Significance
Cancer cells favor high rates of aerobic glycolysis, or the Warburg effect, which is mediated by a key molecule, pyruvate kinase muscle isozyme (PKM)2. PKM2 functions both as a cytosolic enzyme and a nuclear factor in tumor cells. This report shows that PKM2’s nuclear translocation is regulated by Jumonji C domain-containing dioxygenase (JMJD)5 via direct physical binding. JMJD5 hinders the PKM2 tetrameric assembly and facilitates PKM2’s nuclear translocation. Together, they modulate hypoxia-inducible factor 1α-mediated transcriptional reprogramming of metabolic genes. These results reveal a mechanism whereby PKM2’s activity can be modulated by a dioxygenase/demethylase.
Keywords: Warburg effect, aerobic glycolysis, breast cancer, cancer metabolism
Abstract
JMJD5, a Jumonji C domain-containing dioxygenase, is important for embryonic development and cancer growth. Here, we show that JMJD5 is up-regulated by hypoxia and is crucial for hypoxia-induced cell proliferation. JMJD5 interacts directly with pyruvate kinase muscle isozyme (PKM)2 to modulate metabolic flux in cancer cells. The JMJD5-PKM2 interaction resides at the intersubunit interface region of PKM2, which hinders PKM2 tetramerization and blocks pyruvate kinase activity. This interaction also influences translocation of PKM2 into the nucleus and promotes hypoxia-inducible factor (HIF)-1α–mediated transactivation. JMJD5 knockdown inhibits the transcription of the PKM2–HIF-1α target genes involved in glucose metabolism, resulting in a reduction of glucose uptake and lactate secretion in cancer cells. JMJD5, along with PKM2 and HIF-1α, is recruited to the hypoxia response element site in the lactate dehydrogenase A and PKM2 loci and mediates the recruitment of the latter two proteins. Our data uncover a mechanism whereby PKM2 can be regulated by factor-binding–induced homo/heterooligomeric restructuring, paving the way to cell metabolic reprogram.
JMJD5 is a Jumonji C domain-containing dioxygenase shown to be involved in lysine demethylation (1–3) and hydroxylation functions (4). Although the exact cellular substrates and functions of JMJD5 remain unclear, JMJD5 was shown to positively regulate cyclin A1 but negatively regulate p53 and p21 (1–3). Knockdown of JMJD5 in Michigan Cancer Foundation (MCF)-7 cells inhibits cell proliferation (1), and JMJD5−/− embryos showed severe growth retardation, resulting in embryonic lethality at the midgestation stage (3). These data, together with its general overexpression in tumor tissues, implicate a role of JMJD5 in carcinogenesis. In this paper, we define a role of JMJD5 in regulating tumor metabolism under normoxic and hypoxic conditions through its interaction with pyruvate kinase muscle isozyme (PKM)2.
One of the hallmarks of cancer cells is their altered metabolism, referred to as aerobic glycolysis, or the Warburg effect (5). This generally involves an increased uptake of glucose, use of intracellular glucose to pyruvate via glycolysis, and the conversion into lactate in the presence of sufficient oxygen. Along this metabolic flux, PKM1 or its spliced variant, PKM2, which dephosphorylates phosphoenolpyruvate (PEP) into pyruvate, the last step of glycolysis, is an important signal integrator whose activities determine the cytosolic level of pyruvate, thereby affecting subsequent metabolic flow to lactate, tricarboxylic acid cycle or biosynthetic pathway (6). Enzymatically, PKM2, an embryonic isoform found abundantly in tumor cells, is less active than PKM1, which allows the accumulation of glycolytic intermediates and diversion into biosynthetic pathways, demanded by rapid-proliferating cells.
As a pivotal regulator of tumor metabolism, PKM2’s activity is further modulated by allosteric regulation via cofactor bindings, oligomerization, and posttranslational modification to cope with the ever-changing environment for tumor growth. For example, cytosolic PKM2 is allosterically activated by fructose 1,6-bisphosphate (FBP), amino acid serine (7), and SAICAR (succinylaminoimidazolecarboxamide ribose-5′-phosphate), a metabolite of the de novo purine nucleotide synthesis pathway (8). On the other hand, various stimuli including growth factors are known to negatively affect the pyruvate kinase activity of PKM2, thereby diverting the metabolic flow to the anabolic process. This was carried out principally by posttranslational modifications: Y105 phosphorylation by FGFR1 (9, 10), K305 acetylation in the presence of high-glucose concentrations (11), and C358 oxidation upon elevated reactive oxygen species levels (12). However, another way of modulating cytosolic PKM2 activity is its translocation into nucleus, where it serves as a transcriptional coactivator (13, 14) or as a phosphotransferase (15) to modulate transcriptional program. Thus, PKM2 is found to function as a coactivator for Oct4, β-catenin, Stat3, and hypoxia-inducible factor (HIF)-1α (13–16). PKM2 interacts with Oct4 to increase Oct4-mediated transactivation potential (16). Nuclear PKM2 associates with phosphorylated β-catenin upon EGFR activation to promote cyclin D1 (13). PKM2 binds to and phosphorylates Stat3 and histone H3 (17), activating gene transcription and tumorigenesis. Thus, both cytosolic and nuclear PKM2 contribute to altered metabolism and proliferation in cancer. The mechanisms associated with PKM2 translocation are multiple, including phosphorylation by ERK1 at S37 (18) and hydroxylation by PHD3 at P403 and P408 (14). The latter allows PKM2 to bind HIF-1α and hypoxia response elements (HREs) to regulate metabolic gene transcription and promote tumorigenesis under hypoxic conditions.
In this investigation, we describe a role of JMJD5 as a major regulator of PKM2. We describe a mechanism whereby PKM2 is translocated into the nucleus to regulate cancer metabolism. We show that JMJD5 is up-regulated and contributes to hypoxia-induced cell proliferation. We demonstrate that JMJD5 interacts with PKM2, alters the monomer:dimer:tetramer equilibrium, and blocks pyruvate kinase activity. Furthermore, JMJD5 accelerates the translocation of PKM2 into the nucleus and enhances the binding of HIF-1α to the target genes, paving the way to cancer-specific metabolism in response to low-oxygen challenge.
Results
JMJD5 Knockdown Reduces Cell Growth and Glycolytic Genes in MCF-7 Breast Cancer Cells Under Normoxia and Hypoxia.
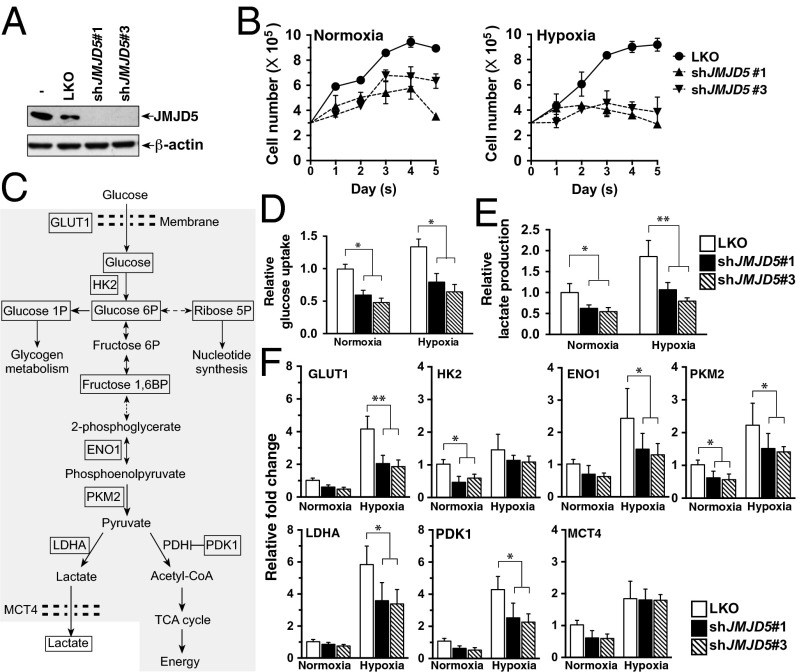
JMJD5 is highly expressed in breast cancer cells and exerts a proliferative function (1). Given that cancer cells frequently suffer from intermittent hypoxia exposure, we sought to test the role of JMJD5 in MCF-7 cells under both normoxic and hypoxic conditions. Two stable JMJD5 knockdown MCF-7 cells with independent target sequences (shJMJD5#1 and shJMJD5#3), as well as mock control (shControl, shLKO; that is, an empty lentiviral vector), were generated. Fig. 1A shows that shJMJD5#1- and shJMJD5#3-incorporated cells had significantly reduced levels of JMJD5 expression compared with that of the control cells. Consistent with our earlier report that knockdown of JMJD5 impeded the growth of MCF-7 under normoxic conditions (1), the two newly established MCF-7–shJMJD5 (shJMJD5) cells also grew more slowly than the shLKO vector-transfected MCF-7–LKO (LKO) (Fig. 1B).
Fig. 1.
JMJD5 is crucial for cell proliferation and promotes glucose metabolism under normoxia and hypoxia. (A) Generation of JMJD5-knockdown MCF-7 cells. MCF-7 cells were infected with lentivirus with control shRNA (LKO) and shJMJD5s, respectively, followed by puromycin selection. Western blotting analysis of the whole-cell lysates from selected clones (LKO, shJMJD5#1, and shJMJD5#3) was performed to evaluate the expression of JMJD5. β-Actin is an internal control. (B) JMJD5-knockdown cells reduce cell growth under normoxia and hypoxia. Cell numbers were counted at indicated time points. Data presented are means ± SD from three independent experiments. (C) A schematic diagram of the glucose metabolic flux. (D and E) The fold changes in glucose uptake (D) and lactate production (E) in MCF-7 LKO and shJMJD5 cells after 24-h culture were measured and expressed as a ratio of LKO normoxia levels. *P < 0.05; **P < 0.01. (F) The relative expression of genes in glucose metabolism of MCF-7 cells (shown in C) was measured by qRT-PCR and shown as a ratio of LKO normoxia levels. PDH, pyruvate dehydrogenase.
To explore the role of JMJD5 in MCF-7 proliferation, we compared the glucose uptake and lactate production (Fig. 1C) in LKO and shJMJD5 cells under normoxic or hypoxic conditions. We found that the JMJD5-knockdown cells significantly reduced the glucose uptake (Fig. 1D) and lactate secretion (Fig. 1E) compared with the control cells either under normoxia or hypoxia. We then tested the expression profiles of genes involved in the Warburg effect using real-time quantitative (qRT)-PCR analysis; they include the glucose transporter (GLUT1), hexokinase II (HK2), enolase 1 (ENO1), PKM2, lactate dehydrogenase A (LDHA), lactate transporter MCT4 (MCT4), and pyruvate dehydrogenase lipoamide kinase isozyme 1 (PDK1) (Fig. 1C). The control LKO cells exhibited a relatively higher level of expression of all these genes than shJMJD5 cells cultured under normoxia, in which statistical significance was seen for HK2 and PKM2 (Fig. 1F). There was even a much higher expression for all these genes in LKO than in shJMJD5 under low-oxygen condition; statistical significance was found for GLUT1, ENO1, PKM2, LDHA, and PDK1. Of note, GLUT1, LDHA, and PDK1 are known to be responsible for increased glucose uptake and consumption via anaerobic glycolysis but not oxidative phosphorylation (7). Additionally, there was a statistically higher production of glucose 6-phosphate (G6P), fructose 1,6 bisphosphate (F1,6BP), glucose-1-phosphate (G1P), and ribose-5-phosphate (R5P) in LKO than in knockdown cells (Fig. S1). These results together suggest that JMJD5 is involved in Warburg metabolism and also reprogramming glucose metabolism under hypoxic conditions to promote cell proliferation.
JMJD5 Physically Interacts with PKM2 and Is Hypoxia-Inducible.
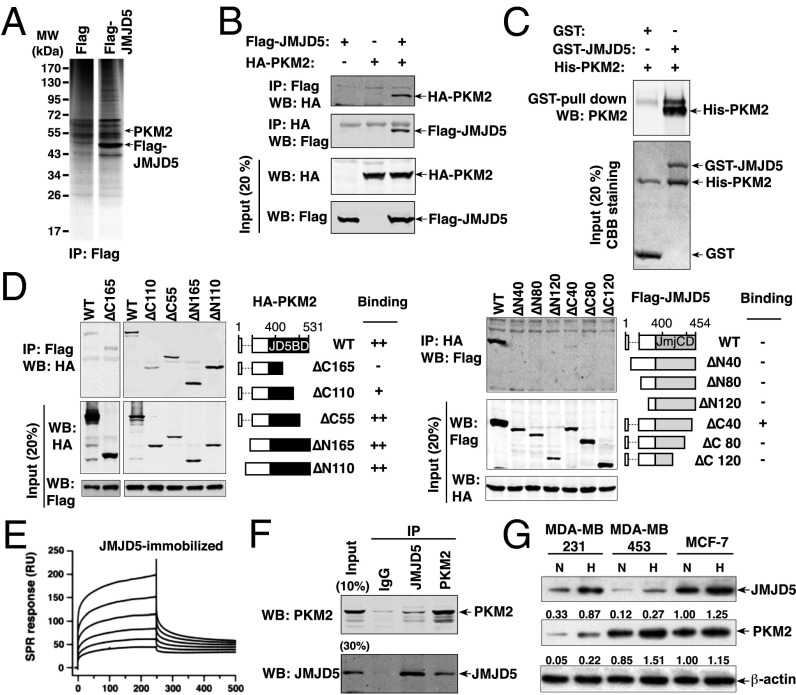
As a first step to understand how JMJD5 affects the expression of proteins in the glycolytic flux, we proceeded to identify cellular interacting partners. To this end, constitutively Flag-JMJD5–expressing HeLa cell lines were generated and verified by immunoblotting analysis (Fig. S2). The global level of H3K36me2 signal was significantly decreased, indicating the overexpressed JMJD5 was functional. Flag-JMJD5 was immunoprecipitated from the nuclear extracts with anti-Flag beads and the associated proteins eluted from the beads by Flag peptides. The eluates were resolved on SDS/PAGE. Several major bands not seen in the control lane were subjected to liquid chromatography–tandem MS (LC-MS/MS) analysis (Fig. S3). As shown in Fig. 2A, a band that gave rise to multiple peptides corresponding to PKM2 was identified.
Fig. 2.
JMJD5 interacts with PKM2. (A) Mass spectrometric analysis identified PKM2 associated with JMJD5. Nuclear extracts were prepared from HeLa-Flag (Flag) or HeLa-Flag-JMJD5 cells (Flag-JMJD5), followed by IP with anti-Flag beads. The protein bands on SDS/PAGE were retrieved and analyzed by MS (Fig. S3). (B) Reciprocal IP were performed in HEK293 cells transfected with the expression vector as indicated. (C) GST pull-down assays were performed with GST alone or GST-JMJD5 fusion protein plus His-PKM2 expressed in bacterial cells. CBB, Coomassie brilliant blue. (D) Determination of minimal PKM2-JMJD5 interaction region. Co-IP assays were performed with an anti-Flag antibody in HEK293T cells transfected with Flag-JMJD5 plus one of a series of N-terminal or C-terminal HA-PKM2 mutants (Upper) or with an anti-HA antibody in HEK293T cells transfected with HA-PKM2 plus one of a series of N-terminal or C-terminal Flag-JMJD5 mutants (Lower). Data was merged from different blots in a series of experiments using WT PKM2 as the positive control. JD5BD, JMJD5 binding domain; JmjCD, the Jumonji C domain of JMJD5. (E) SPR analysis of JMJD5-PKM2 interaction. (F) IP assays were performed with IgG, anti-JMJD5, and anti-PKM2 in MCF-7 cells, followed by Western blotting analysis. (G) JMJD5 and PKM2 are up-regulated in breast cancer cell lines under hypoxic condition.
To confirm the MS/MS-based protein identification results, HEK293T cells were transfected with pcDNA-HA-PKM2 alone or with pcDNA-Flag-JMJD5, followed by immunoprecipitation (IP) using anti-Flag or anti-HA antibodies. Western blotting analysis revealed that HA-PKM2 was present in anti-Flag co-IPs from cells cotransfected with pcDNA-HA-PKM2 and pcDNA-Flag-JMJD5 but not in cells with HA-PKM2 and Flag vector (Fig. 2B). To study whether the interaction between PKM2 and JMJD5 is direct, we used cell-free GST pull-down assays with bacteria-expressed GST-JMJD5 and PKM2. Fig. 2C shows that GST-JMJD5 bound to the recombinant PKM2 compared with GST alone. These results confirmed that PKM2 physically associated with JMJD5.
We next determined the minimal region crucial for the JMJD5-PKM2 interaction. A series of N-terminal– and C-terminal–truncated mutants fused to HA-tag for PKM2 (ΔN110, ΔN165, ΔC55, ΔC110, and ΔC165) were generated. HEK293T cells were cotransfected with the Flag-JMJD5 clone plus each of the truncated PKM2 clones, followed by IP and Western blotting analysis. Fig. 2D, Upper shows that PKM2ΔC55 retained the association with JMJD5. A significant decrease was found for PKM2ΔC110, whereas no signal was detected for PKM2ΔC165. On the other hand, PKM2ΔN110 and PKM2ΔN165 exhibited comparable association with JMJD5. These results revealed that the most critical interaction region in PKM2 resides at the C-terminal region (residues 366−421 and 422−476).
We also performed the reciprocal experiment for JMJD5 and generated truncated mutants fused to Flag-tag for JMJD5 (ΔN40, ΔN80, ΔN120, ΔC40, ΔC80, and ΔC120). As shown in Fig. 2D, Lower, hardly any signal was detected for various mutants (JMJD5ΔN40, JMJD5ΔN80, JMJD5ΔN120, JMJD5ΔC40, JMJD5ΔC80, and JMJD5ΔC120). Only JMJD5ΔC40 exhibited signal. We interpret this to mean that multiple regions, likely the N- and C-terminal domains, are involved in JMJD5’s binding to PKM2. We further measured the binding affinity between PKM2 and JMJD5 using surface plasmon resonance (SPR) spectroscopy, where JMJD5 was immobilized on the sensor chip. A clear binding profile was observed with a binding constant of Kd = 4.6 μM (Fig. 2E).
In addition, the association of endogenous PKM2 and JMJD5 in MCF-7 was studied. IP of lysates from MCF-7 cells showed that endogenous JMJD5 was associated with PKM2 (Fig. 2F). Previously, it was shown that hypoxia induced the expression of PKM2 in HeLa cells (14), which prompted us to evaluate the expression of PKM2 and JMJD5 under hypoxia. Breast cancer cell lines including human MDA-MB-231 adenocarcinoma, MDA-MB-435 carcinoma, and MCF-7 adenocarcinoma cells all exhibited higher expression of PKM2 under hypoxia (Fig. 2G). Interestingly, the JMJD5 expression level in each of these lines was also higher under hypoxia (Fig. 2G). These results, taken together, provide strong evidence that JMJD5 is an interacting partner of PKM2 and that they are coordinately regulated.
JMJD5 Regulates the Nuclear Translocation of PKM2.
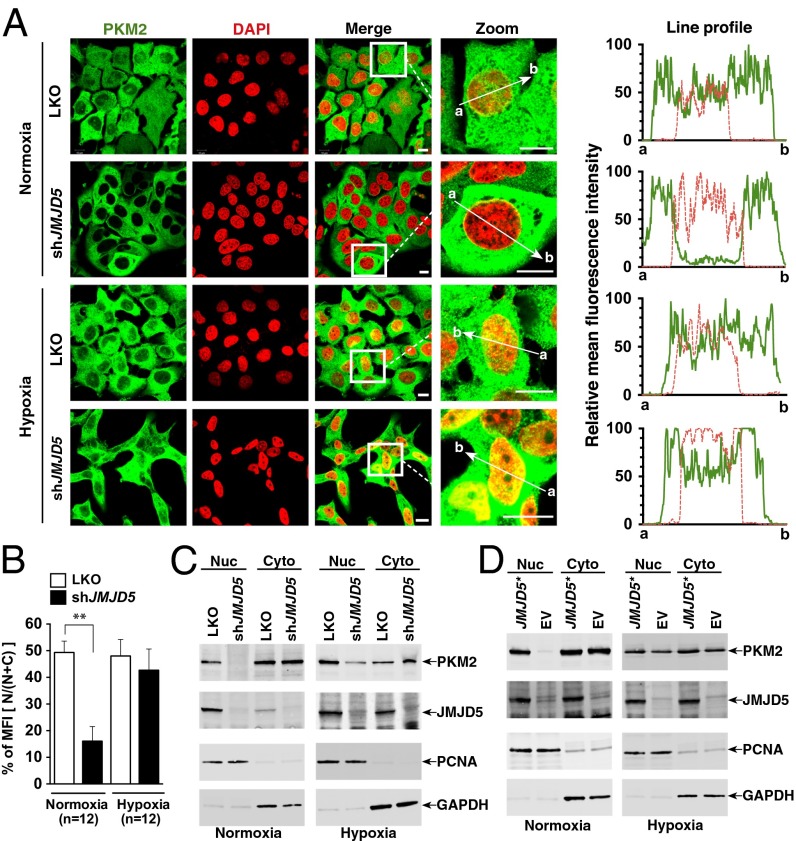
PKM2 has been shown to translocate into the nucleus via PHD3-mediated hydroxylation and serves as a coactivator of HIF-1α under hypoxia (14). Given that JMJD5 is a demethylase/hydroxylase, we asked whether JMJD5 could similarly hydroxylate PKM2 but were unable to demonstrate such a reaction, either based on mass spectrometry or Western blotting analyses with appropriate antibodies (Fig. S4). However, we did find JMJD5’s ability to regulate nuclear translocation of PKM2. The distribution of PKM2 was visualized using confocal microscopic analysis (Fig. 3 A and B). Barely any detectable nuclear PKM2 was seen in shJMJD5 cells compared with that in LKO cells under normoxia. When cells were cultured under hypoxia, shJMJD5 cells retained a lower level of nuclear PKM2, despite detectable nuclear PKM2 signal.
Fig. 3.
JMJD5 regulates the nuclear translocation of PKM2. (A) Subcellular localization of PKM2 in MCF-7 cells. Cells were immunostained with anti-PKM2 (PKM2, green). The nucleus is marked with DAPI (red). Merged images (Merge) are shown. The framed regions are zoomed in the fourth row (Zoom). The line profiles of PKM2 and DAPI signals were measured by ZEN 2011 (Carl Zeiss) software. (Scale bars, 10 μm.) (B) Analysis of the mean fluorescence intensity of PKM2 in the cytosol and nucleus. Mean values from 12 independent cells from three preparations were determined by the ZEN 2011 Histogram program. The relative fluorescence intensity in the nucleus is expressed as percentage of MFI [N/(N + C)]. Statistical significance was evaluated using the paired Student t test. (C) Nuclear and cytosolic lysates were prepared from LKO and MCF-7–shJMJD5 cells exposed to normoxia or hypoxia, followed by immunoblotting analysis. (D) Subcellular localization of JMJD5 and PKM2 in shJMJD5 cells introduced with the shRNA-resistant JMJD5 (JMJD5*) or empty vector (EV). Cyto, cytoplasm; Nuc, nucleus; PCNA, proliferating-cell nuclear antigen; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blotting analysis of nuclear and cytosolic fractions from MCF-7 cells revealed that JMJD5 primarily resided in the nucleus, cultured either under normoxic or hypoxic conditions (Fig. 3C). For PKM2, there were detectable signals in nuclear and cytosolic fractions of LKO cells, cultured either under normoxic or hypoxic conditions (Fig. 3D). A significant decrease of the nuclear PKM2 signal was detected in shJMJD5 cells under normoxia compared with a certain level of the nuclear PKM2 signals in hypoxia-exposed cells, in accordance with the confocal microscopic findings.
To further validate the result that JMJD5 deficiency was responsible for a lower level of PKM2 nuclear translocation, we reintroduced JMJD5* (shRNA#1-resistant JMJD5 cDNA) back into shJMJD5 and were able to restore the nuclear level of PKM2 (Fig. 3D). These results support the notion that JMJD5 is a regulator of PKM2 nuclear translocation.
As a complementary experiment and to generalize the observation to other cell type, we compared the cellular distribution of PKM2 in HeLa cells (which express a trace amount of JMJD5) and the JMJD5-overexpressing HeLa-JMJD5 cells. JMJD5 was predominantly present in the nucleus of HeLa-JMJD5 under normoxia or hypoxia (Fig. S5). A higher fraction of nuclear PKM2 was seen in HeLa-JMJD5 than in HeLa cells under normoxia (30% vs. 10%) and under hypoxia (50% vs. 30%). Together, these results suggest that JMJD5 and hypoxia positively regulated the nuclear translocation of PKM2.
JMJD5 Redirects the Equilibrium of PKM2 Quaternary-Structure Arrangement to Hinder the Tetrameric Assembly and Inhibit PKM2 Pyruvate Kinase Activity.
Our next task was to identify the mechanism associated with JMJD5’s regulation of PKM2 nuclear translocation. As described above, JMJD5’s hydroxylase activity does not seem to be involved; we therefore looked for other mechanism whereby JMJD5 facilitates PKM2 nuclear translocation.
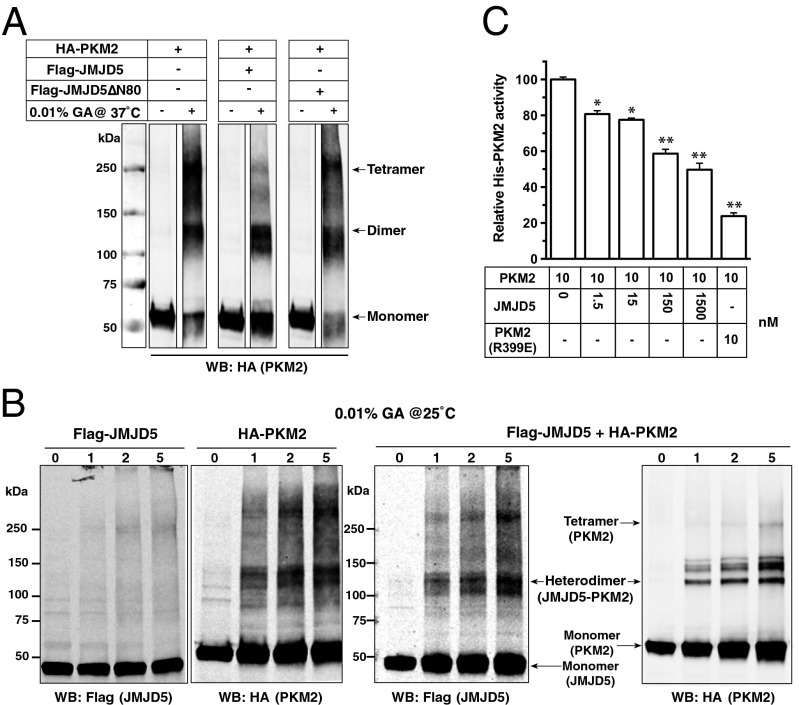
PKM2 has been found to primarily exist in two multimeric forms in tumor cells: an enzymatically active tetramer and a nearly inactive dimer at physiological concentrations of PEP (for a review, see ref. 19). Gao et al. recently showed that the nuclear PKM2 was mainly present as a dimer (15). Notably, Yang et al. (18) found that PKM2 was phosphorylated at the segment involved in multimeric assembly by Erk2, implicating that the monomeric form is crucial for the phosphorylation of PKM2 and its nuclear translocation. Thus, disruption of the tetrameric form is likely to be the key to translocate PKM2 into the nucleus. In light of these findings, we sought to test whether JMJD5 interfered with the monomer:dimer:tetramer equilibrium of PKM2 using cross-linking experiments with glutaraldehyde. Lysates were prepared from HEK293T cells transfected with pcDNA-HA-PKM2 alone or with pcDNA-Flag-JMJD5, followed by treatment with glutaraldehyde at 37 °C. Western blotting analysis with anti-Flag revealed that PKM2 migrated as a single band at ∼60 kDa (monomer) in the absence of cross-linking (t = 0), indicating that SDS treatment resulted in a complete dissociation of potential PKM2 multimers (Fig. 4A). Following glutaraldehyde cross-linking, the monomeric PKM2 was notably reduced and been replaced by dimeric (120 kDa) and tetrameric (240 kDa) forms (Fig. 4A).
Fig. 4.
The quaternary-structure arrangement of PKM2 is influenced by JMJD5. Cell lysates were prepared from HEK293T cells that were transfected with the expression vector(s) as indicated at 37 °C for 1 min (A) or with HA-PKM2 and Flag-JMJD5 vectors at 25 °C for 0−5 min (B), followed by cross-linking experiments using glutaraldehyde and then subjected to Western blotting analysis. Data was prepared from two blots performed at the same time with noncontiguous lanes separated by white spaces. (C) Pyruvate kinase activity of bacteria-expressed PKM2 in the presence of JMJD5 or R399E. The relative activity of wild-type PKM2 was set to 100%.
In the presence of JMJD5, interestingly, there was essentially no tetrameric PKM2. Apart from the monomeric and dimeric forms, an additional band at 110 kDa was found (Fig. 4A), which is likely to be the JMJD5/PKM2 heterodimer (see below). By contrast, a truncated JMJD5-ΔN80 mutant, which can no longer bind PKM2, does not affect PKM2 tetramerization (Fig. 4A). These results suggest that JMJD5 physically hinders the PKM2 tetrameric assembly.
We further used a milder cross-linking condition (0.01% glutaraldehyde at 25 °C) to examine the cross-linking effect. Fig. 4B, Right confirms that there was very little tetrameric PKM2 signal. Western blotting analysis with anti-Flag revealed the presence of signals corresponding to the JMJD5 monomer (50 kDa) and dimer (100 kDa). We also observed signals of higher molecular masses, indicating that JMJD5 might associate with other molecules. Interestingly, an increasing signal for the 110-kDa band that could be recognized by either of the tag antibodies as the cross-linking time increased, suggesting the formation of a JMJD5-PKM2 heterodimer.
Additionally, size-exclusion chromatographic analysis of various combinations of purified recombinant proteins [PKM2/R399E, a dimeric mutant (15); and/or JMJD5/ JMJD5ΔN80 (a binding-defective mutant)] clearly demonstrates that the tetrameric formation of PKM2 is hindered by JMJD5 but not JMJD5ΔN80, confirming that JMJD5 influences the PKM2 oligomerization via a direct binding (Fig. S6).
If JMJD5 truly blocks the formation of PKM2 tetramer, the active form of the enzyme, we should see a decrease of PKM2 pyruvate kinase activity in the presence of JMJD5. In vitro pyruvate kinase reactions were performed with the bacteria-expressed PKM2 and R399E that has been shown to be more tumorigenic than the wild-type PKM2. Fig. 4C shows that sufficient amounts of JMJD5 that interfered with the monomer:dimer:tetramer equilibrium of PKM2 indeed inhibited the pyruvate kinase activity.
JMJD5 Regulates PKM2-Enhanced HIF-1 Transactivation Activity.
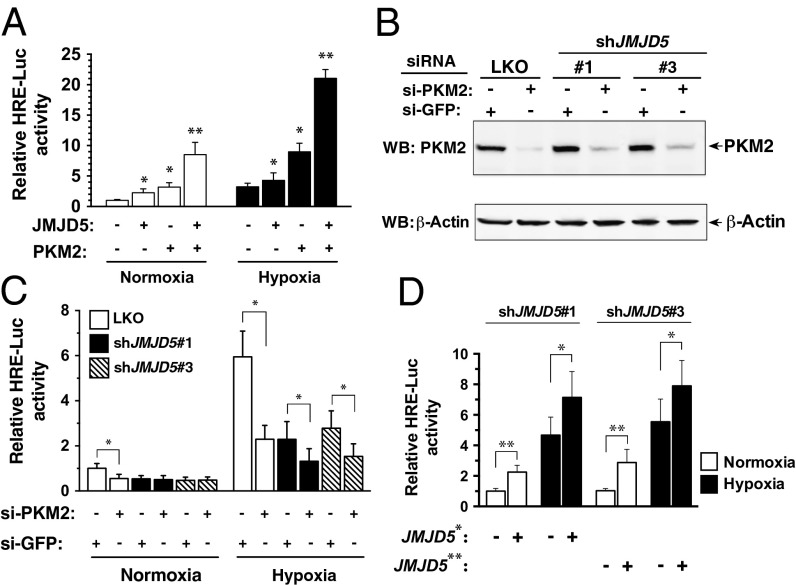
To evaluate whether JMJD5 regulates PKM2-stimulated HIF-1 transactivation activity, we have used a HIF-1α reporter plasmid that consists of three HREs and firefly luciferase coding sequences (pHRE-RLuc) (20) for the promoter-activity assay. LKO cells were cotransfected with the pHRE-FLuc, pTK-Renilla-Luc (an internal control vector), and the empty vector or the JMJD5 or PKM2 or JMJD5 plus PKM2 vectors and cultured under normoxia. Introduction of JMJD5 (twofold) or PKM2 (threefold) or JMJD5 plus PKM2 (ninefold) significantly increased the reporter activity compared with the empty vector (Fig. 5A). When cells were cultured under hypoxia, a threefold increase in transcriptional activity was detected for control cells compared with normoxia-exposed cells. An even higher activity was detected for other combinations: 4-fold for JMJD5, 9-fold for PKM2, and 21-fold for JMJD5 plus PKM2, suggesting that JMJD5 and PKM2 synergistically promote HIF-1 transcriptional activity in MCF-7 cells.
Fig. 5.
JMJD5 is crucial for PKM2-stimulated HIF-1α transactivation activity. (A) Transactivation activity of LKO cells cotransfected with pHRE-Fluc, pTK-Rluc, and expression vectors as indicated. The ratio of Fluc:Rluc activity was normalized to LKO at normoxia (mean ± SD; n = 6). (B) MCF-7 cells were transfected with siRNA-PKM2 as indicated, followed by immunoblotting analysis. (C) Transactivation activity of MCF-7 cells cotransfected with pHRE-Fluc, pTK-Rluc, and siRNA-PKM2 or siRNA-GFP. The ratio of Fluc:Rluc activity was normalized to that with siRNA-GFP (mean ± SD; n = 4). (D) Transactivation activity of shJMJD5 cells cotransfected with a shRNA-resistant JMJD5 vector. *P < 0.05; **P < 0.01. JMJD5* and JMJD5**: the #1 and #3 shRNA-resistant JMJD5 vectors.
Conversely, we asked whether knockdown of PKM2 had any effect on HIF-1 transactivation activity because MCF-7 cells had a relatively high level of endogenous PKM2 and JMJD5. By the use of siRNA-PKM2, the endogenous expression of PKM2 was greatly reduced in MCF-7 cells (Fig. 5B). Introduction with siRNA-PKM2 significantly reduced the transactivation activity for LKO cells either under normoxia or hypoxia. shJMJD5 cells displayed a significantly lower level of activity than did LKO cells without siRNA-PKM2 treatment (P < 0.01), whereas introduction of siRNA-PKM2 led to an additional decrease for hypoxia-exposed shJMJD5 cells (P < 0.05) (Fig. 5C). No further decrease was observed for normoxia-exposed shJMJD5 cells. Depletion of PKM2 therefore led to a great diminution of JMJD5-mediated HIF-1α transactivation activity.
We next addressed whether complementation with JMJD5 in two shJMJD5 lines could restore the transactivation activity using the expression vector JMJD5* and JMJD5** (# 1 and #3 shRNA-resistant JMJD5 cDNAs), respectively (Fig. 5D). A statistically significant increase was indeed seen upon JMJD5 complementation either under normoxia or hypoxia, revealing that complementation of JMJD5 in shJMJD5 cells could restore the transactivation activity. Together, our data suggest that JMJD5 is crucial for PKM2-stimulated HIF-1α transactivation activity.
JMJD5 and PKM2 both Recruit to the HRE Site of the LDHA and PKM2 Loci.
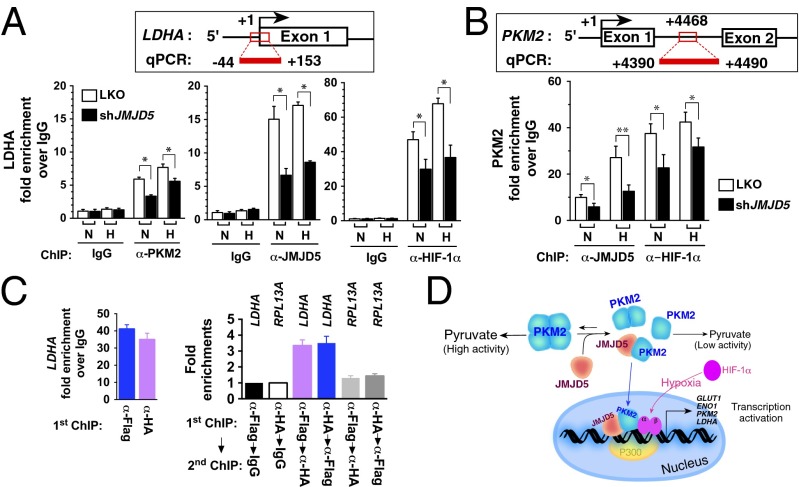
To test whether JMJD5 enhances HIF-1α transactivation by facilitating HIFα and PKM2 recruitment to HRE, we used ChIP analysis and interrogated LDHA and PKM2 loci (14). We first showed that JMJD5 was recruited to the LDHA locus, and its knockdown by shRNA depleted its occupancy of JMJD5 in MCF-7 cells (LKO vs. shJMJD5) [normoxia, 15- vs. 7-fold (P < 0.01); hypoxia, 17- vs. 9-fold (P < 0.01)]. ChIP results with anti–HIF-1α showed that LKO cells exhibited a statistically significant HIF-1α enrichment compared with IgG ChIP results at the LDHA HRE site, either under normoxic or hypoxic conditions (Fig. 6A). A similar trend was also obtained for PKM2 ChIP results, in accordance with the previous finding (14). Interestingly, knockdown of JMJD5 significantly reduced the enrichment of HIF-1α (P < 0.05), and PKM2 (P < 0.05) either under normoxia or hypoxia, indicating that JMJD5 is involved in the recruitment of HIF-1α and PKM2 to the LDHA locus.
Fig. 6.
JMJD5 enhances HIF-1α recruitment to HREs of the LDHA (A) and PKM2 (B) loci. Chromatin was prepared from dimethyloxalylglycine-treated MCF-7 cells cultured under normoxia (N) or hypoxia (H) with anti–HIF-1α, anti-JMJD5, anti-PKM2, or IgG and was analyzed by quantitative (q) PCR. Fold enrichments normalized to IgG are presented (mean ± SD; n = 9 from three independent experiments). *P < 0.05; **P < 0.01. (C) ChIP-reChIP analysis of the JMJD5- PKM2 recruitment to LDHA promoter site. MCF-7 cells cotransfected with Flag-JMJD5 and HA-PKM2 were exposed to hypoxic condition for 24 h. Primary ChIP (first ChIP) and second ChIP were performed sequentially and then analyzed by qPCR. (D) The proposed model that depicts JMJD5 as a major regulator in PKM2-stimulated HIF-1α metabolic reprogramming.
We next tested the PKM2 locus (14) and found that shJMJD5 cells also displayed a significant reduced enrichment of HIF-1α (normoxia, P < 0.05; hypoxia, P < 0.05) and JMJD5 (normoxia, P < 0.05; hypoxia, P < 0.01) to the PKM2 HRE site than did LKO cells (Fig. 6B). These results together suggested that JMJD5, HIF-1α, and PKM2 are corecruited to HREs of LDHA and PKM2.
To directly assess whether JMJD5 and PKM2 were in the HIF-1α transcriptional complex, we further conducted sequential ChIP assay (ChIP-reChIP). MCF-7 cells cotransfected with pcDNA-Flag-JMJD5 and pcDNA-HA-PKM2 were exposed to hypoxia, which was subjected to the first ChIP experiment using anti-Flag or IgG. A subsequent re-ChIP assay with anti-HA was then performed, followed by real-time PCR analysis of the LDHA locus. Fig. 6C shows that JMJD5 had a significant enrichment signal (anti-Flag ChIP) compared with an IgG control. Re-ChIP results with anti-HA, interestingly, revealed the presence of a positive signal at the LDHA locus. A reciprocal experiment was simultaneously performed (first ChIP with anti-HA and second ChIP with anti-Flag), which confirmed the presence of PKM2 and JMJD5 at the LDHA locus. Thus, JMJD5 and PKM2 are confirmed to be corecruited to HREs of LDHA (Fig. 6D) and thereby specifically enhance HIF-1α binding.
Discussion
Embryonic cells, as well as actively proliferating cancer cells, frequently encounter intermittent oxygen shortage, which requires appropriate management of compromised oxygenation for survival and massive growth. In this investigation, we report the connection between two embryogenesis/carcinogenesis-involved molecules, PKM2 and JMJD5, which together regulate HIF-1α. Significantly, knockdown of JMJD5 correlates with the reduced glucose uptake, lactate secretion, and decreased expression of genes in the glycolytic flux either under normoxia or hypoxia, implicating a role in linking PKM2 in glucose metabolism (14). Several lines of evidence strongly suggest that JMJD5 regulates PKM2’s tumorigenic capability: (i) JMJD5 binds to PKM2, hinders PKM2 tetramerization, and blocks pyruvate kinase activity; (ii) JMJD5 positively affects the nuclear translocation of PKM2; (iii) Both proteins are hypoxia-inducible in breast cancer cells and JMJD5 promotes PKM2–HIF-1α–mediated transactivation activity; and (iv) JMJD5 and PKM2 are corecruited to the HRE sites at the LDHA and PKM2 loci, promoting the HIF-1–mediated activity (6).
PKM2 is located at a “gate” position in the glycolytic flux to respond to various stimuli crucial for the Warburg effect (6). Its unique integrative capability relies on a specific alternative-splicing segment encoded by exon 10, which is responsible for multimeric assembly and allosteric regulation to ensure its roles to amalgamate metabolic signals. For instance, FBP and serine can allosterically activate pyruvate kinase activity of PKM2 (7, 21). In this context, interaction of JMJD5 with PKM2 at this region to disrupt the formation of an allosterically controllable tetramer represents a metabolic switch that favors the biosynthetic route such as the pentose phosphate pathway in cells.
Perhaps the most striking finding in this work is that we demonstrate that the expression of JMJD5 correlates with a higher level of nuclear PKM2, positively regulating HIF-1α activity. Previously, JMJD5 was shown to increase the transcription of cyclin A by demethylation of H3K36me2. Additional studies showed that JMJD5 possesses hydroxylase activity (4). We tested the JMJD5-PKM2 binding affinity for catalytically inactive mutants (JMJD5-H321A and PKM2-K367M) (1, 13) and did not observe any significant difference (Fig. S7). Consistently, the JMJD5-H321A mutant is also able to stimulate HIF-1α activity and synergize with PKM2 (either wild type or mutant), albeit at lower levels than the JMJD5 wild type. This suggests that the catalytic activity of JMJD5 is not absolutely required for the observed effects but contributes to some extent (Fig. S7). The physical association of JMJD5 with PKM2 seems to be a dominant factor. In support of this notion, Yang et al. (18) showed that the intersubunit segment of PKM2 was phosphorylated by Erk2, facilitating its translocation into the nucleus. These results together suggest a mechanism whereby PKM2 can be regulated by JMJD5 binding-induced nuclear translocation.
JMJD5 lacks a DNA-binding domain and requires a partner to anchor on chromatin site. Our ChIP and re-ChIP data reveal that JMJD5 increases the occupancy of HIF-1α and PKM2 in the LDHA and PKM2 HRE sites. Coupled with the synergistically JMJD5-PKM2–stimulated HIF-1α transactivation activity, as well as the significant loss of JMJD5-mediated HIF-1α activity upon PKM2 depletion, these results suggest that JMJD5, PKM2, and HIF-1α are corecruited to those loci as a part of complex to modulate gene transcription in the HIF-1 system. The hypoxia-induced up-regulation of JMJD5 and PKM2 should result in an increased abundance of JMJD5/PKM2/HIFα complex working in concert to enhance hypoxia responses, which include the increased glycolysis and anabolic process. This may also explain the involvement of JMJD5 in early embryonic development (2, 3).
Although JMJD5 plays an important role in hypoxia induced response, we found it also increased PKM2 translocation under normoxic conditions, where HIF-1α may not be the most abundant. This suggests that JMJD5/PKM2 may have other HIF-1α–independent targets. Alternatively, even under normoxic conditions in certain contexts, HIF-1α is maintained at a significant level, allowing the JMJD5-PKM2-HIF1α axis to be activated. For instance, Finley et al. reported that under conditions where Sirt3 a tumor suppressor is down-regulated, HIF-1α level is increased under normoxia (22). In the biological systems studied here, we were able to identify JMJD5/PKM2/HIF-1α complex in either condition.
In sum, our studies illustrate that JMJD5 functions as an oncogene, in part by regulating cellular metabolism through PKM2 and HIF-1α (Fig. S8). By the use of both metabolic and nonmetabolic strategies, PKM2 and JMJD5 work in concert to balance survival and cellular growth in a changing oxygen microenvironment. In this regard, JMJD5 joins the increasing list of regulators of PKM2 to control cellular metabolisms. We demonstrate that PKM2 is regulated by direct protein interaction to block its allosteric activation, thereby lowering its metabolic inputs and, at the same time, facilitating nuclear translocation to modulate transcriptional program involved in glucose metabolism.
Materials and Methods
MCF-7, HeLa-S3, HEK293T, MDA-MB-231, and MDA-MB-453 cells were cultured as described (1). JMJD5-expressing HeLa cells were prepared by introducing pcDNA-Flag-JMJD5 into HeLa-S3s. JMJD5 knockdown MCF-7 cells were generated based on lentivirus-mediated shJMJD5s. Detailed information is provided in SI Materials and Methods. LC-MS/MS analysis for identifying JMJD5-PKM2 interaction is described in SI Materials and Methods. ChIP assay was performed with the use of Magna ChIPG assay kit (Millipore). Enrichment of specific gene occupancy onto promoter was detected by qRT-PCR as described in SI Materials and Methods and Tables S1 and S2. Detailed protocols for the reagents, siRNA assays, GST pull-down, immunoblot, confocal microscopic analysis, luciferase activity assay, SPR assay, in vitro kinase assay, and chemical cross-linking assay are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Science Council (NSC), Taiwan Grants NSC101-2811-B-007-002 and NSC101-2325-B-007-001, and the Aim for the Top University Project of National Tsing Hua University and the Ministry of Education, Taiwan, Republic of China. We acknowledge grant support from the US National Institutes of Health (CA114575 and CA165263), as well as National Health Research Institutes, Taiwan Grant 02A1MGPP20-014 and Ministry of Health and Welfare, Taiwan Grant DOH102-TD-M-111-100001 (to H.-J.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311249111/-/DCSupplemental.
References
- 1.Hsia DA, et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107(21):9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh S, Janknecht R. Histone demethylase JMJD5 is essential for embryonic development. Biochem Biophys Res Commun. 2012;420(1):61–65. doi: 10.1016/j.bbrc.2012.02.115. [DOI] [PubMed] [Google Scholar]
- 3.Ishimura A, et al. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development. 2012;139(4):749–759. doi: 10.1242/dev.074138. [DOI] [PubMed] [Google Scholar]
- 4.Youn MY, et al. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J Biol Chem. 2012;287(16):12994–13004. doi: 10.1074/jbc.M111.323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 6.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23(11):560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338(6110):1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2(97):ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 11.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42(6):719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40(5):1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Yang W, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Emerling BM, Weinberg F, Liu JL, Mak TW, Chandel NS. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a) Proc Natl Acad Sci USA. 2008;105(7):2622–2627. doi: 10.1073/pnas.0706790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 22.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.