Abstract
Scope
Furan is a potent hepatotoxicant and liver carcinogen in rodents. However, short-term tests for genotoxicity of furan are inconclusive. The aim of this study was to assess the potential of furan to covalently bind to DNA, and to assess furan genotoxicity in rats in vivo.
Materials and Methods
Accelerator mass spectrometry was used to determine the 14C-content in DNA following administration of [3,4-14C]-furan (0.1 and 2.0 mg/kg bw) to F344 rats. DNA damage, micronuclei, chromosomal aberrations and sister chromatid exchanges were analyzed in F344 rats treated with furan for up to 28 days.
Conclusions
The 14C-content in liver DNA was significantly increased in a dose-dependent manner, with mean concentrations of 7.9 ± 3.5 amol 14C/μg DNA and 153.3 ± 100.2 amol 14C/μg DNA, corresponding to 16.5 ± 7.4 and 325.2 ± 212.7 adducts/109 nucleotides at 0.1 and 2.0 mg/kg bw, respectively. There was no evidence for genotoxicity of furan in peripheral blood and bone marrow cells. However, a dose-related increase in the incidence of chromosomal aberrations in rat splenocytes and some indication of DNA damage in liver were observed. Collectively, results from this study indicate that furan may operate – at least in part – by a genotoxic mode of action.
Keywords: Furan, DNA adducts, liver, carcinogenicity, genotoxicity
1 Introduction
The heat-induced food contaminant furan is a potent hepatotoxicant and liver carcinogen in rodents [1]. In a 2-year bioassay conducted by the National Toxicology Program (NTP), furan was reported to cause a dose-dependent increase in hepatocellular adenomas and carcinomas in male and female B6C3F1 mice and male Fischer 344 rats, and high incidences of cholangiocarcinomas in rats of both sexes [1]. Furan is metabolized by cytochrome P450 2E1 to a toxic metabolite, cis-2-butene-1,4-dial (BDA). This metabolite may interact with proteins to cause cytotoxicity or react with nucleosides to form substituted 1,N6-etheno-2′-deoxyadenosine and 1,N2-etheno-2′-deoxyguanosine adducts [2, 3]. However, available data on genotoxicity of furan and cis-2-butene-1,4-dial are inconsistent and controversial. While furan was not mutagenic in Salmonella typhimurium strains TA100, TA1535, TA1537 and TA98 even in the presence of metabolic activation [1], cis-2-butene-1,4-dial (BDA) was reported to induce mutations in a Salmonella strain sensitive to aldehydes (TA 104) [4] and to cause DNA single-strand breaks and cross-links in mammalian cells [5]. Mutagenicity of cis-2-butene-1,4-dial (BDA) in bacteria and formation of cross-links was not confirmed by an independent study [6]. However, tk+/− gene mutations induced by BDA were observed in a narrow concentration range in L5178Y tk+/− mouse lymphoma cells [6]. In contrast, furan was not genotoxic in L5178Y tk+/- mouse lymphoma cells as evidenced by the comet, micronucleus and tk+/− mutation assays [6], but induced chromosomal aberrations and sister chromatid exchanges (SCEs) in Chinese hamster ovary (CHO) cells [1] and in a cell line stably expressing human cytochrome P450 (CYP) 2E1 (V79-hCYP2E1-hSULT1A1 cells) [7].
In vivo, furan was found to induce chromosomal aberrations but not SCE's in mouse bone marrow cells [1]. Negative results have been reported for furan in the liver UDS (unscheduled DNA synthesis) assay after short-term exposure of rats and mice. While BDA shows chemical reactivity against nucleosides [2, 3], stable DNA-adducts were not detected in a DNA binding study in rats [8]. However, the sensitivity of the study may have been too low to detect furan-derived DNA adducts and thus results from this study were considered inconclusive [9].
To understand the role of direct genotoxicity in furan carcinogenicity and to aid human risk assessment, this study was designed to assess the potential of furan to covalently bind to DNA in rat liver at a known carcinogenic dose and a dose closer to estimated human exposures. Accelerator mass spectrometry (AMS), a highly sensitive method for the detection of 14C which has been used to confirm the presence or absence of DNA-binding of a range of carcinogens [10-16], was applied to analyze 14C associated with DNA following administration of [3,4-14C]-furan to rats. Compounds containing radiolabel were separated by HPLC after enzymatic cleavage and were further analyzed by AMS and LC-MS to obtain structural information and to discriminate between direct binding and metabolic incorporation. In addition to DNA binding, the potential of furan to induce chromosomal aberrations and DNA damage in mammalian cells in vivo was assessed in male F344/N rats treated orally with furan for 5 and 28 days.
2 Materials and Methods
Chemicals
[3,4-14C]-Furan with a specific activity 20 mCi/mmol was purchased from Tjaden Biosciences (Burlington, IA, USA). Furan (Cat. 18,592-2) and corn oil (C8267) were purchased from Sigma-Aldrich (Taufkirchen, Germany). Unless otherwise indicated, all other chemicals were obtained from Sigma-Aldrich Fluka (Taufkirchen, Germany), Merck (Darmstadt, Germany), GE Healthcare (München, Germany), AppliChem (Darmstadt, Germany) or Roth (Karlsruhe, Germany). 2,5-Diacetoxy-2,5-dihydrofuran (purity ≥ 99 %) was prepared as previously described [17].
Animal treatment
All animal experiments were performed according to national animal welfare regulations after authorization by the local authorities (Regierung von Unterfranken, 54-2531-01-74/06). Male F344 rats (200-250 g on arrival, Harlan-Winkelmann GmbH, Borchen, Germany) were housed under standard laboratory conditions (climate cabinets, temperature 22 ± 2°C, relative humidity 30-70%, 12-15 air changes per hour, 12 hour light/dark cycle) in groups of 5 in Makrolon® type-4 cages with wire meshtops (Bayer Makrolon, Leverkusen, Germany) and standard softwood bedding. Rats received pelleted standard rat maintenance diet (SSNIFF Spezialdiäten GmbH, Soest, Germany) and tap-water ad libitum. After acclimatization, animals received [3,4-14C]-furan (specific activity 20 mCi/mmol) in corn oil (4 ml/kg bw) by gavage at a single dose of 0 mg/kg bw, 0.1 mg/kg bw and 2.0 mg/kg bw. The doses were chosen to include a known carcinogenic dose (2.0 mg/kg bw) [1] and a lower dose closer to estimated human exposures, for which no tumor data exist (0.1 mg/kg bw). Rats were sacrificed by cardiac puncture under CO2 anesthesia 2 h after dosing and livers and kidneys were removed, separated into aliquots, flash frozen in liquid nitrogen and stored at − 80°C for further analyses. The early time point of 2 h was chosen to allow distribution and bioactivation of furan based on the rapid excretion of furan metabolites with bile [18], whilst avoiding metabolic incorporation of 14C.
To structurally characterize DNA adducts and to assess in vivo genotoxicity of furan at a known carcinogenic dose (2.0 mg/kg bw) and below (0.1 and 0.5 mg/kg bw), a 28 day toxicity study was conducted in male F344 rats as previously described [19]. Briefly, rats (n=8 per dose and time-point) were administered [12C4]-furan dissolved in corn oil (4 ml/kg bw) at doses of 0, 0.1, 0.5 and 2.0 mg/kg b.w. by gavage for 28 days (9 am, 5 days per week), with an interim sacrifice after 5 days of treatment. An off-dose recovery group (0 and 2.0 mg/kg b.w. dose groups only) was kept for additional two weeks (recovery period) after the end of the 4 weeks treatment period. For cytogenetic analyses in bone marrow cells, 3 animals per group were given an i.p. injection of colchicine (4 mg/kg) three hours prior to sacrifice to accumulate cells in a metaphase-like stage. Animals were killed by CO2 asphyxiation, and blood and tissue samples were harvested and processed as described below. For adduct analysis, liver samples of a previous study in which F344/N rats (n = 4 per group) received a single dose of 0 or 30 mg/kg bw [12C4]-furan dissolved in corn oil (4 ml/kg bw) by oral gavage were used [20]. Animals were sacrificed 24 h after furan administration, liver samples were harvested, flash frozen in liquid nitrogen and stored at − 80°C for further analyses.
DNA extraction and 14C-AMS
DNA was isolated from livers and kidneys of rats treated with [3,4-14C]-furan using the Nucleobond® isolation buffer set IV and AXG 500 columns (Macherey-Nagel, Dueren, Germany) according to the manufacturer's instructions with minor modifications. Briefly, 300-400 mg tissue were homogenized by an ultra-turrax, treated with proteinase K and RNase and loaded onto a Nucleobond® AX G 500 ion exchange cartridge. After washing, DNA was eluted from the cartridge using a modified elution buffer (1.5M NaCl, 0.05M Tris, 15% ethanol, pH 7.0). DNA was precipitated by the addition of 0.7 volumes isopropanol. After washing with 70% ethanol, DNA pellets were freeze dried and stored at –80°C until further analysis by AMS. 14C–AMS was performed at the Lawrence Livermore National Laboratory, Livermore, CA with graphitized DNA samples as described elsewhere [21]. AMS results are given in fraction modern, whereby 1 fraction modern is defined as 97.9 amol 14C/mg C [22], which corresponds to the natural abundance of 14C in contemporary biological material [23].
For adduct characterization, liver DNA from [12C4]-furan treated rats was extracted as described above and stored at – 80°C.
Preparation and characterization of DNA adduct references
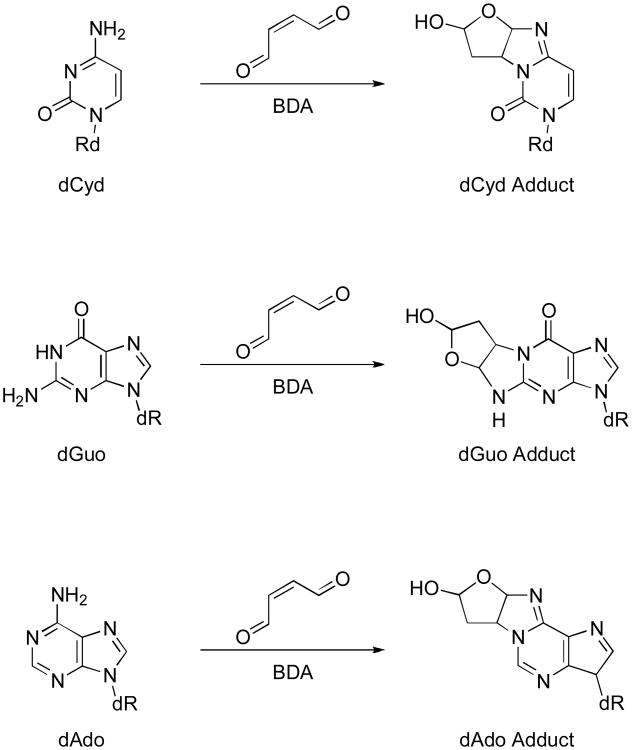
A 25 mM aqueous solution of diacetoxyfuran was shaken for 24 h to synthesize cis-2-butene-1,4-dial (BDA). The BDA-solution was subsequently incubated with 25 mM and 5 mM of 2′-deoxycytidine, 2′-deoxyguanosine, 2′-deoxythymidine and 2′-deoxyadenosine in 50 mM sodium phosphate buffer (pH 7.4) at 37 °C for 2, 4, 6, 8, 24, 28, 31 and 48 h (total volume 5 ml) to establish best reaction conditions of 2′-deoxyribunucleoside adducts. Incubation products were monitored by HPLC-DAD analysis as described below (see Fractionation of DNA by HPLC). Consistent with previous reports [2], reaction with BDA led to the formation of two diastereomeric adducts each in the presence of 2′-deoxycytidine (dCyd), 2′-deoxguanosine (dGuo), and 2′-deoxyadenosine (dAdo), but not 2′-deoxythymidine. Retention times and UV spectral data were as follows: dCyd-BDA: RT 31.5 min/33.8 min, λmax 285; dAdo-BDA: RT 35.9 min/40.5 min, λmax 263, 275 nm; dGuo-BDA: RT 38.8 min/48.8 min, λmax 250, 275 nm. Maximum peak intensities were reached after incubation for 24 h. Formation of adducts was moreover monitored by a Q TRAP 2000 LC-MS/MS mass spectrometer operating in the positive ion mode. Samples (1 μl) were loaded onto a Luna Phenyl-Hexyl column (Phenomenex; 3 μm; 150×4.6 mm). Analytes were separated by a solvent gradient from 100 % 10 mM ammonium acetate pH 6.6 (A) to 15 % methanol (B) within 15 min, then to 30 % B within 5 min and finally to 80 % B within 25 min at a flow rate of 200 μl/min. Data acquisition was performed with a source temperature of 400 °C and an ion spray voltage of 4 200 V. The declustering potential, entrance potential and collision energy were set to 50 V, 10 V or 15 V, respectively. Samples were analyzed in the MRM mode (multiple reaction monitoring). The following mass transitions were analyzed: m/z 312 to 196 and m/z 312 to 178 as quantifier and qualifier, respectively, for dCyd-BDA adducts, m/z 336 to 220 and m/z 336 to 202 for dAdo-BDA adducts, m/z 352 to 236 and m/z 352 to 218 for dGuo-BDA adducts. In addition, MRM-IDA-EPI (multiple reaction monitoring with information dependent acquisition of enhanced product ions) was performed to record EPI spectra in the range of 50 – 700 amu with a scan rate of 4 000 amu/s: dCyd-BDA adducts (RT 33.5 min/34.2 min): m/z 312 [M + H+] +, 196 [M + H+ – deoxyribose] +, 178 [M + H+ – deoxyribose – H2O] +, 136 [etheno-cytosine] +; dAdo adducts (RT 33.2 min/33.9 min): m/z 336 [M + H+] +, 220 [M + H+ – deoxyribose] +, 202 [M + H+ – deoxyribose – H2O] +, 136 [adenine] +; d Guo adducts (RT 34.0 min/35.4 min): m/z 352 [M + H+] +, 236 [M + H+ – deoxyribose] +, 218 [M + H+ – deoxyribose – H2O] +, 152 [guanine] +.
DNA hydrolysis
DNA isolated from [3,4-14C]-furan and [12C4]-furan treated rats and calf thymus DNA incubated with BDA in vitro as described above for 2′-deoxyribonucleosides were enzymatically hydrolyzed by nuclease P1 and alkaline phosphatase. Solutions containing 110 μg DNA in 110 μl distilled water were incubated with 11 units nuclease P1 (Calbiochem) and 2.8 μl 1 M sodium acetate/45 mM zinc chloride buffer pH 4.8 for 60 min at 37°C. After addition of 11 μl 1.5 M Tris-HCl (pH 8.0) and 8.25 units alkaline phosphatase, mixtures were incubated for an additional 30 min at 37°C. Proteins were removed using Microcon® centrifugal filter units.
Fractionation of DNA by HPLC
The liquid handling assembly consisted of a Hewlett Packard 1090 HPLC-DAD system. DNA hydrolyzates (100 μl) were separated on a Luna Phenyl-Hexyl column from Phenomenex (250 × 4.6 mm, 5 μ) with a phenyl guard-column and detected at 254 nm and 280 nm. The analytes were separated by a gradient from 100 % A (10 mM ammonium acetate, pH 6.9) to 5 % B (methanol) within 45 min and to 15 % B within 10 min with a flow rate of 1 ml/min. Initial conditions were reconstituted within 5 min. The fractions were collected at two minute intervals, except for fractions eluting at retention times of 2′-deoxyribonucleosides or DNA adduct references, which were collected separately. The collected fractions were vacuum dried at 40°C and analyzed by AMS as described above.
For detection of BDA derived DNA monoadducts a total of 500 μg DNA of [12C4]-furan treated rats was fractionated. Obtained fractions were concentrated and reconstituted in a final volume of 250 μl H2O before analysis by LC-MS/MS. To characterize unknown peaks, calf thymus DNA incubated with BDA in vitro was fractionated. Fractions containing unknown peaks eluting at about 18 min and 38 min were collected and analyzed with LC-MS/MS in EMS-IDA-EPI mode (enhanced mass spectrometry of information dependent acquisition of enhanced product ions) as described below to record spectra of unknown peaks.
LC-MS analysis of BDA derived DNA adducts in isolated fractions
The liquid handling system consisted of an Agilent 1100 series autosampler and an Agilent 1100 HPLC pump linked to an Applied Biosystems Sciex Instruments API 3000 Quadrupole LC-MS/MS mass spectrometer operating in positive ion mode. Data acquisition was performed in MRM mode with a source temperature of 400 °C and an ion spray voltage of 3 400 V. Declustering potential, entrance potential and collision energy were set to 20 V, 10 V or 30 V, respectively. As solvents, 0.1 % formic acid (A) and acetonitrile (B) were used. Samples (250 μl), i.e. fractions collected from a total of 500 μg hydrolyzed DNA, were loaded onto a Luna Phenyl-Hexyl column (Phenomenex; 3 μm; 150×4.6 m) and analytes were separated by a gradient from 90 % A to 40 % B within 3 min and to 98 % B within 12 min at a flow rate of 300 μl/min. Solvent composition was held for 2 min and initial conditions were reconstituted within 2 min. Spectral data were recorded with N2 as the heater gas at 400°C and as the collision gas (CAD = 4) in the multiple reaction monitoring mode (MRM). The following m/z transitions were analyzed: m/z 312 to 196, m/z 312 to 178 and m/z 312 to 112 for dCyd-BDA adducts, m/z 336 to 220, m/z 336 to 202 and m/z 336 to 136 for dAdo-BDA adducts, m/z 352 to 236, m/z 352 to 218 and m/z 352 to 152 for dGuo-BDA adducts.
Collected fractions of unknown peaks were analyzed by a Q TRAP 2000 LC-MS/MS mass spectrometer in EMS-IDA-EPI mode (enhanced mass spectrometry of information dependent acquisition of enhanced product ions) to record spectra of unknown peaks. LC-MS runs were extracted with MarkerView Software 1.2 (Applied Biosystems/MDS Sciex) to analyze chromatograms for peaks which were present in samples and absent in distilled water which was used as blank. Moreover, identified masses which might present potential BDA derived DNA crosslinks and further theoretical mass transitions of proposed BDA DNA crosslinks based on loss of deoxyribose (M+ - 116) were monitored by an API 3000 LC-MS/MS mass spectrometer in MRM mode in samples obtained from [12C4]-furan treated rats and calf thymus DNA incubated with BDA in vitro.
Comet Assay
Liver was dissected and processed immediately after harvesting. For preparation of cell suspensions, 1/3rd liver lobe was rinsed with cold PBS, placed into a Petri dish containing 3 ml of PBS at +4°C and minced with scissors. Cell homogenates obtained for each animal were filtered through sterile cell strainers (Falcon) with a pore-size of 100 μm to obtain single cell suspensions. Cell suspensions were then centrifuged at 500 rpm for 10 min at 4°C and finally re-suspended in 800 μl cold (+4°C) PBS. For preparation of cell suspensions from bone marrow, both femurs of each animal were rapidly dissected out at sacrifice and cleaned of surrounding tissue. The bone was cut at the distal end and irrigated with fetal calf serum (FCS) using a 2 ml syringe. The suspension of cells was aspirated and this procedure was repeated several times to avoid clumps. The cell suspension was then centrifuged at 1000 rpm for 10 min at + 4°C and re-suspended in a 8 ml cold PBS. Aliquots of heparinized whole blood (10 μl) obtained from each animal were mixed directly with 65 μl of low melting agarose (LMA) kept at 37°C to prepare slides for the comet assay. Comet slides were prepared using the protocol for the alkaline “comet assay” of [24]. Briefly, 10 μl of cell suspension were mixed with 65 μl of 0.7% (w/v) low melting point agarose (Bio-Rad Lab.) and sandwiched between a lower layer of 1% (w/v) normal-melting point agarose (Bio-Rad Lab.) and an upper layer of 0.7% (w/v) low-melting point agarose on microscope slides (Carlo Erba, Milan, Italy). Duplicate slides were prepared from each individual treatment. The slides were then immersed in a lysing solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, pH 10) containing 10% DMSO and 1% Triton × 100 (ICN Biomedicals Inc.) overnight at 4°C. On completion of lysis, slides were placed in a horizontal gel electrophoresis tank with fresh alkaline electrophoresis buffer (300 mM NaOH, 1 mM Na2EDTA, pH ≥ 13) and left in the solution for 25 min at 4°C to allow the DNA to unwind and to express the alkali-labile sites. Electrophoresis was carried out at 4°C for 25 minutes, 30 V (1 V/cm) and 300 mA, using a Bio-Rad power supply. After electrophoresis, the slides were immersed in 0.3 M sodium acetate in ethanol for 30 min. Microgels were then dehydrated in absolute ethanol for 2 h and immersed for 5 min in 70 % ethanol. Slides were air-dried at room temperature. Immediately before scoring, slides were stained with 12 μg/ml ethidium bromide (Boehringer Mannheim, Germany) and examined at 400× magnification with an automatic image analyzer (Comet Assay III; Perceptive Instruments, UK) connected to a fluorescence microscope (Eclipse E400; Nikon). To evaluate the amount of DNA damage, computer generated tail moment values and percentage of migrated DNA were used. For each individual treatment a total of one hundred cells were scored from two different slides. Statistical analysis was performed using ANOVA followed by Dunnett's test.
Micronucleus Test
The femurs of animals were removed and bone marrow cells were obtained by flushing with fetal calf serum. The cells were centrifuged and a concentrated suspension was prepared to make smears on slides (n=3 per animal). Slides were air-dried, stained with haematoxylin and eosin, and embedded in Eukitt. The slides were randomly coded and two thousand polychromatic erythrocytes (PCE's) per animal were examined for the presence of micronuclei at 100 × magnification. For statistical analysis, a modified Chi-squared calculation was employed to compare treated and control groups. The degree of heterogeneity within each group was first calculated and where this was significant it was considered in the comparison between groups. Variance ratios or Chi-squared values were taken to show the significance of any differences between each treated group and controls.
Cytogenetic analyses of bone marrow cells
Three hours after intraperitoneal injection of colchicine (4 mg/kg) to accumulate cells in metaphase-like stage, animals were sacrificed and both femurs of each animal were rapidly dissected out and cleaned of surrounding tissue. The bone was cut at the distal end and irrigated with fetal calf serum (FCS) using a 2 ml syringe. The suspension of cells was aspirated and this procedure was repeated several times to avoid clumps. The cell suspension was centrifuged and the cell pellet was resuspended in 5 ml hypotonic solution (1% tri-sodium citrate) for 15 minutes at room temperature. The cells were then pelleted again and fixed in freshly prepared methanol : acetic acid fixative (3:1 v/v) and washed twice with fixative. A few drops of the cell suspension were dropped onto clean, wet grease-free microscope slides and air-dried to produce metaphase chromosome spreads. Per animal, six slides (three slides each for chromosomal aberration and SCEs analyses) were prepared. Slides to be analyzed for chromosomal aberrations were stained in 5% Giemsa in buffer and rinsed twice with water. Slides to be analysed for SCE's were prepared according to [25, 26]. After staining slides were immersed in xylene for 5 minutes and embedded in Eukitt mounting medium. Slides were randomly coded for slide evaluation. Determination of mitotic indices was based on the number of metaphase observed per 1000 cells and was expressed as percentage. From 100 eligible metaphases the number and types of aberrations and the number of SCE's were recorded. For statistical analysis the Student's T-test (one way) was used.
Cytogenetic analyses in resting G0 splenocytes
At sacrifice, the spleen was removed from each individual animal and placed in RPMI 1640 medium supplemented with 20% fetal calf serum. Spleens were squeezed through a 10 ml syringe without needle to obtain cell homogenates which were then filtered through sterile cell strainers (Falcon) with a pore-size of 100 μm to obtain cell suspensions in 5 ml RPMI 1640 medium. The cell suspensions were carefully layered over Histopaque–1077 (Sigma Chemical, St. Louis, MO U.S.A.) and centrifuged at 400 × g for 25 min at room temperature. Splenocytes were removed from the gradient with a sterile pipette, washed twice with PBS and counted using a haemocytometer. Cells were seeded at 6 × 106 cells per T-75 culture flask in RPMI medium supplemented with 20% FCS (heat inactivated), 200 mM L-glutamine and 20 mM HEPES. Concanavalin A (Sigma) was added to a final concentration of 2 μg/ml culture medium. Cultures were grown for 54 h at 37°C. Colcemid (Gibco BRL) was added 3 h before harvesting at a final concentration of 0.2 μg/ml. At the end of treatment the cells were brought into suspension with trypsin. The cell suspension was centrifuged and resuspended in hypotonic solution. After hypotonic treatment the cells were fixed in freshly prepared methanol-acetic acid fixative and washed three times. Air-dried slides were prepared from the cell suspension, stained in 5% Giemsa in buffer, rinsed twice with water, immersed in xylene for 5 minutes embedded in Eukitt mounting medium. Slides were coded for slide evaluation. Determination of mitotic indices was based on the number of metaphase observed per 1000 cells and was expressed as percentage. For statistical analysis a modified Chi-squared calculation was employed to compare treated and control groups. The degree of heterogeneity within each group was first calculated and where this was significant it was considered in the comparison between groups. Variance ratios or Chi-squared values were taken to show the significance of any differences between each treated group and controls.
3 Results
Covalent binding of [3,4-14C]-furan to DNA
Fraction modern values of liver DNA isolated from control rats ranged from 1.1 to 1.3, which is consistent with the natural isotopic abundance levels of 14C in contemporary biological samples [23]. In contrast, a dose-dependent increase in radiocarbon content was observed in DNA extracted from furan target liver lobes and kidneys of rats treated with [3,4-14C]-furan (Table 1). Based on the assumption that the increase in 14C is due to covalent binding of furan, adduct levels were calculated taking into consideration the specific activity of the administered furan (20 mCi/mmol), the percent labeling (15.6 %), the carbon content of DNA (29 %), and an average molecular weight of 330 g/mol per nucleotide. The 14C-content of livers of rats treated with [3,4-14C]-furan corresponds to mean adduct levels of 1.7 ± 0.7 adducts/108 nucleotides in low dose (0.1 mg/kg bw) and 3.3 ± 2.1 adducts/107 nucleotides in high dose (2 mg/kg bw) animals (Table 1). In the kidney, which is not a target organ for furan carcinogenicity, the 14C-content corresponds to 5.1 ± 0.6 adducts/109 nucleotides and 1.3 ± 0.5 adducts/107 nucleotides in low and high dose animals, respectively (Table 1).
Table 1.
Mean 14C-content ± SD and calculated adduct levels ± SD of DNA isolated from kidney and liver of [3,4-14C]-furan treated and control rats.
| Tissue | Dose Group [mg/kg bw] | 14C-Content [amol/μg DNA] | Adduct Level [Adducts/109 Nucleotides] |
|---|---|---|---|
| 0 | 0.2 ± 0.2 | 0 | |
| Kidney | 0.1 | 2.6 ± 0.3 | 5.1 ± 0.6 |
| 2.0 | 60.2 ± 25.3 | 127.2 ± 53.6 | |
| 0 | 0.1 ± 0.1 | 0 | |
| Liver left lobe | 0.1 | 6.4 ± 1.2 | 13.3 ± 2.5 |
| 2.0 | 208.1 ± 112.7 | 441.5 ± 239.2 | |
| 0 | 0.1 ± 0.1 | 0 | |
| Liver caudate lobe | 0.1 | 9.4 ± 4.5 | 19.7 ± 9.6 |
| 2.0 | 98.6 ± 48.9 | 208.9 ± 103.8 | |
| 0 | 0.1 ± 0.1 | 0 | |
| Liver (mean) | 0.1 | 7.9 ± 3.5 | 16.5 ± 7.4 |
| 2.0 | 153.3 ± 100.2 | 325.2 ± 212.7 |
Discrimination between covalent binding and metabolic incorporation
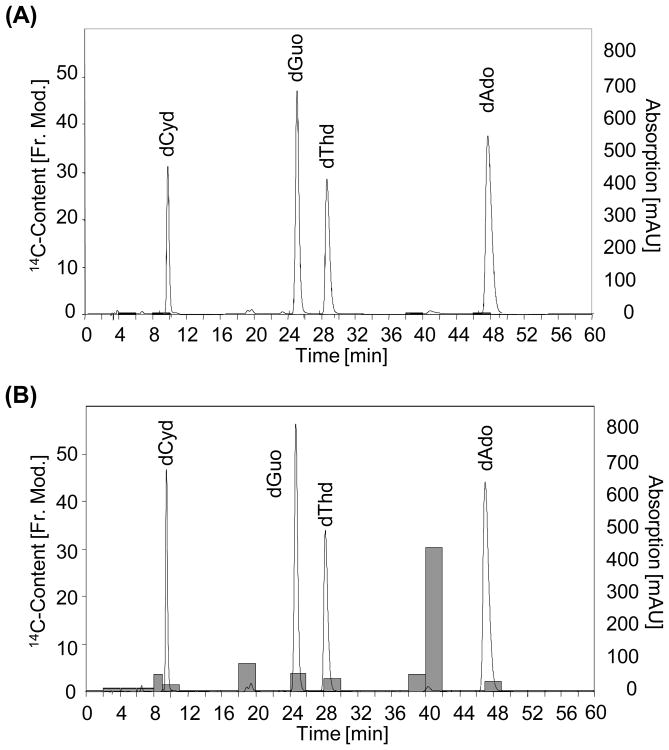
Since 14CO2 has been shown to be a major furan metabolite [8], the increase of radioactivity in DNA obtained from [3,4-14C]-furan treated rats could be also a result of metabolic incorporation of 14C into purines and/or pyrimidines during DNA synthesis. To distinguish between DNA binding and metabolic incorporation of furan, DNA isolated from 14C-furan treated rats was enzymatically hydrolyzed, fractionated via HPLC-DAD and collected fractions were analyzed by AMS. In vitro incubations of cis-2-butene-1,4-dial (BDA) and 2′-deoxyribonucleosides were used as reference compounds for HPLC method development and characterization of radioactive compounds. DNA fractions collected from control rats contained no significant amounts of radiocarbon (Figure 2). As can be seen from Figure 2, the majority of radioactivity present in liver DNA hydrolysates of 14C-furan treated rats did not coelute with normal nucleosides, indicating that metabolic incorporation occurs only to a minor extent. In contrast, the majority of total radioactivity present in the samples eluted at about 18 and 38-40 min (Figure 2).
Figure 2.
14C-content (grey bars) of fractions collected after HPLC-DAD (λ = 254 nm) chromatographic separation of DNA isolated from a control rat (A) and from a [3,4-14C]-furan treated rat (2.0 mg/kg bw) (B).
Characterization of furan-derived DNA adducts
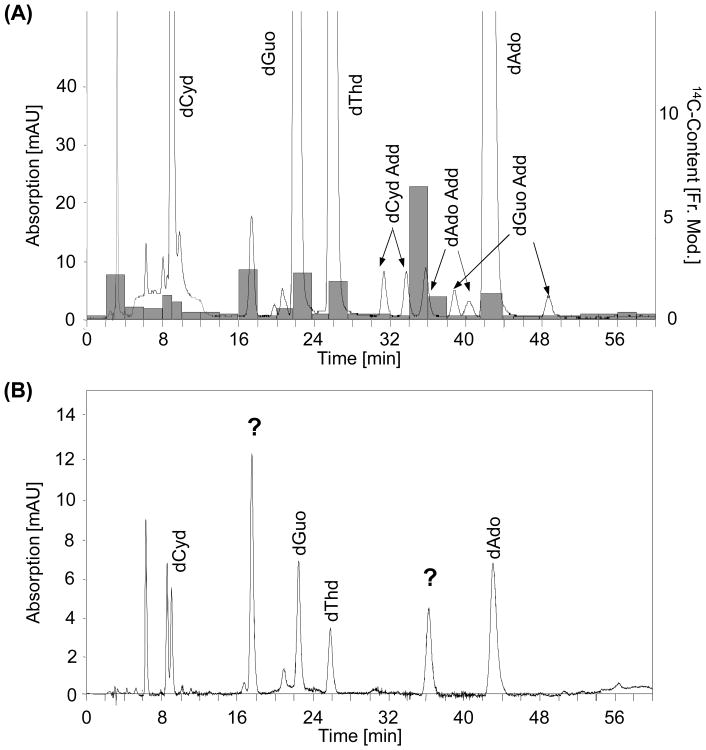
To determine if radioactive compounds coelute with adduct standards, hydrolyzed liver DNA of a 14C-furan treated rat was spiked with a reaction mixture of cis-2-butene-1,4-dial (BDA) and 2′-deoxyribonucleosides, separated by HPLC and analyzed by AMS. These experiments showed that fractions containing radiolabel eluted at retention times close to those of some of the cis-2-butene-1,4-dial-2′-desoxyribonucleoside adduct standards (Figure 3 A). Uncharacterized peaks eluting at these retention times were also observed after incubation of calf thymus DNA with cis-2-butene-1,4-dial (BDA) (Figure 3 B).
Figure 3.
(A) 14C-content of DNA fractions (highlighted in gray) and HPLC chromatogram (λ= 254 nm) of enzymatically hydrolyzed liver DNA from a [3,4-14C]-furan treated rat, spiked with in vitro incubation products of cis-2-butene-1,4-dial and 2′-deoxyribonucleosides.
(B) Separation of nucleosides from calf thymus DNA incubated with cis-2-butene-1,4-dia in vitro, showing the presence of unknown peaks with retention times corresponding to radiolabeled compounds observed in DNA of a [3,4-14C]-furan treated rats.
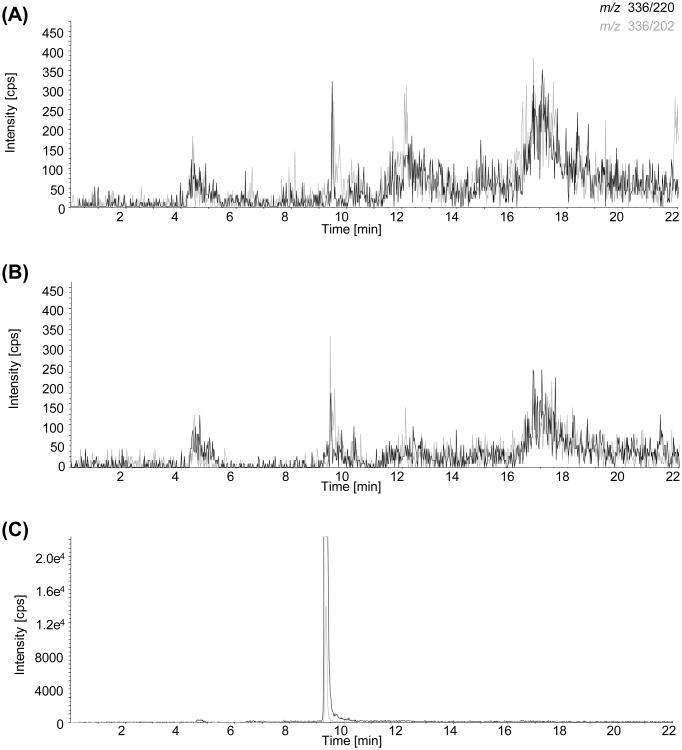
To further characterize compounds carrying the radiolabel, corresponding DNA-fractions obtained from rats treated with a single high dose of furan (30 mg/kg bw) or rats subchronically exposed to furan (2 mg/kg bw for 28 d) were collected and analyzed by LC-MS. Fractionation and subsequent LC-MS analysis of the cis-2-butene-1,4-dial-2′-desoxyribonucleoside adduct standards confirmed that adduct standards are stable during sample preparation. However, none of the adducts formed in vitro by reaction of cis-2-butene-1,4-dial (BDA) with 2′-deoxycytidine, 2′-deoxyadenosine or 2′-deoxyguanosine were detected in DNA isolated from furan treated rats (Figure 4 and data not shown), with detection limits ≤ 50 fmol for dCyd-BDA and ≤ 100 fmol for dGuo- and dAdo-BDA adducts estimated assuming ≤ 100% reaction yield. Based on the absolute LOD and the amount of DNA injected, LODs were thus ≤ 3.3 adducts/108 nucleotides for dCyd-BDA and ≤ 6.6 adducts/108 nucleotides for dGuo- and dAdo-BDA, which is at least an order of magnitude below adduct levels determined by AMS after a single oral dose of furan at 2 mg/kg bw. Unfortunately, further attempts to characterize the structure of DNA modifications using EMS-IDA-EPI failed. In addition, screening for potential DNA crosslinks using theoretical MRMs based on fragmentation of potential crosslink structures by loss of deoxyribose (M+ - 116) did not reveal formation of BDA derived DNA crosslinks in DNA extracted from furan treated rats or in calf thymus DNA incubated with BDA in vitro (data not shown).
Figure 4.
LC-MS MRM analysis of dAdo-BDA (m/z 336 – 220 and 336 – 202) in (A) control rat DNA and (B) DNA from a rat treated with 30 mg/kg bw (single dose study), and (C) calf thymus DNA spiked with dCyd-BDA, dGuo-BDA and dAdo-BDA incubation mixtures.
Furan genotoxicity in rats in vivo
Following repeated oral administration of furan to male F344 rats, no statistically significant increase in the incidence of micronucleated polychromatic erythrocytes (PCEs) was observed (Table 2). Furan did not induce a statistically significant increase in the number of bone marrow cells bearing chromosomal aberrations or in the frequencies of sister chromatid exchanges (SCE's) (Table 2). However, treatment with furan at 2 mg/kg bw resulted in statistically significant increases in the number of splenocytes bearing aberrations at all time points (Table 2). At 0.5 mg/kg bw, a significant increase in chromosomal aberrations in splenocytes was observed after 28 days of treatment.
Table 2.
Determination of the presence of micronuclei (MN) in immature bone marrow erythrocytes, chromosomal aberrations and sister chromatid exchanges (SCE's) in bone marrow cells and Chromosomal aberrations in resting G0 splenocytes of rats after oral administration of furan for up to 28 days.
| Bone marrow | Splenocytes | ||||
|---|---|---|---|---|---|
| Dose-levels mg/kg bw | Treatment time days |
|
|
||
| Group mean incidence of micronucleated PCE's (‰ ± SE) | Group mean incidence of chromosomal aberrations (% ± SE) | Frequencies of SCE's/cell (Group means ± SE) | Group mean incidence of chromosomal aberrations (% ± SE) | ||
| 0.0 | 5 | 0.40 ± 0.18 | 0.33 ± 0.19 | 3.80 ± 2.19 | 0.33 ± 0.19 |
| 0.1 | 0.40 ± 0.18 NS | 1.33 ± 0.77 NS | 4.10 ± 2.37 NS | 2.75 ± 1.59 NS | |
| 0.5 | 0.50 ± 0.22 NS | 1.67 ± 0.96 NS | 3.40 ± 1.96 NS | 3.00 ± 1.73 NS | |
| 2.0 | 0.40 ± 0.18 NS | 0.00 ± 0.00 NS | 3.70 ± 2.14 NS | 5.00 ± 2.89 * | |
|
| |||||
| 0.0 | 28 | 0.30 ± 0.13 | 0.00 ± 0.00 | 4.10 ± 2.38 | 2.00 ± 1.15 |
| 0.1 | 0.40 ± 0.18 NS | 0.00 ± 0.00 NS | 3.70 ± 2.13 NS | 4.00 ± 2.31 NS | |
| 0.5 | 0.50 ± 0.22 NS | 1.67 ± 0.96 NS | 3.20 ± 1.84 NS | 7.63 ± 4.41 ** | |
| 2.0 | 0.40 ± 0.13 NS | 1.33 ± 0.77 NS | 4.30 ± 2.48 NS | 6.60 ±3 .81 * | |
|
| |||||
| 0.0 | 28 + 14 recovery | 0.20 ± 0.09 NS | 0.33 ± 0.19 | 5.30 ± 3.06 | 1.33 ± 0.77 |
| 2.0 | 0.30 ± 0.13 NS | 1.00 ± 0.58 NS | 4.80 ± 2.77 NS | 6.67 ± 3.85 * | |
PCE's: Polychromatic erythrocytes
SE: Standard error of the mean
NS: Not significantly different form concurrent negative control
: Statistically significant at p<0.05
: Statistically significant at p<0.01
Analysis of bone marrow and blood cells by the Comet Assay revealed no significant changes (increases or decreases) in the mean tail moment and tail intensity compared with corresponding untreated control at any dose-level and sampling time assayed (Table 3). In the liver, the target organ of furan carcinogenicity, a slight, although not statistically significant reduction in both tail moment and tail intensity compared to untreated control was observed in all treatment groups after 28 days of furan treatment. In contrast, a statistically significant increase over controls in both tail moment and tail intensity was seen in the recovery group (Table 3).
Table 3.
Comet assay data in bone marrow, blood and liver of F344/N rats orally treated with furan for up to 28 days.
| Bone marrow | Blood | Liver | |||||
|---|---|---|---|---|---|---|---|
| Dose-levels (mg/kg bw) | Treatment time (days) |
|
|
|
|||
| Tail moment (mean ± SD) | Tail intensity (mean ± SD) | Tail moment (mean ± SD) | Tail intensity (mean ± SD) | Tail moment (mean ± SD) | Tail intensity (mean ± SD) | ||
| 0.0 | 5 | 0.85 ± 0.38 | 2.93 ± 1.31 | 0.38 ± 0.17 | 1.99 ± 0.89 | 1.41 ± 0.63 | 4.25 ± 1.90 |
| 0.1 | 0.68 ± 0.30 | 2.65 ± 1.19 | 0.29 ± 0.13 | 1.43 ± 0.64 | 2.04 ± 0.91 | 6.33 ± 2.83 | |
| 0.5 | 0.76 ± 0.34 | 2.82 ± 1.26 | 0.23 ± 0.10 | 1.23 ± 0.55 | 2.15 ± 0.96 | 6.69 ± 2.99 | |
| 2.0 | 0.76 ± 0.34 | 2.63 ± 1.18 | 0.52 ± 0.23 | 1.79 ± 0.80 | 2.60 ± 1.16 | 6.25 ± 2.80 | |
|
| |||||||
| 0.0 | 28 | 1.03 ± 0.46 | 3.86 ± 1.73 | 0.47 ± 0.21 | 2.87 ± 1.28 | 2.09 ± 0.93 | 7.57 ± 3.39 |
| 0.1 | 1.01 ± 0.45 | 3.92 ± 1.75 | 1.00 ± 0.45 | 5.54 ± 2.48 | 1.74 ± 0.78 | 5.72 ± 2.56 | |
| 0.5 | 0.97 ± 0.43 | 3.59 ± 1.61 | 0.48 ± 0.21 | 2.73 ± 1.22 | 1.92 ± 0.85 | 5.90 ± 2.85 | |
| 2.0 | 1.27 ± 0.57 | 4.53 ± 2.03 | 0.36 ± 0.16 | 2.01 ± 0.90 | 1.76 ± 0.79 | 5.69 ± 2.54 | |
|
| |||||||
| 0.0 | 28 + 14 recovery | 0.69 ± 0.31 | 3.04 ± 1.36 | 0.39 ± 0.17 | 2.17 ± 0.97 | 2.34 ± 1.05 | 7.47 3.34 |
| 2.0 | 0.90 ± 0.40 | 3.69 ± 1.65 | 0.50 ± 0.22 | 2.53 ± 1.13 | 5.14 ± 2.30** | 13.40 ± 5.99** | |
: Statistically significant at p<0.01.
4 Discussion
Cis-2-butene-1,4-dial, a reactive metabolite formed by CYP2E1 dependent biotransformation of furan, was shown to form covalent adducts with nucleosides [2, 3], and to induce genotoxic effects in bacteria [4] and in mammalian cells in vitro[5]. In contrast, the potential of furan to covalently bind to DNA in liver in vivo and the contribution of genotoxicity to furan mediated tumor formation are still not fully resolved. Results from the present study demonstrate a significant, dose-dependent increase in the 14C content of DNA in both target and non-target organs following treatment of male F344/N rats with [3,4-14C]-furan in the absence of significant metabolic incorporation, providing clear evidence that furan reacts with DNA in vivo. However, the previously characterized diastereomeric adducts formed by reaction of cis-2-butene-1,4-dial with 2′-deoxycytidine, 2′-deoxyadenosine, and 2′-deoxyguanosine in vitro [2, 3] were not detected in DNA isolated from furan treated rat liver, and further attempts to elucidate the structure of DNA modifications carrying the radiolabel failed. While furan did not induce genotoxic effects in blood and bone marrow cells of rats administered furan for up to 28 days, furan treatment resulted in a dose-dependent increase in the frequency of chromosomal aberrations in splenocytes. In liver, furan induced a reduction of tail moment and tail intensity after 28 days of furan treatment and increased DNA strand breaks after two weeks recovery. Collectively, results from this study suggest that genotoxicity resulting from covalent binding of furan reactive metabolites to DNA may contribute to furan carcinogenicity and needs to be considered in furan risk assessment.
While Burka et al. found no radioactivity associated with DNA following administration of [2,5-14C]-furan to male F344 [8], analysis of DNA extracted from livers and kidneys of rats given a single dose of [3,4-14C]-furan by accelerator mass spectrometry revealed a significant, dose-related increase in 14C bound to DNA. These seemingly contrasting findings may be due to the lower specific activity of [2,5-14C]-furan (90 μCi/mmol) and use of liquid scintillation counting in the Burka study, vs. a specific activity of 20 mCi/mmol and application of accelerator mass spectrometry, a highly sensitive technique for the detection of radiocarbon in biological samples, in our study, resulting in markedly enhanced sensitivity. Importantly, the majority of radiolabel was not associated with normal nucleosides, suggesting covalent binding of a reactive metabolite(s) of [3,4-14C]-furan to DNA. Average adduct levels in liver reached 1.7 ± 0.7 adducts/108 nucleotides and 3.3 ± 2.1 adducts/107 nucleotides in the low (0.1 mg/kg bw) and high (2.0 mg/kg bw) dose group respectively, with no statistically significant differences between furan target lobes. Adduct levels observed after a single dose of furan associated with tumor incidences of 10% for hepatocellular tumors and 86% for cholangiocarcinoma following chronic treatment are thus within the range of adduct levels reported after repeated administration of a range of liver carcinogens at the TD50 (i.e. dose resulting in 50% tumor incidence) [27]. Albeit consistent with renal expression of CYP2E1 and renal toxicity of furan at higher doses [1, 28], the finding that significant DNA binding was also observed in the kidney, which is not a target for furan carcinogenicity, suggests that covalent binding of furan to DNA may not be sufficient for tumor induction.
The fact that we did not detect the previously characterized substituted etheno-adducts despite sufficient sensitivity of our analysis is perhaps not surprising given that the primary adducts of cis-2-butene-1,4-dial with 2′-deoxyadenosine and 2′-deoxyguanosine may rearrange, thereby unmasking an aldehydic moiety with the potential to form cross-links [3]. In the presence of intact DNA, it is also plausible that cis-2-butene-1,4-dial itself may cross-link DNA bases of the same or opposing DNA strand rather than forming single base adducts as in the presence of isolated nucleosides. Although Kellert et al. found no evidence for furan-derived cross-links in L5178Y tk+/- mouse lymphoma cells [6], Marinari et al. observed DNA cross-links in CHO-Kl cells treated with cis-2-butene-1,4-dial using the alkaline elution technique [5]. Moreover, a recent study in turkey fetus liver demonstrating a reduction of % DNA in comet tail and increased DNA strand breaks after treatment with proteinase K, suggested that furan may also form DNA-protein cross links [29]. The latter is consistent with the high reactivity of cis-2-butene-1,4-dial towards cysteine and lysine residues [30, 31]. Due to the bifunctionality of cis-2-butene-1,4-dial, a wide spectrum of covalent DNA modifications may thus be formed, making it difficult to obtain structural information. Although our analyses suggest that 14C in DNA of rats treated with [3,4-14C]-furan is associated with essentially two fractions, which correspond to as yet unidentified peaks observed in rats dosed with furan and/or in DNA treated with cis-2-butene-1,4-dial in vitro, attempts to characterize the structure of these DNA modifications by LC-MS/MS failed.
In a recent 8-week study in transgenic Big Blue rats, Mc Daniel et al. found no evidence for furan genotoxicity in the target organ using the liver cII transgene mutation assay and the comet assay at furan bioassay doses [32], although liver DNA damage was observed at higher doses. Moreover, furan was negative in the peripheral blood micronuclei assay, the Hprt lymphocyte gene mutation assay, and the Pig-a lymphocyte and peripheral red blood cell (RBC) gene mutation assays [32]. These data are largely consistent with the absence of effects of furan on the incidence of micronuclei, chromosomal aberrations and sister chromatid exchanges in bone marrow, and negative findings in the comet assay in blood and bone marrow at any dose and sampling time assayed in our study. The reduction in tail moment and tail intensity in liver after 28 days of furan treatment and significant increases in DNA strand breaks after an additional two week recovery period may indicate that effective repair of cross-links resulting in generation of DNA strand breaks may only occur after furan treatment is discontinued. In contrast, sustained exposure to furan, leading to cross-links and thus reduction in the mean tail moment and intensity, may mask the presence of DNA breaks in the alkaline comet assay. Although further work may be needed to confirm the significance of our rat liver comet data in this low dose range, a recent report on furan induced DNA damage in liver provides clear evidence for a genotoxic potential of furan in the target organ [33]. These authors demonstrated that the extent of direct, endonuclease III (EndoIII)- and formamidopyrimidine glycosylase (FPG)-sensitive strand breakage in rat liver peaked at 1-3 h post-treatment, and returned to control levels 16 h after administration of furan at 16 mg/kg bw for 3 days, suggesting that sampling time may be critical to detect furan-induced DNA damage. Based on the results of their time-course study, the authors were then able to show a dose-related increase in DNA strand breakage in liver, but not bone marrow, of rats given furan at 0, 2, 4, 8, 12, or 16 mg/kg bw for 3 days, whereby statistical significance was reached at ≥ 8, 4 and 12 mg/kg bw for direct, EndoIII- and FPG-sensitive DNA damage [33]. In addition to these findings, the dose-related increase in chromosomal aberrations in rat splenocytes exposed to furan in vivo and stimulated to proliferate in vitro, which is consistent with significant increases of both micronuclei and γ-H2AX foci in splenocytes following subchronic treatment of mice with furan at up to 15 mg/kg/day [34], provides evidence for furan genotoxicity in vivo.
Collectively, the available data demonstrate that furan binds to DNA in the target organ of furan carcinogenicity and has the potential to induce genotoxic effects in the form of DNA strand breaks and chromosomal aberrations in vivo. Although the contribution of furan genotoxicity to the induction of hepatocellular tumors and cholangiocarcinomas may well depend on the tumor type, these data suggest that furan may operate – at least in part - by a genotoxic mode of action.
Figure 1.
Reaction of cis-2-butene-1,4-dial with DNA bases.
Acknowledgments
This work was supported by the 6th Framework Programme of the European Union (SSPE-CT-2006-44393). This Work was partially performed at the Research Resource for Biomedical AMS which is operated at LLNL under the auspices of the U.S. Department of Energy under contract #W-7405-ENG-48 and supported by grants from the National Center for Research Resources (5P41RR013461-14) and the National Institute of General Medical Sciences (8 P41 GM103483-14) from the National Institutes of Health (https://bioams.llnl.gov/). The authors would also like to thank Caroline Kröcher, Elisabeth Rüb-Spiegel, Heike Keim-Heusler, Ursula Tatsch, Michaela Bekteshi, Miriam Kral and Nataly Bittner (Department of Toxicology, University of Würzburg) for excellent technical assistance and animal care.
Abbreviations
- AMS
accelerator mass spectrometry
- BDA
cis-2-butene-1,4-dial
- SCEs
sister chromatid exchanges
References
- 1.NTP. Toxicology and Carcinogenesis Studies of Furan (CAS No. 110-00-9) in F344 Rats and B6C3F1 Mice(Gavage Studies) Natl Toxicol Program Tech Rep Ser. 1993;402:1–286. [PubMed] [Google Scholar]
- 2.Byrns MC, Predecki DP, Peterson LA. Characterization of nucleoside adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 2002;15:373–379. doi: 10.1021/tx0101402. [DOI] [PubMed] [Google Scholar]
- 3.Byrns MC, Vu CC, Peterson LA. The formation of substituted 1,N6-etheno-2′-deoxyadenosine and 1,N2-etheno-2′-deoxyguanosine adducts by cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 2004;17:1607–1613. doi: 10.1021/tx049866z. [DOI] [PubMed] [Google Scholar]
- 4.Peterson LA, Naruko KC, Predecki DP. A reactive metabolite of furan, cis-2-butene-1,4-dial, is mutagenic in the Ames assay. Chem Res Toxicol. 2000;13:531–534. doi: 10.1021/tx000065f. [DOI] [PubMed] [Google Scholar]
- 5.Marinari UM, Ferro M, Sciaba L, Finollo R, et al. DNA-damaging activity of biotic and xenobiotic aldehydes in Chinese hamster ovary cells. Cell Biochem Funct. 1984;2:243–248. doi: 10.1002/cbf.290020411. [DOI] [PubMed] [Google Scholar]
- 6.Kellert M, Brink A, Richter I, Schlatter J, Lutz WK. Tests for genotoxicity and mutagenicity of furan and its metabolite cis-2-butene-1,4-dial in L5178Y tk(+/−) mouse lymphoma cells. Mutat Res. 2008 doi: 10.1016/j.mrgentox.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Glatt H, Schneider H, Liu Y. V79-hCYP2E1-hSULT1A1, a cell line for the sensitive detection of genotoxic effects induced by carbohydrate pyrolysis products and other food-borne chemicals. Mutat Res. 2005;580:41–52. doi: 10.1016/j.mrgentox.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Burka LT, Washburn KD, Irwin RD. Disposition of [14C]furan in the male F344 rat. J Toxicol Environ Health. 1991;34:245–257. doi: 10.1080/15287399109531564. [DOI] [PubMed] [Google Scholar]
- 9.EFSA. Report of the Scientific Panel on Contaminants in the Food Chain on provisional findings on furan in food. The EFSA Journal. 2004;137:1–20. [Google Scholar]
- 10.Coldwell KE, Cutts SM, Ognibene TJ, Henderson PT, Phillips DR. Detection of Adriamycin-DNA adducts by accelerator mass spectrometry at clinically relevant Adriamycin concentrations. Nucleic Acids Res. 2008;36:e100. doi: 10.1093/nar/gkn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creek MR, Mani C, Vogel JS, Turteltaub KW. Tissue distribution and macromolecular binding of extremely low doses of [14C]-benzene in B6C3F1 mice. Carcinogenesis. 1997;18:2421–2427. doi: 10.1093/carcin/18.12.2421. [DOI] [PubMed] [Google Scholar]
- 12.Du HF, Xu LH, Wang HF, Liu YF, et al. Formation of MTBE-DNA adducts in mice measured with accelerator mass spectrometry. Environ Toxicol. 2005;20:397–401. doi: 10.1002/tox.20124. [DOI] [PubMed] [Google Scholar]
- 13.Kwok ES, Buchholz BA, Vogel JS, Turteltaub KW, Eastmond DA. Dose-dependent binding of ortho-phenylphenol to protein but not DNA in the urinary bladder of male F344 rats. Toxicol Appl Pharmacol. 1999;159:18–24. doi: 10.1006/taap.1999.8722. [DOI] [PubMed] [Google Scholar]
- 14.Mally A, Zepnik H, Wanek P, Eder E, et al. Ochratoxin A: lack of formation of covalent DNA adducts. Chem Res Toxicol. 2004;17:234–242. doi: 10.1021/tx034188m. [DOI] [PubMed] [Google Scholar]
- 15.Turteltaub KW, Vogel JS, Frantz C, Felton JS, McManus M. Assessment of the DNA adduction and pharmacokinetics of PhIP and MeIOx in rodents at doses approximating human exposure using the technique of accelerator mass spectrometry (AMS) and 32P-postlabeling. Princess Takamatsu Symp. 1995;23:93–102. [PubMed] [Google Scholar]
- 16.White IN, Martin EA, Mauthe RJ, Vogel JS, et al. Comparisons of the binding of [14C]radiolabelled tamoxifen or toremifene to rat DNA using accelerator mass spectrometry. Chem Biol Interact. 1997;106:149–160. doi: 10.1016/s0009-2797(97)00063-x. [DOI] [PubMed] [Google Scholar]
- 17.Holzapfel CW, Williams DBG. A facile route to 3a,8a-dihydrofuro[2,3-b]benzofuran. Tetrahedron. 1995;51:8555–8564. [Google Scholar]
- 18.Hamberger C, Kellert M, Schauer UM, Dekant W, Mally A. Hepatobiliary toxicity of furan: identification of furan metabolites in bile of male F344/N rats. Drug Metab Dispos. 2010;38:1698–1706. doi: 10.1124/dmd.109.031781. [DOI] [PubMed] [Google Scholar]
- 19.Mally A, Graff C, Schmal O, Moro S, et al. Functional and proliferative effects of repeated low-dose oral administration of furan in rat liver. Mol Nutr Food Res. 2010;54:1556–1567. doi: 10.1002/mnfr.201000064. [DOI] [PubMed] [Google Scholar]
- 20.Moro S, Chipman JK, Antczak P, Turan N, et al. Identification and pathway mapping of furan target proteins reveal mitochondrial energy production and redox regulation as critical targets of furan toxicity. Toxicol Sci. 2012;126:336–52. doi: 10.1093/toxsci/kfs005. [DOI] [PubMed] [Google Scholar]
- 21.Turteltaub KW, Vogel JS, Frantz CE, Fultz E. IARC Sci Publ. 1993. Studies on DNA adduction with heterocyclic amines by accelerator mass spectrometry: a new technique for tracing isotope-labelled DNA adduction; pp. 293–301. [PubMed] [Google Scholar]
- 22.Vogel JS, Love AH. Quantitating isotopic molecular labels with accelerator mass spectrometry. Methods Enzymol. 2005;402:402–422. doi: 10.1016/S0076-6879(05)02013-6. [DOI] [PubMed] [Google Scholar]
- 23.Turteltaub KW, Felton JS, Gledhill BL, Vogel JS, et al. Accelerator mass spectrometry in biomedical dosimetry: relationship between low-level exposure and covalent binding of heterocyclic amine carcinogens to DNA. Proc Natl Acad Sci U S A. 1990;87:5288–5292. doi: 10.1073/pnas.87.14.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 25.Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974;251:156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- 26.Wolff S, Perry P. Differential Giemsa staining of sister chromatids and the study of chromatid exchanges without autoradiography. Chromosoma. 1974;48:341–353. doi: 10.1007/BF00290991. [DOI] [PubMed] [Google Scholar]
- 27.Otteneder M, Lutz WK. Correlation of DNA adduct levels with tumor incidence: carcinogenic potency of DNA adducts. Mutat Res. 1999;424:237–247. doi: 10.1016/s0027-5107(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 28.Hamadeh HK, Jayadev S, Gaillard ET, Huang Q, et al. Integration of clinical and gene expression endpoints to explore furan-mediated hepatotoxicity. Mutat Res. 2004;549:169–183. doi: 10.1016/j.mrfmmm.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Jeffrey AM, Brunnemann KD, Duan JD, Schlatter J, Williams GM. Furan induction of DNA cross-linking and strand breaks in turkey fetal liver in comparison to 1,3-propanediol. Food Chem Toxicol. 2011 doi: 10.1016/j.fct.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Chen LJ, Hecht SS, Peterson LA. Characterization of amino acid and glutathione adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 1997;10:866–874. doi: 10.1021/tx9700174. [DOI] [PubMed] [Google Scholar]
- 31.Lu D, Peterson LA. Identification of furan metabolites derived from cysteine-cis-2-butene-1,4-dial-lysine cross-links. Chem Res Toxicol. 2010;23:142–151. doi: 10.1021/tx9003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel LP, Ding W, Dobrovolsky VN, Shaddock JG, Jr, et al. Genotoxicity of furan in Big Blue rats. Mutat Res. 2012;742:72–78. doi: 10.1016/j.mrgentox.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Ding W, Petibone DM, Latendresse JR, Pearce MG, et al. In vivo genotoxicity of furan in F344 rats at cancer bioassay doses. Toxicol Appl Pharmacol. 2012 doi: 10.1016/j.taap.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Leopardi P, Cordelli E, Villani P, Cremona TP, et al. Assessment of in vivo genotoxicity of the rodent carcinogen furan: evaluation of DNA damage and induction of micronuclei in mouse splenocytes. Mutagenesis. 2010;25:57–62. doi: 10.1093/mutage/gep043. [DOI] [PubMed] [Google Scholar]