Significance
This work explores the endogenous functions of two disease- and RNA-associated proteins, FMRP (fragile X protein) and Ataxin-2, in memory, synaptic plasticity, RNA regulation, and messenger ribo-nucleoprotein particle assembly. By documenting an array of common phenotypic consequences from loss of Atx2 or dFMR1, the results argue that both proteins have similar in vivo functions and that differences in spinocerebellar ataxia 2 and fragile X disease pathologies may arise from the distinct features of the respective disease causative mutations. This work provides insight into the in vivo mechanisms of long-term memory (LTM)-associated synaptic plasticity and into the roles of FMRP and Atx2 in neuronal translational control.
Keywords: synapse plasticity, olfactory memory, neural circuits, neurological disease
Abstract
Fragile X mental retardation protein (FMRP) and Ataxin-2 (Atx2) are triplet expansion disease- and stress granule-associated proteins implicated in neuronal translational control and microRNA function. We show that Drosophila FMRP (dFMR1) is required for long-term olfactory habituation (LTH), a phenomenon dependent on Atx2-dependent potentiation of inhibitory transmission from local interneurons (LNs) to projection neurons (PNs) in the antennal lobe. dFMR1 is also required for LTH-associated depression of odor-evoked calcium transients in PNs. Strong transdominant genetic interactions among dFMR1, atx2, the deadbox helicase me31B, and argonaute1 (ago1) mutants, as well as coimmunoprecitation of dFMR1 with Atx2, indicate that dFMR1 and Atx2 function together in a microRNA-dependent process necessary for LTH. Consistently, PN or LN knockdown of dFMR1, Atx2, Me31B, or the miRNA-pathway protein GW182 increases expression of a Ca2+/calmodulin-dependent protein kinase II (CaMKII) translational reporter. Moreover, brain immunoprecipitates of dFMR1 and Atx2 proteins include CaMKII mRNA, indicating respective physical interactions with this mRNA. Because CaMKII is necessary for LTH, these data indicate that fragile X mental retardation protein and Atx2 act via at least one common target RNA for memory-associated long-term synaptic plasticity. The observed requirement in LNs and PNs supports an emerging view that both presynaptic and postsynaptic translation are necessary for long-term synaptic plasticity. However, whereas Atx2 is necessary for the integrity of dendritic and somatic Me31B-containing particles, dFmr1 is not. Together, these data indicate that dFmr1 and Atx2 function in long-term but not short-term memory, regulating translation of at least some common presynaptic and postsynaptic target mRNAs in the same cells.
A large number of mRNA-binding proteins are associated with neurological disease (1, 2). Although many are associated with posttranscriptional control of mRNAs and/or with cytoplasmic maternal ribo-nucleoprotein particle (mRNP) aggregates such as stress granules, their function in normal brain function and maintenance remain unclear (3–9).
The fragile X syndrome, seen in 1 in 4,000 male children, is caused by triplet CGG expansions in the 5′ untranslated region of the fragile X mental retardation gene (FMR1) (10, 11). This results in reduced production of fragile X mental retardation protein (FMRP) and a variety of pathologies, including mental retardation and autism (12–15). Current work suggests multiple origins for the resulting pathologies, including increased global protein synthesis and enhanced mGluR5-induced long-term depression (16–21). Two other neurological disorders, type II spinocerebellar ataxia and a form of amyotrophic laterosclerosis, are caused by similar triplet expansions (CAG) in the gene encoding the candidate RNA-binding protein Ataxin-2 (Atx2) (22, 23). However, in these instances CAG repeats, being present in coding sequence of Atx2, result in the inclusion of polyglutamine repeats in the mutant protein, the formation of intracellular inclusion bodies enriched in the mutant protein, and eventually, age-dependent degeneration of specific subsets of neurons (24–27).
Despite the apparent differences in the effects of disease-causing mutations and disease pathologies, functional connections between dFmr1 and Atx2 have emerged from several recent lines of data. First, both proteins are candidate RNA-binding proteins, with Atx2 having the like SM (LSM) domain and dFmr1 having the K homology (KH) domain that mediates RNA binding (28–30), and are implicated in the microRNA (miRNA) pathway (16, 31–35). Second, in cultured cells, both proteins are present on cytoplasmic mRNP aggregates (36–42); and third, recent work in Drosophila indicates that both proteins may function in the consolidation of different forms of long-term memory (LTM) (16, 31, 43–45). These observations lead to several important questions. First, do both proteins function in the same cells for mRNA regulation and long-term memory formation? Second, do they regulate translation of the same or of different mRNAs? Third, do they have identical or widely different mechanisms of action in mRNP assembly and translational control in vivo?
The Drosophila antennal lobe, with its relatively well-understood neural circuitry, is a uniquely convenient preparation in which to address the above questions. Here, a simple Atx2-dependent form of long-term memory, long-term habituation (LTH), arises from the plasticity of inhibitory transmission between two cell types, local circuit interneurons (LNs) and output projection neurons (PNs) (31, 46, 47). This potentially allows us to ask whether dFmr1 and Atx2 function in the same cells for LTH formation. In addition, the translation of the Ca2+/calmodulin-dependent protein kinase II (CaMKII) mRNA in PN dendrites, previously shown to be activity-regulated in vivo, is conveniently quantified using GFP-based translational reporters under control of the CaMKII 3′ UTR (48, 49). Finally, mRNP assemblies containing the translational repressor Me31B/RCK may be visualized and quantified in cell bodies and synapses in vivo (49).
Here we show that dFmr1 is required for LTH, a process in which it functions together with Atx2, Me31B, and Argonaute 1. dFmr1 and Atx2 are required not only in PNs but also in LNs, suggesting that translational control in both pre- and postsynaptic elements of the LN–PN synapse is required for this form of long-term synaptic and behavioral plasticity. Consistent with this interpretation, we find that dFmr1, Atx2, and several miRNA pathway components, Dicer1, the miRNA-pathway protein GW182, and Ago1, are required for repression of a CaMKII translational reporter both in PN and in LN processes in the antennal lobe. However, although the numbers of somatic and synaptic foci marked by Me31B are substantially dependent on Atx2, they are largely independent of dFmr1. We propose a model to account for these shared and distinct functions of dFmr1 and Atx2 in the specific in vivo contexts analyzed here.
Results
dFmr1 Is Required for Long-Term but Not Short-Term Olfactory Habituation.
The shared association of dFmr1 and Atx2 with stress granules and miRNA function led us to ask whether dFmr1 was required like Atx2 for LTH but not short-term habituation (STH). This issue is of interest because the simple and genetically accessible circuitry involved in olfactory habituation would allow us to compare the cell type-specific requirements for both proteins (31, 46).
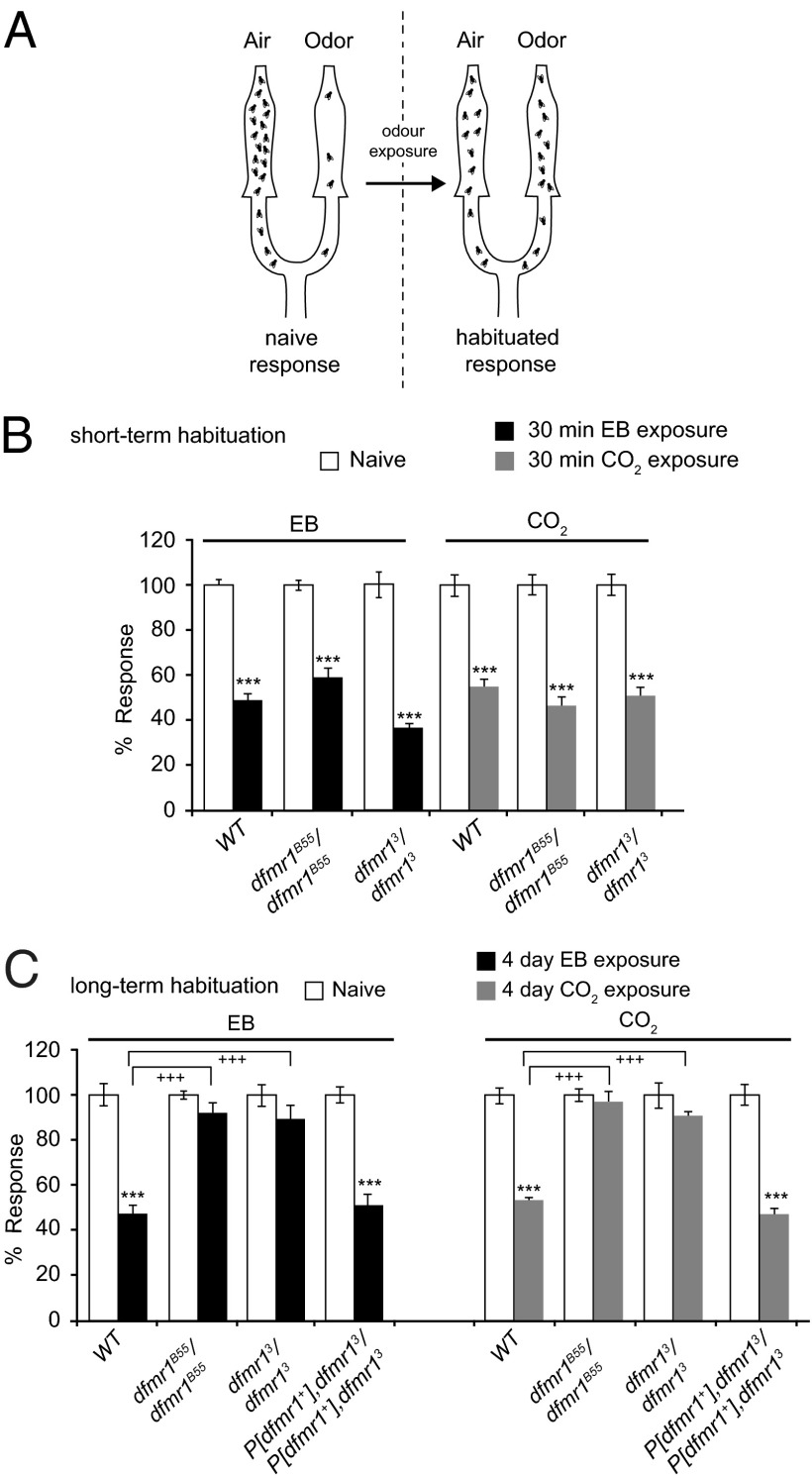
Habituation is measured by a transiently reduced avoidance to CO2 or ethyl butyrate (EB) induced by a prolonged exposure to the respective odorant (Fig. 1A). In contrast to STH that lasts for approximately 60 min, LTH caused by 4-d-long exposure to CO2 or EB persists for several days (46) (Materials and Methods). We found that dfmr1B55/dfmr1B55and dfmr13/dfmr13 mutants showed normal levels of avoidance to both EB and CO2 and normal STH to both odorants, which were at levels indistinguishable from control wild-type flies (Fig. 1B). In contrast, 4-d odorant-exposed dfmr1B55/dfmr1B55and dfmr13/dfmr13 mutants, tested 1 d after exposure, showed no significant LTH, with olfactory avoidance scores comparable to those of naïve, unexposed flies (Fig. 1C). Transgenic rescue experiments showed that the presence of a wild-type genomic dfmr1+ transgene in the mutant background completely restored normal LTH to both EB and CO2 to dfmr13/dfmr13 flies (Fig. 1C). Thus, dFmr1 is required for normal LTH but dispensable for STH.
Fig. 1.
dFmr1 mutants show normal STH but defective LTH. (A) Cartoon illustrating the Y-maze assay used for behavioral testing (46). (B) Flies were exposed to EB (black bars) or CO2 (gray bars) for 30 min or not exposed to any odor (white bars). Control flies (WT) and dFmr1 mutants both show comparable and normal levels of STH. (C) Reduction in odor avoidance in WT flies compared with dfmr1B55 and dfmr13 mutants. Flies were exposed to EB (black bars) or CO2 (gray bars) for 4 d or were exposed to mock odors paraffin oil or air, respectively (white bars). There is a significant defect in LTH in the mutants compared with control flies after exposure to EB/CO2. In comparison with control WT flies exposed to EB/CO2, which show reduced avoidance to the exposed odor, dfmr1B55 [q = 9.841, ***P < 0.001 (EB); q = 10.729, +++P < 0.001 (CO2), Student–Newman–Keuls (SNK) test] and dfmr13 [q = 910.776, ***P < 0.001 (EB); q = 9.927, +++P < 0.001 (CO2), SNK test] mutant flies fail to habituate. The behavioral phenotype of dFmr1 mutants was rescued by a genomic dFmr1 rescue construct P[dfmr1+] (16). Error bars show ±SEM (n > 8 sets). Raw RI values and n numbers are given in Table S1. ***P < 0.001 (Student t test in B and two-way ANOVA in C).
dFmr1 Is Required in PNs for LTH and Associated Physiological Plasticity.
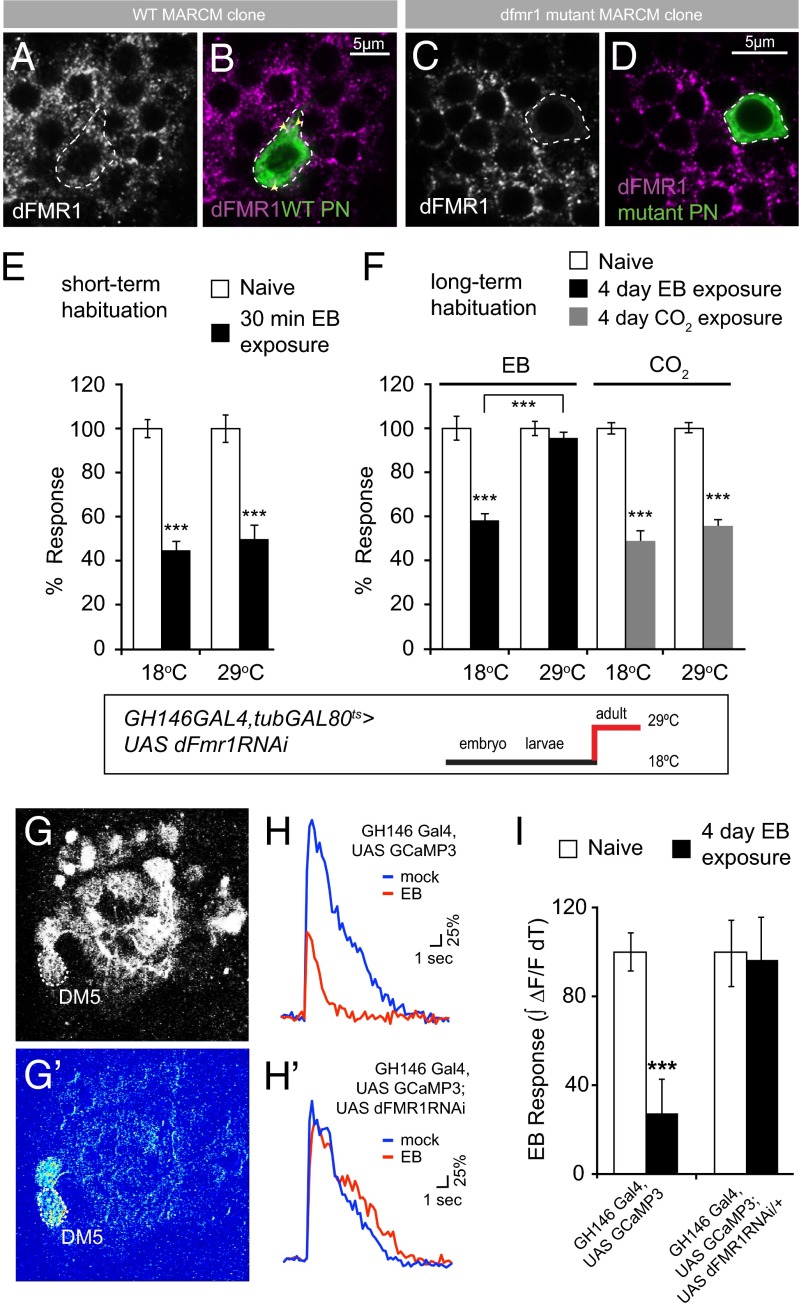
In additional experiments, we asked whether dFmr1, like Atx2, was required in PNs for LTH as well as for associated synaptic plasticity assessed through functional brain imaging. We first confirmed that the protein was expressed in PNs. An antibody against dFmr1 labeled large cytoplasmic foci (particles) in wild-type, but not dfmr1-mutant, PN cell bodies, which were visualized using the MARCM (mosaic analysis with a repressible cell marker) technique (Fig. 2 A–D).
Fig. 2.
dFmr1 is expressed in PNs innervating the antennal lobe and is required in PNs for LTH formation, which is associated with physiological changes. (A and B) Expression of dFmr1 (A, magenta in B) in a wild-type PN cell body (B, green). A confocal slice of a single PN clone generated using the MARCM technique (95) is shown. (C and D) Generating dFmr1 mutant PN cell bodies by MARCM, using the dfmr13 allele, depletes dFmr1 expression (C, magenta in D). Genotypes: (A and B) hsFLP,UAS-mCD8-GFP; GH146-Gal4; FRT82B/tub-gal80,FRT82B. (C and D) hsFLP,UAS-mCD8-GFP; GH146-Gal4; dfmr13,FRT82B/tub-gal80,FRT82B. (E and F) Adult-specific depletion of dFmr1 in PNs by RNAi (using tubGal80ts) does not affect STH but blocks LTH formation. (E) Flies exposed to EB (black bars) or paraffin oil (white bars); 30-min exposure at 18 °C or 29 °C shows normal STH to EB. (F) Flies were exposed to EB (black bars) or CO2 (gray bars) and to mock odors paraffin oil or air, respectively (white bars) at 18 °C or 29 °C. At 18 °C GH146-GAL4,tubGAL80ts>dFmr1RNAi flies show normal habituation, whereas at 29 °C, when dFmr1 is knocked down after eclosion and during exposure, these flies show a block in habituation to EB (q = 11.223, ***P < 0.001). The k/d of dFmr1 in GH146 subset of PNs does not show any effect on habituation to CO2. Genotype and schematic showing the developmental temperature profile is also given. Error bars show ±SEM (n > 8 sets). Raw RI values and n numbers are given in Table S1. ***P < 0.001 (Student t test except in F, which uses two-way ANOVA). (G–I) LTH to EB is accompanied by a dFmr1-dependent decrease in EB evoked responses in PNs. (G and G′) EB-evoked Ca2+ responses in the DM5 glomerulus; (G) baseline expression, (G′) pseudocolored image showing a response to a 2-s pulse of EB. (H) Representative traces of mean fluorescent change (∆F/F) from DM5 in paraffin oil- (blue lines) or EB-exposed (red lines) flies; (H) GH146-Gal4, UAS-GCaMP3/+; tub-Gal80ts/+; (H′) GH146-Gal4, UAS-GCaMP3/+; tub-Gal80ts/UAS-Fmr1-RNAi. (I) Mean area under the curve of the first 5.5 s of responses expressed as percentage of mock odor-evoked PN responses ± SEM. White bars indicate responses in mock-exposed flies, black bars indicate responses in EB-exposed flies. ***P < 0.001 (ANOVA).
To knock down (k/d) levels of dFmr1 in these PNs, we drove a transgenic RNAi construct in adult GH146-expressing PNs. We achieved this by using the Gal4/Gal80ts system, in which GH146-Gal4 provided spatial specificity of Gal4 expression in EB-responsive PNs and a Tub-Gal80ts transgene allowed temporal specificity. Temporal control was achieved by inactivation of Gal80 (a Gal4 inhibitor) by experimentally controlled shifts of flies from temperature permissive for Gal80ts function (18 °C) to one (29 °C) at which the Gal80ts protein is nonfunctional (50). Thus, flies of the genotype GH146-Gal4,Tub-Gal80ts>UASdFmr1(1-7)RNAi reared at 18 °C and shifted as adults to 29 °C (Fig. 2 E and F) developed normally through larval and pupal stages, but were specifically reduced for dFmr1 function in GH146-positive, adult PNs (Materials and Methods). Adult-specific k/d of dFmr1 in PNs had no effect on STH to EB but resulted in a complete block of LTH to this odorant (Fig. 2 E and F). As expected, the same flies show normal LTH to CO2 (Fig. 2 E and F), because GH146-Gal4 marks PNs that innervate EB-responsive glomeruli but not those that innervate the CO2-responsive V glomerulus (46, 51). Thus, dFmr1 is required in adult PNs to mediate odorant-selective LTH.
EB LTH is associated with a reduction in the EB-evoked physiological response of the DM5 PN, measured using functional imaging of odor-evoked responses through PN-expressed GCaMP, a genetically encoded calcium sensor that reports calcium fluxes in vivo (31, 46, 52). We tested whether this LTH-associated form of neural plasticity was dependent on dFmr1 function in PNs.
The EB-evoked calcium fluxes in DM5 glomeruli of control GH146-Gal4,UAS-GCaMP3/+;Tub-Gal80ts/+ flies shifted as adults from 18 °C to 29 °C were substantially decreased after 4-d EB exposure compared with siblings exposed to paraffin oil (the carrier solvent for EB). In contrast, EB-evoked fluxes in experimental GH146-Gal4,UAS-GCaMP3/+; Tub-Gal80ts/UASdFmr1(1-7)RNAi flies, in which dFmr1 levels have been knocked down specifically in adult PNs, were not reduced by identical 4-d EB exposure (Fig. 2 G–I). This clearly indicates that dFmr1 function in PNs is required for behavioral as well as physiological changes associated with LTH, which closely matches previous findings on Atx2 (31).
dFmr1 Interacts with Atx2 and Other miRNA Pathway Components for LTH.
The strong similarities between dfmr1 and atx2 mutant phenotypes in LTH suggested that the two proteins might work together for underlying activity-dependent changes in antennal lobe function. This suggestion is further strengthened by previous reports implicating both proteins (independently) in Ago-1 mediated, miRNA-dependent repression of neuronal mRNAs (16, 31, 33, 35, 53, 54). We therefore examined, first, whether dfmr1 and axt2 mutants showed transdominant phenotypic interactions, which strongly indicate closely linked functions in vivo; and second, whether dfmr1 mutations also showed transdominant genetic interactions in LTH with me31b and ago1 mutations, which show similar interactions with atx2 (46).
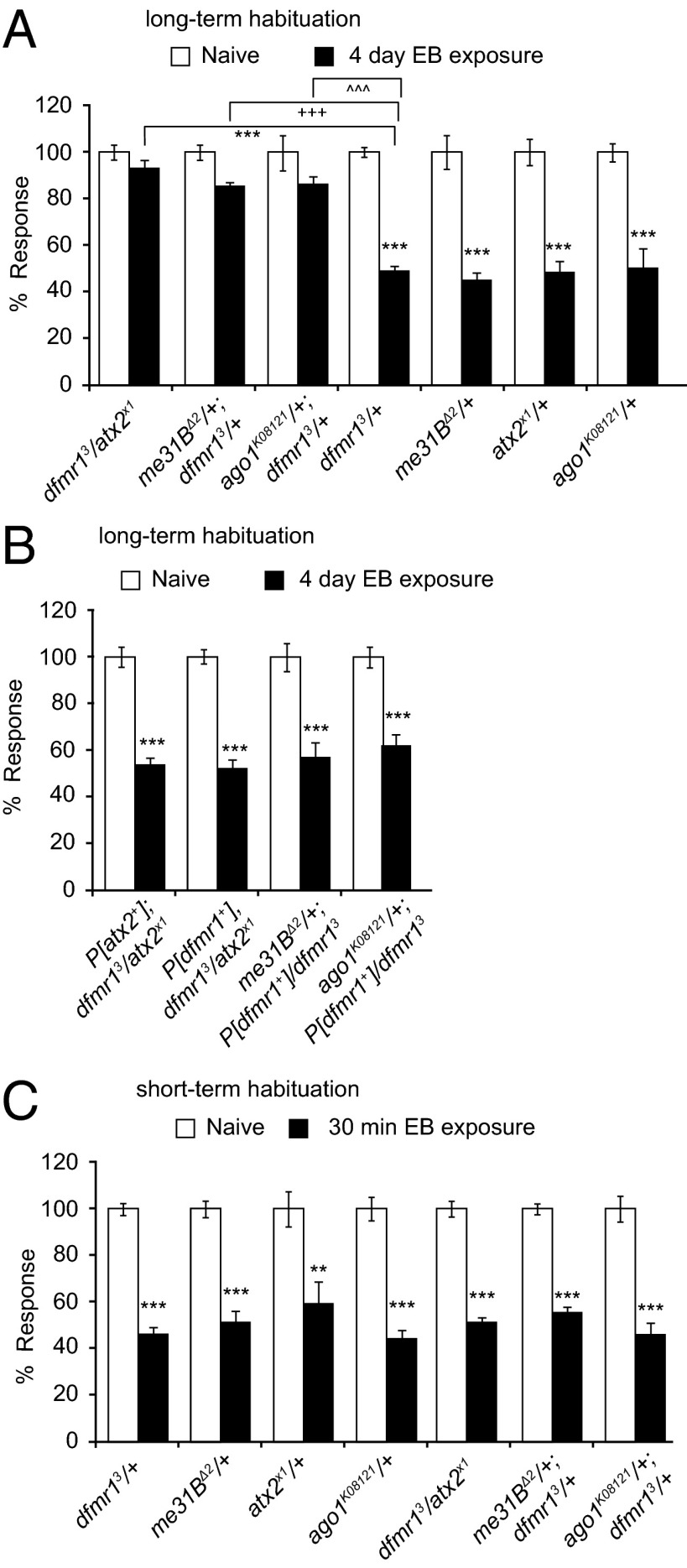
Compared with dfmr13/+ or atx2x1/+ heterozygotes, atx2x1/+;dfmr13/+ double heterozygote flies show strongly reduced LTH to EB or CO2 (Fig. 3A). In contrast, STH to these odorants remains unaffected (Fig. 3C). Similar strong and specific transdominant interactions are shown by transheterozygote combinations of dfmr13 with the atx2X1 interacting me31BΔ2 and ago1K08121 mutations (Fig. 3A). The same phenotype is shown by these flies when tested for LTH to CO2 (Fig. S1).
Fig. 3.
dFmr1 interacts with Atx2, Me31B, and Ago1 for LTH. (A) Compared with dFmr1 in heterozygous condition (dfmr13/+), transheterozygote flies for dfmr13/atx2x1 (q = 8.8220, ***P < 0.001), me31BΔ2/+ ;dfmr13/+ (q = 7.846, +++P < 0.001), and ago1K08121/+;dfmr13/+ (q = 6.383, ^^^P < 0.001) show a defect in LTH formation after 4-d exposure to EB (black bars). Flies heterozygous for Atx2, Me31B, and Ago1 (atx2x1/+, me31BΔ2/+, and ago1K08121/+) show normal LTH after exposure to EB. (B) The expression of an Ataxin2 genomic rescue transgene (P[atx2+]) (31) restores the LTH defect in dfmr13/atx2x1 flies, and LTH defects observed in dfmr13/atx2x1, me31BΔ2/+ ;dfmr13/+, and ago1K08121/+;dfmr13/+ flies are rescued by the expression of a dFmr1 genomic rescue transgene (P[dfmr1+]) (16). (C) Transheterozygote dfmr13/ atx2x1, me31BΔ2/+;dfmr13/+, and ago1K08121/+;dfmr13/+ flies, as well as heterozygous mutations, do not show a defect in STH; (A–C) black bars indicate EB-exposed and white bars indicate mock (parafin oil)-exposed flies. Error bars show ±SEM (n > 8 sets). Raw RI values and n numbers are given in Table S1. Student t test performed (**P < 0.01, ***P < 0.001) except in A, which uses two-way ANOVA.
To confirm that these double-heterozygote phenotypes truly reflected interactions between the relevant genes (and not unknown second-site mutations), we tested whether a genomic, wild-type atx2+ transgene in the dfmr13/atx2x1 flies or a genomic, wild-type dfmr1+ transgene in the other transheterozygote backgrounds would restore normal olfactory LTH. Consistent genetic rescue observed with these transgenes (Fig. 3B) provided strong support for the hypothesis that dFmr1 functions in long-term synaptic plasticity that underlies LTH in a processes that also involved Atx2, Me31B, and Ago1, known translational control factors that likely act to control activity-dependent RNA translation in vivo.
dFmr1, Atx2, and miRNA Pathway Components Repress Pre- and Postsynaptic CaMKII Expression.
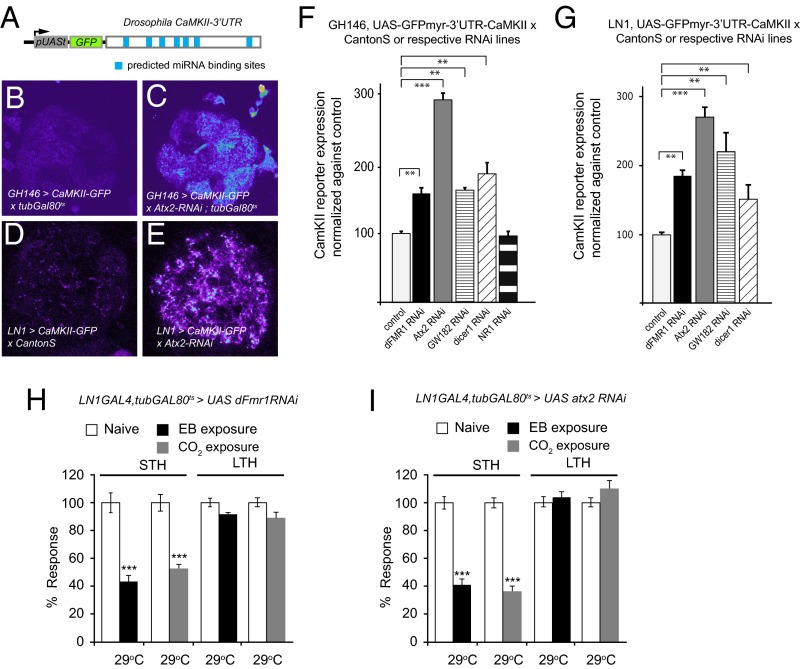
To directly test whether the dFmr1 interactions with Atx2 and Ago1 reflected a shared role in miRNA-mediated neuronal translational control, we tested whether dFmr1 or its interactors regulated synaptic expression of a GFP-based translational reporter under control of the CaMKII 3′ UTR (Fig. 4A). The CaMKII mRNA is dendritically localized, subject to activity-dependent translational activation, and contains, binding sites for several miRNAs and translational regulators in its 3′ UTR (48, 55, 56). We tested whether expression of this reporter mRNA is subject to common repression by dFmr1, and Atx2 in dendrites of PNs, as well as whether the reporter is subject to miRNA regulation in vivo, an issue that has not been clearly established by previous work (48, 49). For these experiments, we used RNAi to k/d target proteins in PNs expressing the CaMKII translational reporter. We used adult-specific k/d only in the case of atx2, where chronic expression of the atx2-RNAi construct in GH146-positive PNs caused reduced viability at 25 °C.
Fig. 4.
Regulation of CaMKII expression by dFmr1 and other microRNA pathway components in PNs and local interneurons (LNs) and requirement for dFmr1 and Atx2 in LNs for LTH formation. (A) Cartoon showing the CaMKII-reporter (UAS-myrGFP-CaMKII-3′UTR) used to assay CaMKII expression in PNs. (B–E) Pseudocolored representative confocal images of CaMKII-reporter expression by GH146-Gal4 in a subset of PNs (B and C) and by LN1-Gal4 in a subset of LNs (D and E). The depletion of atx2 in PNs and LNs by RNAi elevates expression levels of the CaMKII-reporter (C and E). (F) UAS-myrGFP-CaMKII-3′UTR fluorescence quantified in GH146 subset of PNs in the DA1 glomerulus of antennal lobes of WT and experimental flies (dFmr1, Aatx2, GW182, Dicer1, and NMDA receptor depleted by RNAi). Because chronic expression of the Atx2 RNAi construct in GH146 cells causes substantial inviability, adult-specific k/d of Atx2 using tub-Gal80ts was necessary. For Atx2, crosses were kept at 18 °C until late pupal stages, and Gal4 expression was induced by switching the flies to 30 °C for 4 d. (G) UAS-myrGFP-CaMKII-3′UTR fluorescence in LNs quantified in the whole-antennal lobes of WT and experimental flies (dFmr1, Atx2, GW182, and dicer1 depleted by RNAi). Error bars show ±SEM; **P < 0.01, ***P < 0.001 (Student t test). Measurements correspond to the average mean intensity of CaMKII reporter fluorescence. (H and I) Adult-specific depletion of dFmr1 and Atx2 in LNs by RNAi affect LTH formation. Flies exposed to EB (black bars) or CO2 (gray bars) and paraffin oil or air, respectively (white bars). Flies were shifted to 29 °C after eclosion and during odor exposure to specifically k/d dFmr1 and Atx2 in adults. (H) LN1-GAL4,tubGAL80ts>dfmr1RNAi flies show a defect in LTH to EB as well as CO2, whereas they show normal STH to both odors. (I) Adult-specific k/d of Atx2 in LNs shows similar defects in LTH; LN1-GAL4,tubGAL80ts>Atx2RNAi flies show a defect in LTH, whereas they show normal STH. Error bars show ±SEM (n > 8 sets). **P < 0.01, ***P < 0.001 (Student t test).
An important result was that k/d of dFmr1 or Atx2 in GH146-positive PNs causes a strong increase in CaMKII reporter expression (Fig. 4 B and C). The CaMKII reporter is also significantly up-regulated in PNs after k/d of key components of the miRNA machinery, including GW182, required for Ago-1–mediated translational control, and Dicer1, essential for miRNA biogenesis in Drosophila. Thus, as in GH146,UAS-myrGFP-CaMKII-3′UTR/UAS-dFmr1-RNAi1-7 flies, PN dendrites of GH146,UAS-myrGFP-CaMKII-3′UTR/UAS-GW182-RNAiVDRC and GH146,UAS-myrGFP-CaMKII-3′UTR/UAS-Dcr1RNAi flies showed enhanced levels (∼1.5-fold) of CaMKII reporter expression compared with control GH146,UAS-myrGFP-CaMKII-3′UTR/+ animals. This effect was specific: GH146,UAS-myrGFP-CaMKII-3′UTR/UAS-dsNR1flies, expressing an RNAi construct that targets the NMDA receptor not expected to repress mRNA translation, caused no detectable up-regulation of the reporter (Fig. 4F). These data are entirely consistent with dFmr1, Atx2, and other miRNA pathway components functioning together at the CaMKII 3′ UTR to repress its translation in PN dendrites.
dFmr1 and Atx2 Are Required in Local Interneurons for CaMKII Reporter Repression and LTH.
We observed that dFmr1 was expressed in central neurons, including LNs that send GABAergic processes to glomeruli in the antennal lobe. This was particularly interesting when taken in context of two recent observations indicating, first, a presynaptic role for FMRP in mammalian neurons (57), and second, roles for both pre- and postsynaptic translation in regulation of some forms of long-term synaptic plasticity in Aplysia (58). Thus, we were motivated to examine potential functions for dFmr1 and Atx2 in LN1 interneurons that are presynaptic to PNs.
Knockdown of dFmr1 or Atx2 in LN1 neurons expressing the CaMKII reporter results in considerable increase of reporter expression in LN processes in the antennal lobe (Fig. 4 D and E). Thus, LN1,UAS-myrGFP-CaMKII-3′UTR/UAS-dFmr1RNAi1-7 and LN1,UAS-myrGFP-CaMKII-3′UTR/UASAtx2 RNAi flies showed two- to threefold increase in the reporter expression in the antennal lobe compared with control LN1,UAS-myrGFP-CaMKII-3′UTR/+ animals (Fig. 4G). Likewise, k/d of Dicer1 or GW182 in LN1 also caused significant enhancement in reporter expression in the antennal lobe, indicating that CaMKII in LN1 processes was also repressed by a dFMR1, Atx2, and miRNA-mediated pathway similar to the one used in PN dendrites (Fig. 4G).
In light of the evidence for dFmr1 and Atx2-dependent translational control in LN1 neurons, we tested whether dFmr1 was also required in LNs for LTH. We found that adult-specific k/d of dFmr1 in LN1 neurons caused a block in LTH to both EB and CO2 after 4 d exposure (Fig. 4H), whereas the k/d has no effect on STH to both odors (Fig. 4H), indicating that dFmr1 mediates local translational regulation of CaMKII or other targets in LN1 neurons. Because Atx2 regulates CaMKII levels in LN processes as well, we tested whether k/d of Atx2 in LNs would affect LTH. We found that adult-specific k/d of Atx2 in the LN1 subset of interneurons also caused similar to dFmr1 k/d specific block in LTH to both EB and CO2 (Fig. 4I). Similar k/d of Me31B in LNs also selectively inhibited LTH (Fig. S2).
Neuronal Atx2 and dFmr1 Proteins Physically Associate and Bind to CaMKII mRNA.
Robust transdominant interactions between atx2 and dfmr1 mutations, similar transdominant interactions with me31b and ago1 mutations, as well as their common requirement for repressing CaMKII reporter expression in dendrites, are strongly suggestive of a direct interaction between dFmr1 and Atx2, as well as their physical presence on UTRs of CaMKII and other common mRNA targets. We used biochemical methods to test and thereby confirm this prediction.
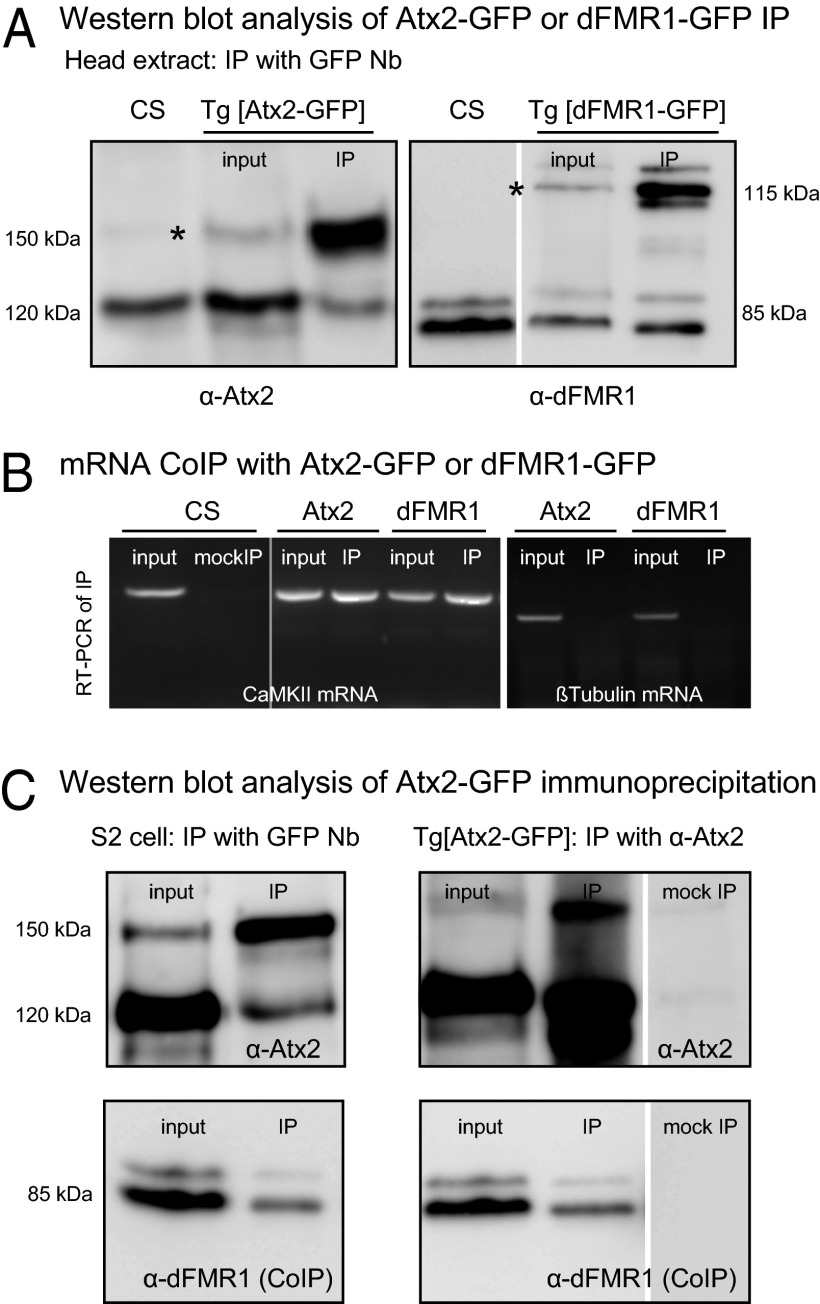
We developed an efficient method to immunoprecipitate (IP) Atx2 and dFMR1 proteins from fly heads by combining the use of a very robust and high-affinity llama antibody against GFP (59) (which we refer to as a GFP nanobody or GFP-Nb) with newly made transgenic lines (transgenome or Tg lines) that express functional forms of GFP- and epitope-tagged Atx2 or dFmr1 under endogenous cis regulatory control. Thus, we analyzed GFP-Nb immunoprecipitates from head extracts of Tg [Atx2GFP] or Tg [dFmr1GFP] flies, expressing tagged Atx2GFP or dFmr1GFP.
GFP-Nb coupled beads efficiently and selectively immunoprecipitated the GFP-tagged form of Atx2 (150 kDa) or dFmr1 (115 kDa) from appropriate fly head lysates (Fig. 5A). To test whether Atx2 and dFmr1 proteins were physically associated with CaMKII in brains, the RNA bound to GFP-Nb beads incubated with Tg [Atx2GFP] or Tg [dFmr1GFP] fly head lysate was extracted, and RT-PCR was performed with primers specific for either CaMKII-3′ UTR-specific or the also-abundant β-tubulin UTR. CaMKII mRNA was found to be present in the GFP-Nb immunoprecipitates of both Atx2 and dFmr1. In contrast, Nb coupled beads that were similarly treated with head extracts from CS flies (mock IP) did not yield any CaMKII upon RT-PCR, showing that the interaction between CaMKII and Atx2/dFmr1 seen in IPs with Tg [Atx2GFP] or Tg [dFmr1GFP] flies is specific and occurs via the respective tagged proteins. In contrast to CaMKII mRNA, RT-PCR of the same RNA using β-tubulin 3′ UTR-specific primers did not yield any product. Thus, the pulldown of CaMKII mRNA with Atx2 or dFMR1 reflects selective association (Fig. 5B).
Fig. 5.
Functional interaction between Atx2, dFMR1, and CaMKII mRNA. (A) IP of Atx2 (Left) and dFmr1 (Right) from head extracts of Tg [Atx2GFP] and Tg [dFmr1GFP] flies using GFP Nanobody (Nb). In comparison with the Input, there is specific enrichment of the tagged forms of Atx2 (150 kDa) and dFmr1 (115 kDa) in the IP sample, indicated by asterisks (*). (B) RT-PCR data (after RNA IP). cDNA reverse transcribed from the RNA extracted from GFP Nb coupled beads used to IP Atx2-GFP or dFmr1-GFP were amplified using CaMKII or β-tubulin–specific primers. Both Atx2-GFP and dFmr1-GFP immunoprecipitates yield CaMKII mRNA (Left) but not β-tubulin mRNA (Right; control). A mock IP, using GFP Nb beads with CS extracts, does not yield any CaMKII mRNA. (C) dFmr1 Co-IP with Atx2. IP of Atx2 from extracts of S2 cells transfected with Tg [Atx2-GFP] (Left), using GFP Nb coupled beads, contain tagged Atx2 (Upper) as well as dFmr1 (Lower). Similarly, immunoprecipitates of Atx2 from Tg [Atx2GFP] fly head extracts using Atx2 antibody (Right) contain not only Atx2 (Upper) but also co-IPed dFmr1 (Lower). A mock IP with just the beads without the Atx2 antibody does not yield any Atx2 or dFmr1. The interaction of Atx2 with dFmr1 is RNA independent because the IPs (for co-IP) were performed in the presence of RNase-A.
To test whether Atx2 and dFmr1 might function as a complex in vivo to regulate the translation of target transcripts, we tested whether Atx2 can co-IP dFmr1. IP from S2 cell extracts transfected with Tg [Atx2GFP] constructs, which showed a specific enrichment of the tagged Atx2, showed co-IP of dFmr1. IP of endogenous Atx2 from head extracts of the Tg [Atx2GFP] flies using a polyclonal antibody to Atx2 also showed co-IP of dFmr1 (Fig. 5C). These results indicate that dFmr1 and Atx2 interact to regulate common targets like CaMKII.
Differential Roles for dFmr1 and Atx2 k/d in Me31B Foci Formation.
dFmr1 and Atx2 orthologs in other species are present on Me31B-positive cytoplasmic mRNP aggregates related to stress granules (36, 60–63). In addition, Atx2 function seems to be essential for the formation of Me31B-containing mRNP assemblies both in yeast as well as in mammalian cultured cells (39). Given the potential significance of these assemblies for translational control, we examined how loss of dFmr1 or Atx2 affected Me31B foci that can be visualized and quantified in both cell bodies and synapses of single genetically marked projection neurons in vivo (49) (Fig. 6 A and F). For these studies, we used the MARCM technique to generate, in heterozygous animals, GFP-marked PNs that were homozygous for atx2X1 and dfmr13 mutations. Mosaic lobes were stained for GFP to visualize mutant PNs and with anti-Me31B antibodies to visualize and quantify Me31B foci in these cells. Control animals were identical but for the genotype of the GFP-marked PN, which was either wild type or atx2X1 mutant in the background of a wild-type “rescuing” atx2+ transgene.
Fig. 6.
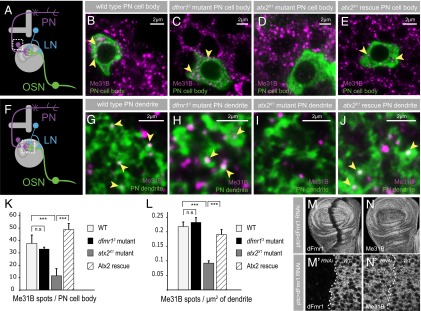
mRNP particles in dfmr1 and atx2 mutants. (A and F) Cartoons illustrating the neuronal circuitry in the adult antennal lobe; the boxed areas mark PN-cell bodies in A and the neuropil region containing PN dendrites in F. The effect of dfmr1 and atx2 mutants on mRNP particles, marked by Me31B (magenta), in projection neuron cell bodies (B–E, green) and dendrites (G–J, green). Mutant PN clones for dfmr13 and atx2X1 were generated using the MARCM technique (95). WT projection neurons show expression of Me31B mRNP particles (magenta) in the PN cell bodies (B) and dendrites (G) (green). The depletion of dFmr1 has no effect on the expression of Me31B in PN cell bodies (C) or dendrites (H). In contrast, the depletion of Atx2 reduces Me31B particles in PN cell bodies (D) and dendrites (I). The reduction of Me31B particles in atx2x1 mutant PN cell bodies and dendrites is rescued by a genomic Atx2 rescue construct (E and J). (K and L) Quantification of Me31B mRNP particles in PN cell bodies and dendrites. Error bars show ±SEM; ***P < 0.001 (Student t test). Measurements correspond to the average number of Me31B spots per PN cell body (K) or per µm2 of dendritic PN area in the neuropil (L). (M and N) dFmr1 depletion in a defined region of the larval wing imaginal disk by RNAi, using ptc-Gal4, does not effect Me31B expression (N). dFmr1 expression, in contrast, is depleted (M). (M′ and N′) Confocal image of an area in the wing imaginal disk scanned at a higher magnification.
Levels of Me31B particles were not visibly altered by loss of dFmr1. Thus, the number of Me31B particles in cell bodies and dendrites of dfmr13 mutant PNs (hsFLP,UAS-mCD8-GFP;NP225-Gal4;dfmr13,FRT82B/tub-Gal80,FRT82B) were not different compared with control wild-type PN cell bodies and dendrites (hsFLP,UAS-mCD8-GFP;NP225-Gal4;FRT82B/tub-Gal80,FRT82B) (Fig. 6 B and C, G and H, respectively). In contrast, Me31B particles were substantially reduced in cell bodies and the distant dendrites of PNs homozygous for atx2X1 (hsFLP,UAS-mCD8-GFP;NP225-Gal4;atx2X1,FRT82B/tub-Gal80,FRT82B) (Fig. 6 B and D, G and I). This reduction in Me31B foci numbers in atx2x1 mutant PNs was restored to normal levels by the expression of a fully functional Atx2+ genomic transgene in the background (hsFLP,UAS-mCD8-GFP;NP225-Gal4,P[atx2+]atx2X1,FRT82B/tub-Gal80,FRT82B) (Fig. 6 B and E, G and J), as quantified in Fig. 6K (PN cell body) and Fig. 6L (PN dendrite).
The distinctive effects of dFmr1 and Atx2 loss on Me31B foci were not confined to the nervous system. Thus, the k/d of dFmr1 in patched-Gal4 marked regions of the larval wing imaginal disk had no visible effect on Me31B levels or distribution (Fig. 6 M and N). However, similar k/d of Atx2 greatly reduced levels of Me31B foci in imaginal, epithelial cells (31).
The observations above are consistent with recent independent experiments indicating that dFMR1 is not required for the assembly of stress granules in Drosophila nonneuronal cells (64). Taken together, the data indicate that although dFmr1 and Atx2 function together with Me31B in the translational regulation of the dendritic CaMKII reporter mRNA, dFmr1 is dispensable but Atx2 broadly necessary for the presence of Me31B foci that represent likely sites of RNA regulation in neurons. Although these observations also indicate that dFmr1 is not essential for the dendritic transport of the majority of Me31B-containing mRNPs in vivo, it remains possible that dFmr1 contributes to transport dynamics or for trafficking of a subset of RNAs (65–67).
Discussion
Observations presented here lead to three significant insights into the endogenous functions of dFmr1 and Atx2 in the nervous system and their contribution to long-term synaptic plasticity. First, the data strongly indicate that both proteins function in the same pathway, namely translational control, to mediate the form of long-term memory analyzed here. Second, the remarkably similar effects of knocking down these proteins in LNs and PNs provide in vivo support for an emerging idea that translational control of mRNAs in both presynaptic and postsynaptic compartments of participating synapses is necessary for long-term synaptic plasticity (57, 68–72). Finally, although both dFmr1 and Atx2 have isoforms containing prion-like, Q/N domains (44), the different effects of loss of Atx2 and dFmr1 on neuronal Me31B aggregates indicate important differences in the mechanisms by which the two proteins function in translational control.
dFmr1 and Atx2 Function Together in the Nervous System.
The different molecular and clinical consequence of pathogenic mutations in FMRP and Atx2 encoding genes has led to largely different perspectives on their functions. Fragile X causative mutations cause reduced levels of the encoding mRNA and lower levels of FMRP, leading to increased protein synthesis and a range of pathologies evident in children and young adults. These pathologies importantly do not include the formation of inclusion bodies. In contrast, SCA-2 and amyotrophic laterosclerosis causative mutations in Atx2 result in the dominant formation of inclusion body pathologies and age-dependent degeneration of the affected neuronal types (27, 73). Observations made in this article indicate that the distinctive pathologies of the two diseases have obscured common molecular functions for the two proteins in vivo.
The genetic, behavioral, and biochemical observations show (i) shared roles of the two proteins in olfactory neurons for long-term but not short-term habituation, and (ii) striking transdominant genetic interactions of dfrm1 and atx2 mutations with each other as well as with miRNA pathway proteins, which is not only consistent with prior genetic and behavioral studies of the two respective proteins (16, 31) but also strongly indicative of a common role for the two proteins in translational repression of neuronal mRNAs. This conclusion is supported at a mechanistic level by (iii) the finding that both proteins are required for efficient repression mediated by the 3′ UTR of CaMKII, a 3′ UTR that we show to be repressed by the miRNA pathway, and (iv) strong evidence for in vivo biochemical interaction among dFmr1 and Atx2 and for binding of these regulatory proteins with the UTR of the CaMKII transcript that they jointly regulate. Thus, dFMR1 and Atx2 function with miRNA pathway proteins for the regulation of a dendritically localized mRNA in identified olfactory neurons.
Multiple Synaptic Sites for Translational Control Necessary for Long-Term Habituation.
An unexpected observation was that dFMR1 and Atx2 seemed to be necessary for LTH as well as for CaMKII reporter regulation in both inhibitory LNs and excitatory PNs of the antennal lobe.
Until recently mammalian FMRP was regarded as a postsynaptic protein, consistent with the view that translational control of mRNAs essential for long-term plasticity occurs exclusively in postsynaptic dendrites. In contrast, work in Aplysia indicated that translational control of mRNAs is required in presynaptic terminals for long-term synaptic plasticity. This conflict between vertebrate and invertebrate perspectives is beginning to be resolved by findings that (i) mammalian FMRP is present in axons and presynaptic terminals; and that (ii) translational control of both presynaptic and postsynaptic mRNAs is essential for long-term plasticity of cultured Aplysia sensorimotor synapses (57, 58, 68, 74).
Prior studies at the Drosophila neuromuscular junction have strongly indicated presynaptic functions for dFmr1 and translational control but have also pointed to their significant postsynaptic involvement in neuromuscular junction maturation, growth, and plasticity (75, 76). More direct studies of experience-induced long-term plasticity have been performed in the context of Drosophila olfactory associative memory, wherein a specific dFmr1 isoform in particular and translational control in general are necessary for long-term forms of memory (16, 44). However, the incomplete understanding of the underlying circuit mechanism has made it difficult to conclude presynaptic, postsynaptic, or dual locations for dFmr1 function in long-term memory. In contrast, recent work showing an essential role for Atx2 and Me31B in PNs for LTH more strongly indicate a postsynaptic requirement for translational control mediated by these proteins; however, this did not address a potential additional presynaptic function.
Our finding that dFmr1 and Atx2 are necessary in both LNs and PNs for LTH, a process driven by changes in the strength of LN–PN synapses, provides powerful in vivo support for a consensus model in which translational control on both sides of the synapse is necessary for long-term plasticity (68). A formal caveat is that the anatomy of LN–PN synapses in Drosophila antennal lobes remains to be clarified at the EM level. If it emerges that these are reciprocal, dendrodendritic synapses, similar to those between granule and mitral cells in the mammalian olfactory bulb (77–79), then a clear assignment of the terms “presynaptic” and “postsynaptic” to the deduced activities of dFmr1 and Atx2 in this context may require further experiments.
Different Mechanisms of Action for dFmr1 and Atx2.
Previous studies in Drosophila have indicated a broader role for Atx2 than dFmr1 in miRNA function in nonneuronal cells (31, 80). Although Atx2 was necessary for optimal repression of four miRNA sensors examined in wing imaginal disk cells, dFmr1 was not necessary for repression of any of these sensors (31, 80). The resulting conclusion that dFmr1 is required only for a subset of miRNAs to function in context of specific UTRs is consistent with the observation that only a subset of neuronal miRNAs associate with mammalian FMRP (54) and that the protein shows poor colocalization with miRNA pathway and P-body components in mammalian cells. Parallel studies have shown that Atx2 in cells from yeast to man is required for the formation of mRNP aggregates termed stress granules, which in mammalian cells also contain Me31B/RCK and FMRP (39, 63, 81). In addition, biochemical interactions between these proteins and their mammalian homologs with each other as well as with other components of the miRNA pathway have been reported (35, 36, 82–84). However, neither the mechanisms of Atx2-driven mRNP assembly, nor the potential role for FMRP in such assembly, have been tested in molecular detail (42).
Our demonstration that loss of Atx2 in neurons results in a substantial depletion of Me31B-positive foci in PN cell bodies and in dendrites is consistent with Atx2 being required for the assembly of these two different (somatic and synaptic) in vivo mRNP assemblies. Thus, the mechanisms that govern their assembly, particularly of synaptic mRNPs in vivo, overlap with mechanisms used in P-body and stress granule assembly in nonneuronal cells.
The finding that loss of dFmr1 has no visible effect on these Me31B-positive foci can be explained using either of two models. A simple model is that dFmr1 is not required for mRNP assembly, a function mediated exclusively by Atx2. This would suggest that Atx2 contains one or more functional domains missing in dFmr1 that allow the multivalent interactions necessary for mRNP assembly. This is most consistent with the observation that that although dFMR1 is a component of stress granules in Drosophila nonneuronal cells, it is not required for their assembly (64). An alternative model would allow both dFmr1 and Atx2 to mediate mRNP assembly but posit that dFmr1 is only present on a small subset of mRNPs, in contrast to Atx2, which is present on the majority. In such a scenario, loss of dFmr1 would only affect a very small number of mRNPs, too low to detect using the microscopic methods we have used here. In the context of these models, it is interesting that both dFmr1 and Atx2 contain prion-like Q/N domains, potentially capable of mediating mRNP assembly. It is to be noted here that the dFmr1 Q/N domain, although lacking prion-forming properties, is capable of serving as a protein interaction domain enabling the assembly of dFmr1 into RNP complexes (44). This observation would support the view that dFmr1 may be involved in the formation of only a subset of cellular mRNP complexes. Future studies that probe the potential distinctive properties of these assembly domains may help discriminate between these models. In addition, potential interaction of Atx2 with other proteins that are involved in mRNP formation across species, like Staufen, could help to understand the mechanisms behind Atx2-dependent function in mRNP assembly.
However, the observations presented here clearly show that despite the remarkable similarities in the roles of dFmr1 and Atx2 for repression of CaMKII expression at synapses and the control of synaptic plasticity that underlies long-term olfactory habituation, both proteins also have distinctive molecular functions in vivo.
Concluding Remarks.
Mutations that affect neuronal translational control are frequently associated with neurological disease, particularly with autism and neurodegeneration (1, 85, 86). Although these clinical conditions differ substantially in their presentation, a broadly common element is the reduced ability to adapt dynamically to changing environments, a process that may require activity-regulated translational control at synapses. Taken together with others, our observations suggest that there may be two routes to defective activity-regulated translation. First, as in dFmr1 mutants, the key mRNAs are no longer sequestered and repressed, leading to a reduced ability to induce a necessary activity-induced increase in their translation. Second, we suggest that increased aggregation of neuronal mRNPs (indicated by the frequent occurrence of TDP-43 and Atx2-positive mRNP aggregates in neurodegenerative disease) (87, 88) may result in a pathologically hyperrepressed state from which key mRNAs cannot be recruited for activity-induced translation. Thus, altered activity-regulated translation may provide a partial explanation not only for defects in memory consolidation associated with early-stage neurodegenerative disease but also for defects in adaptive ability seen in autism spectrum disorders.
Materials and Methods
Drosophila Stocks.
Fly stocks were raised at 25 °C unless mentioned otherwise on standard cornmeal and agar media. Wild-type stocks (Canton-S), atx2 genomic-rescue P[atx2+] (31), and UAS-myrGFP CaMKII 3′ UTR (49) were from the Ramaswami stock collection. tubGal80ts2X was from V. Rodrigues (National Centre for Biological Sciences, Bangalore, India). dfmr13, dfmr1B55, UAS-dFmr1RNAi1-7 (II), and dFmr1 genomic rescue P[dFmr1+] were from F. Bolduc (University of Alberta, Canada) (16). UAS-Atx2RNAi (II) (34955), UAS-GW182RNAi (II) (45772), and UAS-dicer1RNAi (II) (24666) were from the Vienna Drosophila RNAi Center stock collection; UAS-dsNR1 was from T. Tully [Cold Spring Harbor Laboratory, New York (89, 90)]; UAS-Me31B RNAi 4916R-1 [on X (36)] was from the NIG-FLY collection. atx2X1,FRT82B was from L. Pallanck (University of Washington, Seattle). dfmr13,FRT82B was from D. Zarnescu (University of Arizona, Tucson, AZ). Me31BΔ2was from A. Nakamura (Riken Center, Kobe, Japan). ago1K08121 was from J. Dubnau (Cold Spring Harbor Laboratory). UAS-GCaMP3 was from V. Jayaraman and L. Looger (Janelia Farm Research Campus, Ashburn, VA) (91). hsFLP,mCD8-GFP was from G. Jefferis [Laboratory of Molecular Biology (LMB), Cambridge, United Kingdom]. GH146-Gal4, LN1-Gal4, ptc-Gal4, dfmr150M, FRT82B, and FRT82B,tubGal80 were from the Bloomington stock center.
GFP-Tagged Genomic Transgenes.
GFP sequence was cloned in-frame onto the 3′ end of the Atx2 and dFMR1 ORFs by Red/ET recombineering, resulting in a C-terminal GFP tag. FlyFos031746 and FlyFos019181 (92), respectively, were used as target plasmids. For Atx2 the isoforms RB, RC, and RD were tagged, for dFMR1 isoforms RA, RC, RD, RE, RF, and RG. Transgenes for these constructs were generated using phiC31-mediated recombinase-mediated cassette exchange using standard techniques (93).
Immunohistochemistry.
Drosophila adult flies were decapitated and heads transferred to chilled adult haemolymph (AHL) solution. The brains were removed from the head capsule, and any attached trachea and surrounding tissues were carefully removed. The dissected brains were transferred into a 0.5-mL tube containing 4% (wt/vol) paraformaldehyde diluted in PBS containing 0.2% Triton-X100 (PTX) and fixed for 20 min at room temperature. Fixed samples were washed four times for 15 min in PTX, blocked with blocking solution [PBS + 0.2% Triton + 5% (vol/vol) normal goat serum] for 1 h and then incubated in primary antibody diluted in blocking solution overnight at 4 °C, followed by incubation with secondary antibodies for 3 h at room temperature diluted in blocking solution.
Larval wing imaginal discs were dissected in PBS and fixed in 4% (wt/vol) formaldehyde diluted in PBS for 20 min at room temperature. Fixed samples were washed four times for 15 min each in PTX, blocked with blocking solution [PBS + 0.2% Triton + 5% (vol/vol) normal goat serum], and then incubated in primary antibody diluted in blocking solution overnight at 4 °C, followed by incubation with secondary antibodies for 3 h at room temperature diluted in blocking solution.
Primary antibodies used: mouse anti-Me31B (1:100) (94), rabbit anti-GFP (1:1,000) (Molecular Probes), chicken anti-GFP (1:1,000) (Abcam), mouse anti-dFmr1 5A11 (1:100; primary antibody incubation over two nights at 4 °C) (Developmental Studies Hybridoma Bank), and mouse anti-dFmr1-sp(507) (1:300; primary antibody incubation over two nights at 4 °C; B. Hassan, Katholieke Universiteit Leuven, Leuven, Belgium).
Secondary antibodies used were Alexa 488- and Alexa 555-conjugated anti-rabbit and anti-mouse IgG (1:1,000) (Molecular Probes). Preparations were mounted in Vectashield Mounting Medium (Vector Laboratories) on slides using coverslips (thickness no. 1) as spacers and imaged on a Zeiss LSM 510-meta confocal microscope.
Inducing and Measuring Olfactory Habituation.
The olfactory response was assayed, and LTH and STH were induced as previously described (46). Briefly, olfactory responses were measured in sets of ∼25–40 flies using the Y-Maze apparatus. The response index (RI) was defined as the fractional avoidance of an odorant-containing arm of the maze compared with a control arm containing air [RI = N(odor) − N(air)/N(total)]. N equals number of flies. Successful STH shows a reduced RI after 30-min exposure to specific odors [20% (vol/vol) EB or 5% (vol/vol) CO2]. In LTH flies show a reduced RI after 4-d exposure. Either ANOVA or Student’s t test was used to estimate significance of all pairwise comparisons examined. Raw data and “n” values for the experiments are provided in Table S1.
Image Acquisition.
All images were taken on a Zeiss LSM 510 confocal microscope. To image Me31B mRNP particles in fixed brain tissue, we used a 63× objective (Zeiss Plan-Apochromate, 1.4 Oil Ph3) and the “digital zoom” software feature of the LSM 510 software (zoom factor between 3 and 4) that allows scanning of a region of interest at a higher pixel resolution, constrained of course by the normal ∼300-nm resolution limit of light microscopy. Regions of particular interest were selected in Photoshop (Adobe Systems) and presented in a way to allow better evaluation by the reader.
MARCM to Create and Visualize Single PNs of Defined Genotype.
For the induction of MARCM clones Drosophila (95), crosses of the respective genotype were raised at 25 °C, and the flies were transferred regularly to a new vial for timed egg collections. A heat-shock pulse at 37 °C in a water bath for 1–2 h was given at optimal time points to induce single-cell clones in PNs projecting to the DL-1 glomerulus. The time points (between 0 and 60 h after larval hatching) were chosen after G. Jefferis (96). Heat-shocked larvae were transferred back to 25 °C, raised to adulthood, and the adult flies were dissected 1–4 d after eclosion.
Immunoprecipitation.
For each genotype, 300 fly heads were homogenized in 600 µL lysis buffer [25 mM Tris·HCl (pH 7.5), 150 mM NaCl, 10% (vol/vol) glycerol, 1 mM EDTA, 1 mM DTT, 0.5% Nonidet P-40, and Complete Protease Inhibitor Tablets from Roche] and incubated for 20 min at 4 °C. Soluble fraction was incubated with 30 µL of GFP-TrapA beads (ChromoTek) for 2 h at 4 °C. After washes, bound proteins were extracted from beads using SDS sample buffer and analyzed by Western blot. For IP from S2 cells, the cell pellet was lysed in 500 µL 150 mM NaCl lysis buffer, and cytoplasmic fraction was obtained. Further IP was performed similarly using GFP Nanobody beads (59). For IP with Atx2 antibody from fly heads, 600 µL precleared head lysate was incubated with 3 µL Atx2 overnight at 4 °C . Protein G beads (Millipore) were then added for 2 h at 4 °C, followed by washes and elution as mentioned above. Western blots were performed using guinea pig anti-Atx2 [1:4,000 (97)] and mouse anti-dFMR1 (6A15, Abcam; 1:1,000). Secondary antibodies conjugated with HRP from Jackson Immunoresearch were used at 1:10,000 dilution. For co-IP experiments RNase-A (Invitrogen) was added to the soluble fraction and incubated at room temperature for 15 min before IP.
For RNA immunoprecipitation 300 fly heads were lysed in 600 µL lysis buffer [25 mM Tris·HCl (pH 7.5), 300 mM NaCl, 10% (vol/vol) glycerol, 1 mM EDTA, 1 mM DTT, 0.5% Nonidet P-40, and Complete Protease Inhibitor Tablets and Protector RNase Inhibitor (100 U/mL), both from Roche]. IP was performed as mentioned above. After washes, RNA was extracted using phenol-chloroform reagent (pH 4.5; Ambion) followed by alcohol precipitation. cDNA synthesis was done using the oligo dT primers and SuperScript III (Invitrogen Life Kit), followed by RT-PCR using CaMKII/tubulin-specific primers. Each experiment was repeated thrice.
CaMKII Reporter Assay.
For the quantification of myrGFP-CaMKII 3′ UTR reporter levels, adult Drosophila brains of the respective genotype were dissected in AHL, fixed for 20 min in 4% (wt/vol) paraformaldehyde diluted in PTX, and washed in PTX three times for 20 min. Preparations were mounted immediately in Vectashield Mounting Medium (Vector Laboratories) and imaged on a Zeiss LSM 510-meta confocal microscope. The obtained images where quantified for mean fluorescence of CaMKII-GFP in specific areas of interest.
Quantification of mRNP Particles.
Somatic spots were analyzed by “Spotnik,” a MATLAB plug-in developed in collaboration with Kangyu Pan and Anil Kokaram (49). Dendritic spots were analyzed in Photoshop. Images were double blinded and quantified for the amount of Me31B spots in certain regions of interest.
Imaging Calcium Dynamics in Vivo.
For two-photon imaging of calcium dynamics in PNs we used the calcium sensor GCaMP3 (91) expressed in PNs under the control of GH146-Gal4. Isolated brain preparation, odor stimulations, and image analysis was performed as previously described (46).
Supplementary Material
Acknowledgments
We thank Madhumala Sadanandappa, John Lee, and Isabell Twick for advice and help throughout the course of these experiments, and L. Pallanck, D. Zarnescu, A. Nakamura, J. Dubnau, G. Jefferis, T. Tully, V. Jayaraman, L. Looger, and Bassem Hassan (Vlaams Instituut voor Biotechnologie, Flanders, Belgium), and also Bloomington and Vienna Stock Centers, for fly stocks and reagents. This work was funded by grants from Science Foundation Ireland (to M.R.), the Government of India Department of Biotechnology (to K.V.), and Core funds from the National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore, India. I.P.S. was supported by a Government of India Council of Scientific and Industrial Research (CSIR) postgraduate fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309543111/-/DCSupplemental.
References
- 1.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31(45):16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7(12):1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: An overview. J Neurosci. 2006;26(27):7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeitelhofer M, et al. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28(30):7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5(4):635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Mikl M, Vendra G, Doyle M, Kiebler MA. RNA localization in neurite morphogenesis and synaptic regulation: Current evidence and novel approaches. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2010;196(5):321–334. doi: 10.1007/s00359-010-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bramham CR, Wells DG. Dendritic mRNA: Transport, translation and function. Nat Rev Neurosci. 2007;8(10):776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 10.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 11.Oberlé I, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann WE, et al. Autism spectrum disorder in fragile X syndrome: Communication, social interaction, and specific behaviors. Am J Med Genet A. 2004;129A(3):225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez RN, et al. Autism spectrum disorder in fragile X syndrome: A longitudinal evaluation. Am J Med Genet A. 2009;149A(6):1125–1137. doi: 10.1002/ajmg.a.32848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25(1):315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 15.Swanson MS, Orr HT. Fragile X tremor/ataxia syndrome: Blame the messenger! Neuron. 2007;55(4):535–537. doi: 10.1016/j.neuron.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11(10):1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009;19(3):319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenough WT, et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci USA. 2001;98(13):7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24(11):2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 21.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27(7):370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lastres-Becker I, Rüb U, Auburger G. Spinocerebellar ataxia 2 (SCA2) Cerebellum. 2008;7(2):115–124. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 24.Orr HT. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22(3):251–255. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Ramahi I, et al. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007;3(12):e234. doi: 10.1371/journal.pgen.0030234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessing D, Bonini NM. Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biol. 2008;6(2):e29. doi: 10.1371/journal.pbio.0060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 28.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100(3):323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 29.Neuwald AF, Koonin EV. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J Mol Med (Berl) 1998;76(1):3–5. doi: 10.1007/s001090050184. [DOI] [PubMed] [Google Scholar]
- 30.Achsel T, Stark H, Lührmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc Natl Acad Sci USA. 2001;98(7):3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci USA. 2011;108(36):E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16(19):2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin P, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7(2):113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 36.Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52(6):997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1(1):109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, et al. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19(1):105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18(4):1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA. 2009;15(10):1814–1821. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15(16):2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 42.Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell. 2009;20(1):428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dockendorff TC, et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34(6):973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee P, et al. Short- and long-term memory are modulated by multiple isoforms of the fragile X mental retardation protein. J Neurosci. 2010;30(19):6782–6792. doi: 10.1523/JNEUROSCI.6369-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride SM, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45(5):753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 46.Das S, et al. Plasticity of local GABAergic interneurons drives olfactory habituation. Proc Natl Acad Sci USA. 2011;108(36):E646–E654. doi: 10.1073/pnas.1106411108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudhakaran IP, et al. Plasticity of recurrent inhibition in the Drosophila antennal lobe. J Neurosci. 2012;32(21):7225–7231. doi: 10.1523/JNEUROSCI.1099-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Hillebrand J, et al. The Me31B DEAD-box helicase localizes to postsynaptic foci and regulates expression of a CaMKII reporter mRNA in dendrites of Drosophila olfactory projection neurons. Front Neural Circuits. 2010;4:121. doi: 10.3389/fncir.2010.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 51.Suh GS, et al. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431(7010):854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- 52.Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112(2):271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 53.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42(5):673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65(3):373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA. 1996;93(23):13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30(2):489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 57.Akins MR, Berk-Rauch HE, Fallon JR. Presynaptic translation: Stepping out of the postsynaptic shadow. Front Neural Circuits. 2009;3:17. doi: 10.3389/neuro.04.017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Till SM, Li HL, Miniaci MC, Kandel ER, Choi YB. A presynaptic role for FMRP during protein synthesis-dependent long-term plasticity in Aplysia. Learn Mem. 2011;18(1):39–48. doi: 10.1101/lm.1958811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7(2):282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell. 2009;36(6):932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hillebrand J, Barbee SA, Ramaswami M. P-body components, microRNA regulation, and synaptic plasticity. Scientific WorldJournal. 2007;7:178–190. doi: 10.1100/tsw.2007.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183(3):441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson P, Kedersha N. Stress granules: The Tao of RNA triage. Trends Biochem Sci. 2008;33(3):141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Gareau C, et al. Characterization of fragile X mental retardation protein recruitment and dynamics in Drosophila stress granules. PLoS ONE. 2013;8(2):e55342. doi: 10.1371/journal.pone.0055342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: The roots of fragile X syndrome. Nat Rev Neurosci. 2005;6(5):376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 66.Estes PS, O’Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol Cell Neurosci. 2008;39(2):170–179. doi: 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14(6):926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glanzman DL. Common mechanisms of synaptic plasticity in vertebrates and invertebrates. Curr Biol. 2010;20(1):R31–R36. doi: 10.1016/j.cub.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324(5934):1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin KC, et al. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell. 1997;91(7):927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 71.Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol. 2005;64(1):116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- 72.Chen CC, et al. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335(6069):678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- 73.Orr HT. Cell biology of spinocerebellar ataxia. J Cell Biol. 2012;197(2):167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai D, Chen S, Glanzman DL. Postsynaptic regulation of long-term facilitation in Aplysia. Curr Biol. 2008;18(12):920–925. doi: 10.1016/j.cub.2008.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135(8):1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107(5):591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 77.Rall W, Shepherd GM, Reese TS, Brightman MW. Dendrodendritic synaptic pathway for inhibition in the olfactory bulb. Exp Neurol. 1966;14(1):44–56. doi: 10.1016/0014-4886(66)90023-9. [DOI] [PubMed] [Google Scholar]
- 78.Rall W, Shepherd GM. Theoretical reconstruction of field potentials and dendrodendritic synaptic interactions in olfactory bulb. J Neurophysiol. 1968;31(6):884–915. doi: 10.1152/jn.1968.31.6.884. [DOI] [PubMed] [Google Scholar]
- 79.Isaacson JS, Strowbridge BW. Olfactory reciprocal synapses: Dendritic signaling in the CNS. Neuron. 1998;20(4):749–761. doi: 10.1016/s0896-6273(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 80.Cziko AMJ, et al. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182(4):1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ralser M, et al. An integrative approach to gain insights into the cellular function of human ataxin-2. J Mol Biol. 2005;346(1):203–214. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 82.Sutton MA, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125(4):785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 83.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee EK, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17(6):732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493(7432):327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelleher RJ, 3rd, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 87.Ginsberg SD, et al. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998;96(5):487–494. doi: 10.1007/s004010050923. [DOI] [PubMed] [Google Scholar]
- 88.Couthouis J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu CL, et al. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10(12):1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia S, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15(7):603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ejsmont RK, Sarov M, Winkler S, Lipinski KA, Tomancak P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat Methods. 2009;6(6):435–437. doi: 10.1038/nmeth.1334. [DOI] [PubMed] [Google Scholar]
- 93.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128(17):3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 95.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24(5):251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 96.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414(6860):204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340(6134):879–882. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.