- 43.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151(2):182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Polin M, Tuval I, Drescher K, Gollub JP, Goldstein RE. Chlamydomonas swims with two “gears” in a eukaryotic version of run-and-tumble locomotion. Science. 2009;325(5939):487–490. doi: 10.1126/science.1172667. [DOI] [PubMed] [Google Scholar]
- 45.Dornic I, Chaté H, Muñoz MA. Integration of Langevin equations with multiplicative noise and the viability of field theories for absorbing phase transitions. Phys Rev Lett. 2005;94(10):100601. doi: 10.1103/PhysRevLett.94.100601. [DOI] [PubMed] [Google Scholar]
- 46.Bonachela J, Muñoz MA, Levin SA. Patchiness and demographic noise in three ecological examples. J Stat Phys. 2012;148(4):723–739. [Google Scholar]
- 47.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond, B. 1952;237(641):37–72. [Google Scholar]
- 48.Holyoak M, Lawler SP. Advances in Ecological Research. Vol 37. London: Academic; 2005. Population dynamics and laboratory ecology; pp. 245–271. [Google Scholar]
- 49.Altermatt F, Bieger A, Carrara F, Rinaldo A, Holyoak M. Effects of connectivity and recurrent local disturbances on community structure and population density in experimental metacommunities. PLoS ONE. 2011;6(4):e19525. doi: 10.1371/journal.pone.0019525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altermatt F, Schreiber S, Holyoak M. Interactive effects of disturbance and dispersal directionality on species richness and composition in metacommunities. Ecology. 2011;92(4):859–870. doi: 10.1890/10-1095.1. [DOI] [PubMed] [Google Scholar]
- 51.Gardiner C. Stochastic Methods. 4th Ed. Berlin: Springer; 2006. [Google Scholar]
- 52.Moro E. Numerical schemes for continuum models of reaction-diffusion systems subject to internal noise. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70(4 Pt 2):045102. doi: 10.1103/PhysRevE.70.045102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.