Abstract
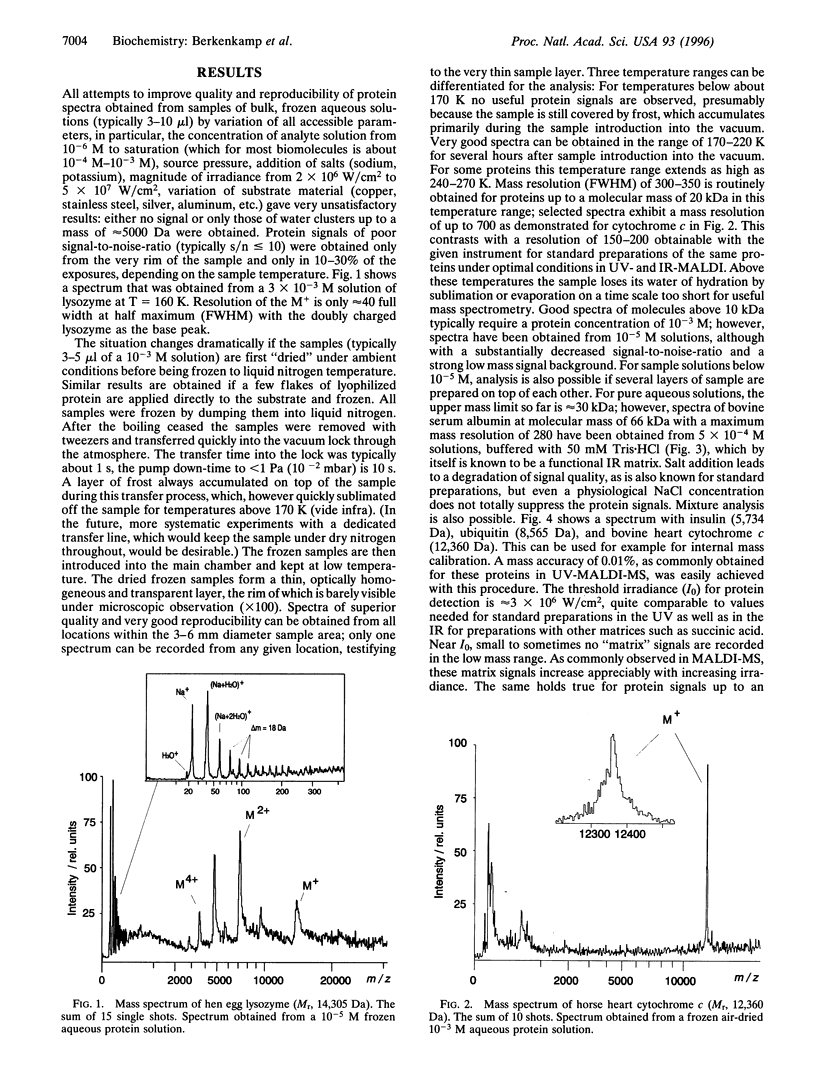
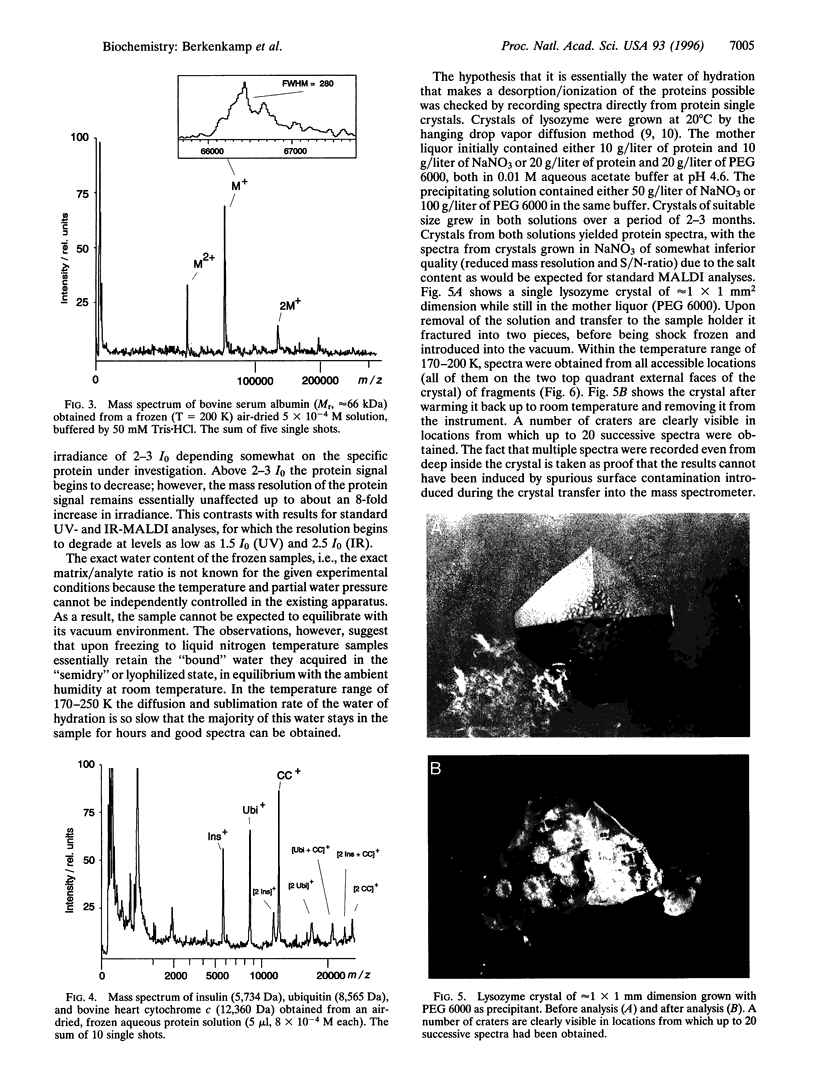
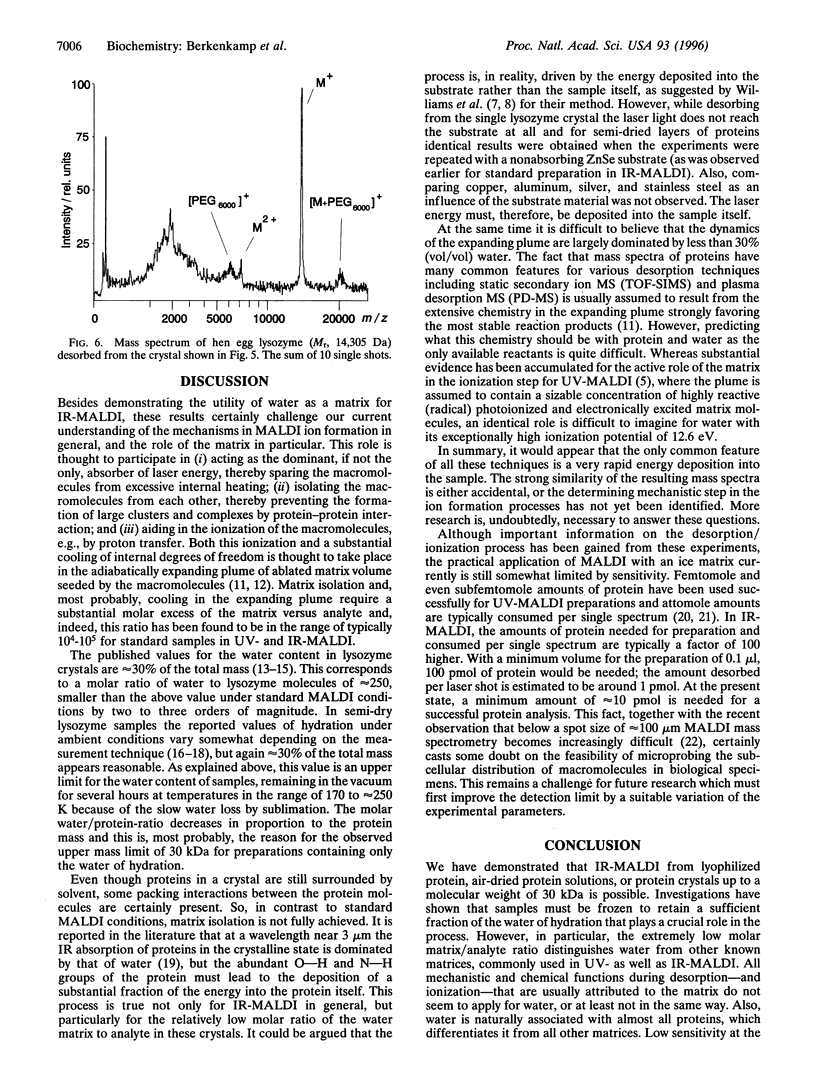
Lasers emitting in the ultraviolet wavelength range of 260-360 nm are almost exclusively used for matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) of macromolecules. Reports about the use of lasers emitting in the infrared first appeared in 1990/1991. In contrast to MALDI in the ultraviolet, a very limited number of reports on IR-MALDI have since been published. Several matrices have been identified for infrared MALDI yielding spectra of a quality comparable to those obtained in the ultraviolet. Water (ice) was recognized early as a potential matrix because of its strong O-H stretching mode near 3 microm. Interest in water as matrix derives primarily from the fact that it is the major constituent of most biological tissues. If functional as matrix, it might allow the in situ analysis of macromolecular constituents in frozen cell sections without extraction or exchanging the water. We present results that show that IR-MALDI of lyophilized proteins, air dried protein solutions, or protein crystals up to a molecular mass of 30 kDa is possible without the addition of any separate matrix. Samples must be frozen to retain a sufficient fraction of the water of hydration in the vacuum. The limited current sensitivity, requiring at least 10 pmol of protein for a successful analysis needs to be further improved.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bull H. B., Breese K. Protein hydration. I. Binding sites. Arch Biochem Biophys. 1968 Nov;128(2):488–496. doi: 10.1016/0003-9861(68)90055-6. [DOI] [PubMed] [Google Scholar]
- Harata K. X-ray structure of a monoclinic form of hen egg-white lysozyme crystallized at 313 K. Comparison of two independent molecules. Acta Crystallogr D Biol Crystallogr. 1994 May 1;50(Pt 3):250–257. doi: 10.1107/S0907444993013290. [DOI] [PubMed] [Google Scholar]
- Hillenkamp F., Karas M., Beavis R. C., Chait B. T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991 Dec 15;63(24):1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D., Jr, Kauzmann W. Hydration of proteins and polypeptides. Adv Protein Chem. 1974;28:239–345. doi: 10.1016/s0065-3233(08)60232-6. [DOI] [PubMed] [Google Scholar]
- Kurachi K., Sieker L. C., Jensen L. H. Structures of triclinic mono- and di-N-acetylglucosamine: lysozyme complexes--a crystallographic study. J Mol Biol. 1976 Feb 15;101(1):11–24. doi: 10.1016/0022-2836(76)90063-2. [DOI] [PubMed] [Google Scholar]
- Nelson R. W., Rainbow M. J., Lohr D. E., Williams P. Volatilization of high molecular weight DNA by pulsed laser ablation of frozen aqueous solutions. Science. 1989 Dec 22;246(4937):1585–1587. doi: 10.1126/science.2595370. [DOI] [PubMed] [Google Scholar]
- Ramanadham M., Sieker L. C., Jensen L. H. Refinement of triclinic lysozyme: II. The method of stereochemically restrained least squares. Acta Crystallogr B. 1990 Feb 1;46(Pt 1):63–69. doi: 10.1107/s0108768189009195. [DOI] [PubMed] [Google Scholar]
- SOPHIANOPOULOS A. J., RHODES C. K., HOLCOMB D. N., VAN HOLDE K. E. Physical studies of lysozyme. I. Characterization. J Biol Chem. 1962 Apr;237:1107–1112. [PubMed] [Google Scholar]



