Abstract
Mitochondrial outer membrane is a major site of apoptosis regulation across phyla. Human and C. elegans Bcl-2 family proteins and Drosophila Hid require the C-terminal Tail Anchored (TA) sequence to insert into the mitochondrial membrane, but it remains unclear if cytosolic proteins actively regulate the mitochondrial localization of these proteins. Here, we report that cdk7 regulates the mitochondrial localization of Hid and its ability to induce apoptosis. We identified cdk7 through an in vivo RNAi screen of genes required for cell death. Although CDK7 is best known for its role in transcription and cell cycle progression, a hypomorphic cdk7 mutant suppressed apoptosis without impairing these other known functions. In this cdk7 mutant background, Hid failed to localize to the mitochondria and failed to bind to recombinant IAPs. These findings indicate that apoptosis is promoted by a novel function of CDK7, which couples the mitochondrial localization and IAP-binding of Hid.
Introduction
Apoptosis is a specific form of cell death that helps to eliminate superfluous and dangerous cells in animal tissues. In Drosophila, the Inhibitor of Apoptosis (IAP)-antagonists, Reaper, Grim, Hid and Sickle, initiate apoptosis in this organism by binding to, and inhibiting Drosophila Inhibitor of Apoptosis 1 (DIAP1), which allows caspases to become active and execute apoptosis (Ryoo and Baehrecke, 2010).
Across phyla, proteins that localize to the mitochondrial outer membrane play important roles in regulating apoptosis. For example, mammalian Bcl-2 family proteins at the mitochondrial outer membrane regulate the release of cytochrome c into the cytoplasm, which initiates caspase activation for execution of cell death (Wang, 2001). Although Bcl-2 homologs do not play an obvious role in Drosophila apoptosis, the role of mitochondria in the regulation of cell death is conserved. Specifically, a number of independent studies have now established that Drosophila IAP-antagonist proteins initiate apoptosis after localizing to the outer membrane of the mitochondria (Abdelwahid et al., 2007; Claveria et al., 2002; Haining et al., 1999; Olson et al., 2003; Sandu et al., 2010). One of these proteins, Hid, has a mitochondrial membrane insertion sequence within the last 20 amino acids of the coding sequence, similar to Bcl-2. Such membrane insertion sequences close to the C-terminal end are collectively referred to as Tail Anchors (TA) (Borgese et al., 2007; Haining et al., 1999). Deletion of the TA sequence impairs the pro-apoptotic function of Hid (Abdelwahid et al., 2007; Sandu et al., 2010). The requirement of IAP-antagonists to localize to the mitochondrial outer membrane has remained a puzzle, as their main target, DIAP1 is not associated with the mitochondria.
Recent studies have elucidated new features in the trafficking mechanism of a subset of TA proteins that are destined to the endoplasmic reticulum (ER) membrane. It has been found that these TA proteins are recognized by a chaperone complex, and delivered to the ER membrane through the assistance of an ATPase and ER docking proteins (Hegde and Keenan, 2011). On the other hand, it remains unclear whether TA proteins that are destined to the mitochondrial outer membrane are subject to regulation by cytosolic factors (Colombo et al., 2009).
Here, we report the identification of cdk7 as a gene required for Hid’s mitochondrial localization. In addition, we show that mitochondrial localization of Hid is coupled with its ability to bind to DIAP1. These observations support the idea that cells actively regulate apoptosis through controlled localization of pro-apoptotic proteins to the mitochondrial outer membrane.
Results
Identification of CDK7 and MAT1 as mediators of the Rhodopsin-1G69D overexpression phenotype
To identify genes that mediate cell death, we used a Drosophila model for Retinitis Pigmentosa, in which cell death is caused by a mutant Rhodopsin-1 (Rh-1) allele, G69D, that fails to fold properly (Colley et al., 1995; Kurada and O’Tousa, 1995). As rhodopsins are membrane proteins that are synthesized and undergo folding in the endoplasmic reticulum (ER), these alleles cause stress in the ER (Ryoo et al., 2007).
To establish a more facile screening method, we overexpressed this mutant Rh-1 allele in the developing larval eye discs, using the eye specific Glass Multimer Repeat (GMR) promoter. Under these conditions, massive cell death occurs during development, and as a result, the adults eclose with partially ablated eyes that are readily noticeable. Specifically, these adult fly eyes showed a glassy surface, due to the ablation of the ommatidial structures that are normally in regular arrays (Figure 1a, b, e, f). In this background, we performed an in vivo RNAi screen to identify suppressors of this phenotype (see Experimental Procedures). We specifically targeted kinases and phosphatases, and those implicated in ubiquitin-mediated protein degradation.
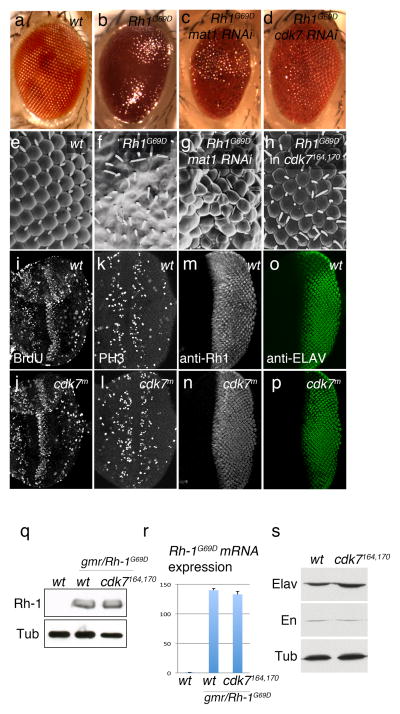
Figure 1. CDK7 and MAT1 mediate the mutant Rh-1G69D overexpression phenotype.
External eyes imaged by light microscopy (a–d) or Scanning EM (e–h). Wild type adult fly eyes show regular array of ommatidia (a, e), which is destroyed when Rh-1G69D is overexpressed during eye development through the eye-specific GMR promoter (b, f). In vivo RNAi screen for suppressors of this phenotype identified lines that target mat1 (c, g) and cdk7 (d). GMR-Rh-1G69D –induced ommatidial disruption phenotype was also suppressed in a cdk7S164A,T170A background (h). (i–l) eye imaginal discs were labeled for BrdU incorporation (i, j), anti-phospho Histone H3 antibody (k, l). (m,-p) GMR>Rh-1G69D eye discs were labeled with anti-Rh-1 antibody (m, n) and anti-ELAV antibody (o, p). These signals appear similar between cdk7 wild type (i, k, m, o) and cdk7S164A,T170A discs (j, l, n, p). (q) Western blots for Rh-1 and Beta-Tubulin (Tub) from wild type (lane 1) and GMR>Rh-1G69D (lanes 2 and 3) eye imaginal disc extracts. Lane 3 shows overexpressed Rh-1G69D levels in the cdk7164, 170background. (r) Quantitative RT-PCR against Rh-1G69D transcripts in genotypes equivalent to that of (q). (s) Western blots for ELAV, Engrailed (En) and Beta-Tubulin (Tub) in wild type and cdk7S164A,T170A imaginal disc extracts.
We identified three hits from the initial screen, which included CDK5, which we recently reported (Kang et al., 2012). The other two hits were lines VDRC10442 and VDRC106780, which target cdk7 and mat1 respectively (Figure 1c, d, g, h). We found this significant, as their encoded proteins are both subunits of the CDK Activating Kinase (CAK) complex (Fisher, 2005). MAT1 contains a RING domain, which is a signature ubiquitin-ligase motif, and CDK7 is a kinase. Knockdown of cdk7 not only rescued the eye size, but also restored the regular array of ommatidial structures (Figure 1d). The mat1 knockdown phenotype was less obvious, but the Scanning Electron Microscopy image showed that the disruption of ommatidial repeats by Rh-1G69D overexpression was partially rescued (Figure 1c, g). We further confirmed the role of cdk7, by employing a hypomorphic cdk7 mutant allele, S164A, T170A. These mutations reduce the kinase activity significantly, destabilizes the CAK complex and show temperature sensitivity (Larochelle et al., 2001). When GMR-Rh-1G69D was crossed into this cdk7S164A,T170A background and reared in 25°C, or even in lower temperatures, adult flies eclosed with a restored array of ommatidial repeats, validating the role of CDK7 in mediating the Rh-1G69D overexpression phenotype (Figure 1h).
CDK7 is best known for its role in general transcription and cell cycle progression. Specifically, CDK7 is part of the TFIIH complex that regulates general transcriptional initiation and mRNA processing (Fisher, 2005). In addition, CDK7 phosphorylates other CDKs to stimulate their enzymatic activity, and is required for general cell cycle progression (Larochelle et al., 1998).
To assess whether cdk7S164A,T170A mutants reared in 25°C have noticeable differences in the populations of S and M phase cells, we examined the incorporation of BrdU and anti-phospho-histone H3 antibody labeling. We found a similar pattern of these markers in the wild type and cdk7S164A,T170A mutant tissues (Figure 1i–l). Neither did we see an obvious difference in gene expression patterns of select genes, including Rh-1G69D overexpressed by the GMR promoter, ELAV, Engrailed and Beta-Tubulin (Figure 1m–s). We conclude that the hypomorphic cdk7S164A,T170A mutants reared in 25°C, still retains enough CDK7 activity for global gene expression and cell cycle progression, while strongly suppressing cell death.
CDK7 is required for DIAP1-antagonist-induced cell death
In Drosophila, most developmental or stress-induced apoptosis is mediated by the induction of four genes, grim, hid, reaper and sickle, whose main role is to antagonize Drosophila Inhibitor of Apoptosis Protein 1 (DIAP1) (Ryoo and Baehrecke, 2010) (Figure 2a). One of the pro-apoptotic proteins that induce these DIAP1-antagonists is p53 (Brodsky et al., 2000; Ollmann et al., 2000). Overexpression of these pro-apoptotic genes in the eye using the GMR promoter caused a partial ablation of the eye, due to excessive cell death during eye development (Figure 2b–f). Interestingly, these eye cell death phenotypes were significantly suppressed in the cdk7S164A,T170A background(Figure 2h–l), indicating that CDK7 is required for apoptosis initiated by DIAP1 antagonists.
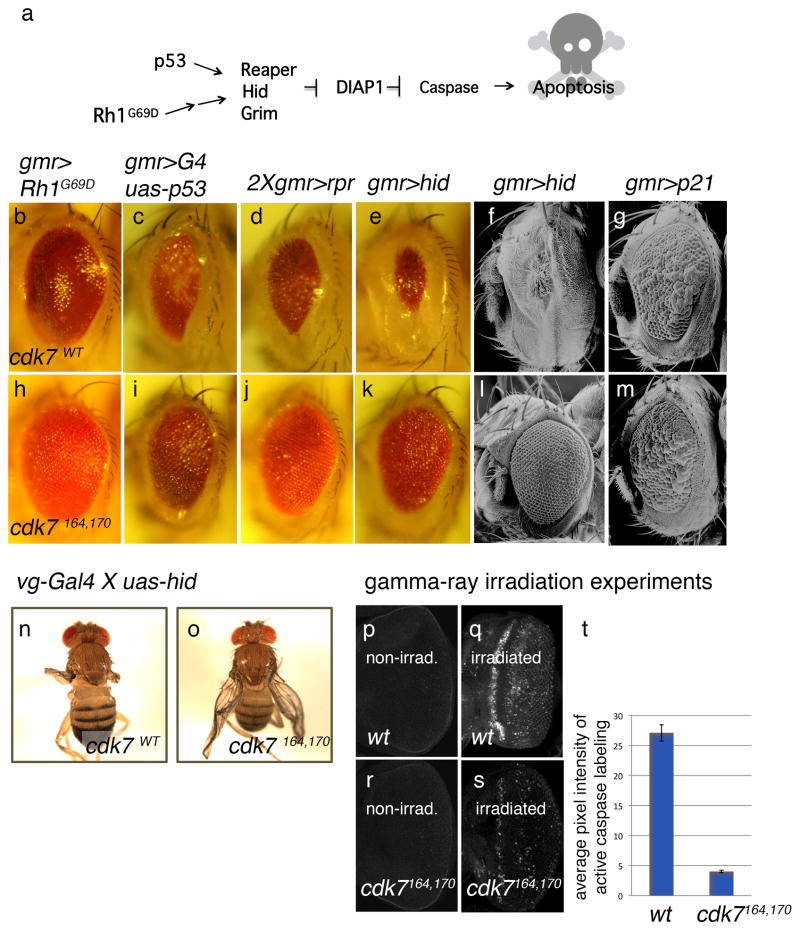
Figure 2. cdk7S164A,T170A suppresses DIAP1-anatagonist-induced cell death.
(a) Summary of the apoptosis pathway in Drosophila. The DIAP1 antagonists, reaper, hid and grim are induced by diverse apoptotic stimuli, such as in response to the overexpression of mutant Rh-1G69D or p53. These genes were overexpressed during eye development using either the eye-specific GMR promoter (b–m), or the wing-specific vg-Gal4 line (n, o), either in backgrounds of cdk7+(b–g, n), or in a cdk7S164A,T170A (h–m, o). As a control, the cell cycle inhibitor p21 was overexpressed through the GMR promoter (g, m). (p–t) Gamma-ray-induced caspase activation is suppressed in a cdk7S164A,T170A background. Larval eye imaginal discs were labeled with anti-cleaved caspase antibody (white) to assess cell death. Shown are non-irradiated discs (p, r), gamma-ray irradiated wild type discs (q), and gamma-ray irradiated cdk7S164A,T170A discs (s). Quantification of caspase activation is shown in (t).
Several pieces of evidence indicate that the effect of cdk7 is not due to an artifact of the eye-ablation based experimental setup. First, the GMR promoter driven overexpression of the cell cycle inhibitor, p21 (de Nooij and Hariharan, 1995), caused ommatidial fusion in adult eyes (Figure 2g), but this phenotype was not affected in the cdk7S164A,T170A background (Figure 2m), indicating that cdk7’s effect is specific to apoptosis. Second, the effect of cdk7S164A,T170A on apoptosis was also observed in the developing wing. When the DIAP1-antagonist, Hid, was overexpressed in this tissue using the vestigial-Gal4 driver, most flies emerged with ablated wings. However, the degree of wing ablation was strongly suppressed in the cdk7S164A,T170A background (Figure 2n, o).
To further validate the role of CDK7 in general apoptosis regulation, we subjected wild type and cdk7S164A,T170A imaginal discs to gamma-ray irradiation. Whereas non-irradiated imaginal discs showed very few cells undergoing apoptosis, as assessed by anti-cleaved caspase-3 antibody labeling (Figure 2p), gamma-ray irradiation induced massive apoptosis in the tissue (Figure 2q). Such gamma ray-induced caspase activation was significantly suppressed in cdk7S164A,T170A flies reared in 25°C (Figure 2r – t). These data indicate that CDK7 is required for cell death in diverse tissues of Drosophila.
cdk7S164A,T170A does not affect the transcriptional induction of DIAP1-antagonist
Since CDK7 plays a role in general transcription, we examined whether cdk7S164A,T170A tissues affect the transcription of key pro-apoptotic genes, reaper and hid. Ionizing irradiation triggers cell death, in part, by transcriptionally inducing these genes(Chen et al., 1996; Grether et al., 1995; White et al., 1994). However, we did not see a significant difference in gamma-ray induced reaper and hid transcript levels in the cdk7S164A,T170A embryos and larvae (Figure 3a, b). Rh-1G69D overexpression in eye imaginal discs also induced reaper transcripts (Figure 3c), but quantitative RT-PCR results did not detect differences in the cdk7S164A,T170A mutant background. These results argue against the effect on Hid transcription. However, CDK7 is also implicated in the processing of mRNA transcripts (Kanin et al., 2007), and we cannot fully rule out the possibility that the observed effect occurs at the level of Hid’s mRNA maturation.
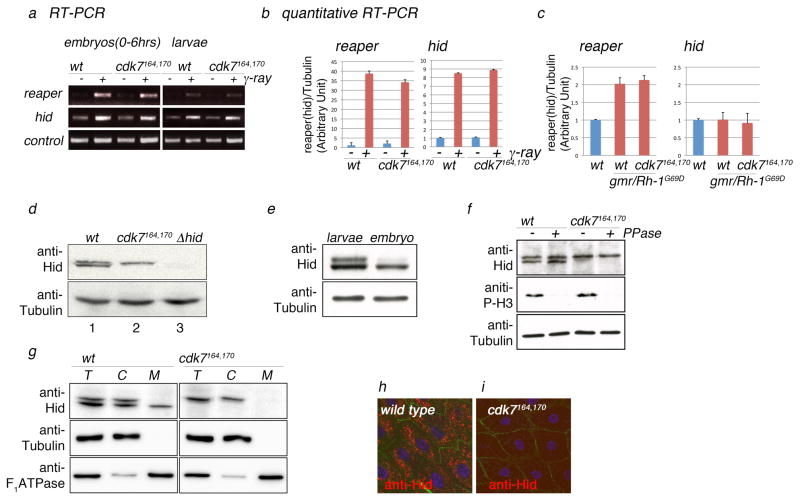
Figure 3. CDK7 is required for the mitochondrial localization of Hid protein.
(a) Gamma-ray induced reaper and hid transcripts assessed by semi-quantitative RT-PCR. Left gel shows induction in 0–6 hr old embryos. The right gel shows results from 3rd instar larval tissues. RT-PCRs against ubcD1 transcripts are shown as controls (bottom gel). (b) Quantitative RT-PCR against reaper and hid transcripts with or without gamma ray irradiation in larval eye imaginal discs. (c) Quantitative RT-PCR against reaper and hid transcripts in larval eye imaginal discs overexpressing Rh-1G69D (lanes 2 and 3), compared to that of the wild type controls (lanes 1). Lanes 3 show results from cdk7S164A,T170A tissues overexpressing Rh-1G69D. (d) Anti-Hid western blot reveals two closely migrating subspecies of Hid, and the lower band does not appear in the cdk7S164A,T170A background (lane 2). These anti-Hid bands migrated near the 70 kDa marker, which were absent from extracts of hid mutants (lane 3). Anti-beta tubulin western is shown as the loading control (bottom gel). (e) The ratio between the faster and slower migrating band changes during development. Larval extracts show roughly similar ratio of the two bands, whereas the embryonic extract predominantly shows the faster migrating band. (f) Phosphatase treatment under denaturing conditions in embryo extracts before (untreated; −) and after (+phosphatase; +PPase) treatment. Anti-phopho histone H3 antibody (anti-P-H3) was used as a positive control and anti-beta Tubulin antibody as a loading control. (g) Subcellular fractionation followed by anti-Hid western blots. Shown are Hid bands from total extracts (T lanes), cytosolic fraction (C lanes), and mitochondrial fraction (M lanes). Anti-beta-tubulin was used as the cytosolic marker, and anti-F1ATPase was used as the mitochondrial marker. (h, i) Anti-Hid immunohistochemistry (red) of larval proventriculus cells exposed to gamma-rays show a punctate pattern of labeling indicative of mitochondria in wild type tissues (h), but not in cdk7S164A,T170A mutants (i). Anti-armadillo antibody labeling (green) was used to mark cell boundaries, and anti-TO-PRO 3 to mark nuclei (blue).
CDK7 is required for the mitochondrial localization of Hid
Next, we examined possible effects of CDK7 on Hid protein. Interestingly, anti-Hid western blots from larval extracts revealed two closely migrating bands in wild type tissues, but only one from cdk7S164A,T170A extracts (Figure 3d). Furthermore, the ratio of the two Hid bands changed dynamically during development. Whereas larval extracts showed two bands of comparable amount, extracts from early embryos had mostly the faster migrating band (Figure 3e). To test if the two bands arise are a result of protein phosphorylation, we subjected Hid extracts to lambda phosphatase treatment (see Experimental Procedures). Whereas such treatment dephosphorylated Histone H3 efficiently, they did not affect the migration of Hid bands on Western blots (Figure 3f).
As Hid is a protein that localizes to the mitochondria for its function (Abdelwahid et al., 2007; Haining et al., 1999; Sandu et al., 2010), we explored the possibility that the two protein isoforms have distinct subcellular distribution patterns by performing subcellular fractionation assays. We found that the faster migrating band of Hid protein from the larval extracts fractionated with the mitochondrial marker, whereas the slower migrating form did not (Figure 3g). Consistently, extracts from cdk7S164A,T170A did not show any Hid protein fractionating with the mitochondria. The result was further validated by immunohistochemistry of larval proventriculus cells. Due to their large cell size, we were able to observe grainy patterns of anti-Hid labeling indicative of mitochondrial localization in wild type, but not in cdk7 mutant tissues (Figure 3h, i). Although cdk7164, 170 cells appear to have a generally reduced anti-Hid labeling, this is most likely due to the diffuse nature of Hid distribution or epitope masking, as Western blots detect a comparable level of Hid in the cytoplasmic fraction. We interpret that CDK7 is mostly required for producing the smaller Hid subspecies that localizes to the mitochondria, and cdk7S164A,T170A has impaired apoptosis because DIAP1-antagonists, such as Hid, fail to localize to that subcellular compartment.
Hid’s mitochondrial localization is linked with its ability to bind DIAP1
Because Hid triggers cell death by binding to DIAP1, and since mitochondrial localization is required for Hid’s cell killing activity (Abdelwahid et al., 2008; Sandu et al., 2010; Ryoo and Baehrecke, 2010), we examined whether the two isoforms of Hid showed different binding properties to DIAP1. Previous structural and biochemical studies have established that the DIAP-binding motif of Reaper and Hid consists of 4 to 8 amino acid residues at the N-terminus. However, the first methionine residue interferes with its high affinity interactions with DIAP1 (Wu et al., 2000; Wu et al., 2001), and it is speculated that this methionine is cleaved by a methione aminopeptidase in order to allow DIAP1 binding and initiation of apoptosis (Zachariou et al., 2003) (also, see Figure 4a).
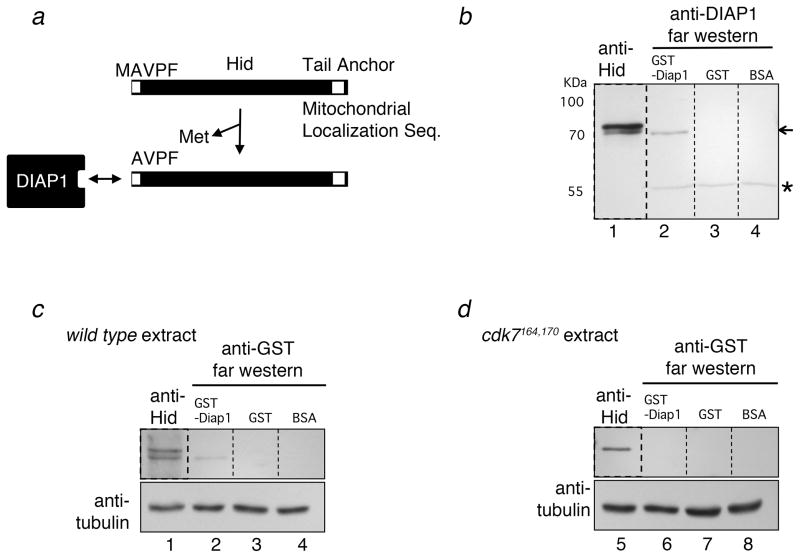
Figure 4. The smaller form of Hid binds to DIAP1.
(a) A schematic diagram of Hid’s primary structure and its interaction with DIAP1. The DIAP1-binding motif is near the N-terminus of Hid. High affinity interaction with DIAP1 requires the loss of the initiation Methionine, thereby exposing an N-terminal IAP binding motif of the sequence, AVPF (see text for reference). (b–d) Far-western blots show that only the faster migrating Hid band can bind to recombinant DIAP1. Larval extracts were run on SDS-PAGE, followed by membrane transfer. Each lane was cut (shown with the dash line) in order to incubate with the designated proteins and antibodies. These blots were later aligned, prior to exposure to film. Lanes 1 of all panels show anti-Hid western blots. Lanes 2–4 of all panels show blots that were preincubated with recombinant GST-DIAP1 (lane 2), GST (lane 3) and BSA (lane 4). The GST-DIAP1 binding bands were visualized by anti-DIAP1 (b) or anti-GST antibodies (c, d). Only GST-DIAP1 treated lanes show bands that correspond to the faster migrating Hid band. The arrow in (b) shows the Hid band visualized by GST-DIAP1 Far western. The asterisk in (b) shows the endogenous DIAP1 band near the 55 kDa marker. Extracts prepared from cdk7S164A,T170A larvae do not show the Hid band on Far western (d), indicating that Hid fails to bind DIAP1 in these mutants.
To assess the binding between DIAP1 and Hid, we performed ‘Far western’ blots. Specifically, extracts that were separated by SDS-Polyacrylamide gel and transferred to membranes, and those proteins were examined for the ability to bind recombinant GST-DIAP1 protein (See Experimental Procedures). The blots were aligned with anti-Hid western blots to determine which Hid band(s) can bind to DIAP1 in vitro. Under these conditions, only the faster migrating Hid band was detected by GST-DIAP1 Far-Western (Figure 4b, c). Blots that were incubated with GST or BSA did not show these bands, indicative of specific binding between recombinant DIAP1 and the faster migrating Hid isoform. Moreover, the interaction between GST-DIAP1 and the Hid band was no longer detected in the blots from the cdk7S164A,T170A larval extracts (Figure 4d). These results indicate that the faster migrating form of Hid, which localizes to the mitochondria and requires CDK7 for its generation, has a high affinity to DIAP1.
Discussion
Here, we report a previously unexpected mechanism of cell death regulation in Drosophila, in which the mitochondrial localization of a pro-apoptotic TA protein is regulated by CDK7. Moreover, we report that the mitochondrial localization of Hid is coupled with its ability to bind to DIAP1. Our finding provides an explanation for the mitochondrial requirement of IAP-antagonists.
Future studies are required to understand the structural nature of these Hid subspecies, and how this can be generated in a CDK7-dependent manner. Since only the faster migrating form binds to DIAP1, we favor the idea that the two isoforms differ in their N-terminus. In one speculative model, the faster migrating form may represent the proteolytically processed form that exposes critical N-terminal alanine that is responsible for DIAP1-binding. Alternatively, it is also possible that the slower migrating form receives a modification that inhibits DIAP1-binding.
Recent studies indicate that dedicated trafficking machinery exists for other TA proteins destined to the endoplasmic reticulum (Hegde and Keenan, 2011). However, the equivalent trafficking factors for mitochondria destined TA proteins have not yet been found, and it is widely assumed that those TA proteins insert into the mitochondrial outer membrane without active assistance. By contrast, our finding indicates that the Hid’s mitochondrial localization can be regulated in cells, suggesting the existence of an active trafficking machinery for the mitochondrial TA protein, Hid.
Experimental Procedures
Drosophila Genetics
Fly culture and crosses were carried out according to standard procedures. Gal4/UAS was used (Brand and Perrimon, 1993) to express the RNAi lines and a number of other transgenes. Unless otherwise noted, adult flies and embryos were kept at 25°C.The in vivo RNAi screen was performed as described previously (Kang et al., 2012). In brief, in vivo RNAi lines from the Vienna Drosophila Stock Center were crossed to the flies of the following genotype: GMR-Gal4; ey-Gal4, GMR-Rh-1G69D/CyO; uas-dicer. The cdk7S164A/T170A mutant flies [wDf(1)JB254, Pw+(snf+,dhd+)/wDf(1)JB254, Pw+(snf+,dhd+); +/+; Pw+(cdk7 S164A/T170A)/Pw+ (cdk7 S164A/T170A)] was a gift from Beat Suter and John Lis (Larochelle et al., 1998; Schwartz et al., 2003). Since cdk7 is on the X-chromosome, cdk7 S164A/T170A females or control wild type flies were crossed to relevant male flies, and their male progeny were analyzed. The following fly lines were described previously: GMR-Rh1G69D (Kang, 2012), GMR-Gal4,uas-p53 (Brodsky, 2000), uas-2xGMR>reaper (White et al., 1996), GMR-hid (Grether et al., 1995), GMR-p21 (de Nooij and Hariharan, 1995).
Measurement of RNA Levels
Total RNA was isolated using Trizol Reagent (Invitrogen) and reverse transcription was performed from 2ug of total RNA using SuperScript First-Strand Synthesis System (invitrogen). Real-time PCR was performed using a Power SYBR green kit and the applied ABI 7900HT Sequence Detection Systems machine (Applied Biosystems, Foster City, CA. USA). Beta-tubulin, Histon H3, Rpl15 were chosen as controls for Real-time PCRs. All experiments were repeated at least three times. Primer sequences are
Reaper-F; AGTCACAGTGGAGATTCCTGG
Reaper-R; TGCGATATTTGCCGGACTTTC
Hid-F; ACGGCCATCCGAATCCGAAC
Hid-R; TGCTGCTGCCGGAAGAAGAAGTT
beta-Tubulin-F; CTCAGTGCTCGATGTTGTCC
beta-Tubulin-R; GCCAAGGGAGTGTGTGAGTT
Histon H3-F; AGAGCACCGAGCTTCTAATCC
Histon H3-R; TTCGCTAGCTTCCTGCAGAG
Rpl15-F; AGGATGCACTTATGGCAAGC
Rpl15-R; GCGCAATCCAATACGAGTTC
Gamma-ray irradiation
To induce apoptosis, embryos or feeding third instar larvae were exposed to 40Gy of gamma-ray prior to dissection. Discs were dissected in PBS 2–3 hours post irradiation.
Scanning electron microscopy of adult fly eyes
Scanning electron microscopy (SEM) of adult flies was carried out after fixation in 2% glutaraldehyde (EM grade, Electron Microscopy Sciences), dehydration through a graded series of ethanol and HMDS solvent.
Immnohistochemistry
Antibody staining of imaginal discs was performed using the following antibodies; rat anti-ELAV antibody (U of Iowa, DSHB), guinea pig anti-Hid antibody (Ryoo, 2004), monoclonal anti-rhodopsin-1 (U of Iowa, DSHB, 4C5), mouse anti-engrailed antibody (U of Iowa, DSHB, 4D9), mouse anti-beta-tubulin antibody (Sigma), rabbit anti-active caspase-3 antibody (Cell Signaling Technology, D175), mouse anti-BrdU antibody (BD PharMingen), rabbit anti-phospho-HistonH3 antibody (Upstate), mouse anti-ATP5A antibody (MitoSciences, 15H4C4). All fluorescent images were captured on a Zeiss LSM510 confocal microscope under identical conditions. Adjustment of contrast through Photoshop was applied to every pixel in the image, and to the same extent as control panels. BrdU was labeled for 30min and standard protocols were followed thereafter (de Nooij, 1995).
Western Blot and Far Western Blot Analysis
Larval extracts were made by dissecting imaginal discs and brains from each genotype, which were lysed in ice-cold lysis buffer (10mM Tris-HCl[pH 7.5], 150mM NaCl, protease inhibitor cocktail [Roche], 1mM EDTA, 1% SDS). Mitochondrial fractionation of embryos and larval tissues was performed as described previously (Fernandez-Moreno et al., 2007). Primary and secondary antibodies were used at the following dilution: rat anti-ELAV antibody (U of Iowa, DSHB; 1:1000), mouse anti-engrailed antibody (U of Iowa, DSHB, 4D9; 1:1000), mouse anti-beta-tubulin antibody (Sigma; 1:10000), guinea pig anti-Hid antibody (1:800), mouse anti-ATP5A antibody (MitoSciences, 15H4C4; 1:5000).
For Far Western Blot, recombinant GST or GST-DIAP1 was purified from BL21 E. coli cells. Immunoblots were blocked and then incubated with the following purified proteins in blocking solution; GST-Diap1 protein (0.2mg/ml). GST (0.8mg/ml) protein and BSA (0.8mg/ml) served as negative controls. After washing, bound proteins were detected with rabbit anti-GST antibody (sigma; 1:7500) or rabbit anti-Diap1 antibody (1:5000) (Ryoo, 2002). The binding was visualized by the ECL system.
Phosphatase treatment
Embryos were collected overnight and lysed in ice-cold PMP buffer (NEB) with 0.1% SDS. Supernatants were heated for 40 sec at 95°C, then added 1% NP-40. The samples were incubated for 30 min at 30°C with 400U of Lambda Protein Phosphatase (NEB) and subjected the Western Blot analysis.
Highlights.
CDK7 is required for apoptosis in Drosophila.
CDK7 is required for the mitochondrial localization of Hid.
Mitochondrial localization of Hid is coupled with its ability to bind DIAP1.
Acknowledgments
We thank J. Lis and R. Fisher for the cdk7S164A,T170A flies and antibody, P. Cho, E. Robbins and M. Ren for technical advice, and R. Fisher, H. Funabiki and Y. Ye for critically reading the manuscript. This work was supported by the NIH grant R01EY020876.
Footnotes
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley NJ, Cassill JA, Baker EK, Zuker CS. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SF, Longhi R, Borgese N. The role of cytosolic proteins in the insertion of tail-anchored proteins into phospholipid bilayers. J Cell Sci. 2009;122:2383–2392. doi: 10.1242/jcs.049460. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- Fernandez-Moreno MA, Farr CL, Kaguni LS, Garesse R. Drosophila melanogaster as a model system to study mitochondrial biology. Methods Mol Biol. 2007;372:33–49. doi: 10.1007/978-1-59745-365-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005;118:5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:4936–4941. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Chung J, Ryoo HD. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat Cell Biol. 2012;14:409–415. doi: 10.1038/ncb2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurada P, O’Tousa JE. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Larochelle S, Chen J, Knights R, Pandur J, Morcillo P, Erdjument-Bromage H, Tempst P, Suter B, Fisher RP. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 2001;20:3749–3759. doi: 10.1093/emboj/20.14.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S, Pandur J, Fisher RP, Salz HK, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, Duyk G, Friedman L, Prives C, Kopczynski C. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, Gan EC, Colon-Ramos DA, Kaplan B, Kornbluth S. A GH3-like domain in reaper is required for mitochondrial localiztion and induction of IAP degradation. J Biol Chem. 2003;278:44758–44768. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Baehrecke EH. Distinct death mechanisms in Drosophila development. Curr Opin Cell Biol. 2010;22:889–895. doi: 10.1016/j.ceb.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded Protein Response in a Drosophila Model for Retinal Degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Sandu C, Ryoo HD, Steller H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J Cell Biol. 2010;190:1039–1052. doi: 10.1083/jcb.201004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Larochelle S, Suter B, Lis JT. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol Cell Biol. 2003;23:6876–6887. doi: 10.1128/MCB.23.19.6876-6886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]