Abstract
Over the past three decades of intense research on the contribution of viral and host factors determining the variability in HIV-1 infection outcome, HIV pathogenesis is still a fascinating topic that requires further study. An understanding of the exact mechanism of how these factors influencing HIV pathogenesis is critical to the development of effective strategies to prevent infection. Significant progress has been made in identifying the role of CCR5 (R5) and CXCR4 (X4) HIV strains in disease progression, particularly with the persistence of R5 HIV-1 strains at the AIDS stage. This indicates that R5 strains are as fit as X4 in causing CD4+ T cell depletion and in contribution to disease outcome, and so questions the prerequisite of the shift from R5 to X4 for disease progression. In contrast, the ability of certain HIV strains to readily use CXCR4 for infection or entry into macrophages, as the case with viruses are homozygous for tropism by CCR5delta32. This raises another major paradox in HIV pathogenesis about the source of X4 variants and how do they emerge from a relatively homogeneous R5 viral population after transmission. The interactions between viral phenotypes, tropism and co-receptor usage and how they influence HIV pathogenesis are the main themes addressed in this review. A better understanding of the viral and host genetic factors involved in the fitness of X4 and R5 strains of HIV-1 may facilitate development of specific inhibitors against these viral populations to at least reduce the risk of disease progression.
Key words: HIV/AIDS, tropism, coreceptor, SI/NSI
HIV primary infection and disease progression
There are several viral and host factors determining the variability in HIV-1 infection outcome and in rates of disease progression in HIV-1 infected individuals. Cellular tropism which defines viral phenotype and receptorcoreceptors which determine viral entry into various cell types are the major factors influencing HIV pathogenesis. Despite the intense research for the last 25 years, the exact mechanism of how these factors contribute to the dramatic loss of CD4+ T cells and the persistence of R5 and X4 strains during the AIDS status is still not well identified.1
Infection with HIV starts without symptoms or ill-feeling and is accompanied by slight changes in the immune system. This stage spans up to three months after infection until seroconversion where HIV-specific antibodies can be detected in individuals following recent exposure. The outcome of infection and duration for disease progression with clinical symptoms may vary greatly between individuals, but often it progresses fairly slowly.2 It takes several years from primary infection to the development of symptoms of advanced HIV diseases and immunosuppression.
During primary infection, although individuals may look healthy, the virus is actively replicating in the lymph nodes and blood stream of infected individuals. As a result, the immune system may get slowly damaged by the burst of viral load in their bodies.3
Symptomatic stage of disease indicates the late phase of HIV disease (AIDS) where individuals may be susceptible to other opportunistic infections (OIs),4 such as infections with Mycobacterium avium, Mycobacterium tuberculosis, Pneumocystis carinii, CMV, toxoplasmosis and candidiasis. It is agreed that infected individuals develop an AIDS status when their plasma HIV load is high and the CD4+ T count is less than 200 mm3 (Figure 1). The availability of the highly active antiretroviral therapy (HAART) may question the dilemma as to whether everyone who seroconverts to HIV will develop AIDS.
Figure 1.
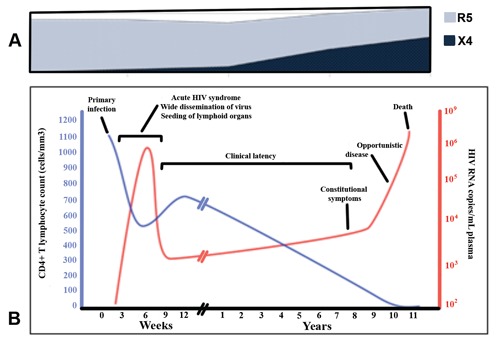
Role of R5 and X4 HIV strains in disease progression. A) Persistence R5 HIV-1 strains at all stages of HIV infection particularly at the late stages of HIV infection whereas X4 HIV-1 strains emerge at the advanced disease stage only; B) the course of HIV infection and disease. Levels of CD4 and viral load are shown to correlate with the progress of HIV infection and CD4+ T cell depletion.
One mechanism HIV weakens the immune system is by infecting and destroying CD4+ T cells, which in turn leads to immunodeficiency at later stage of disease.5 HIV attaches to the CD4+ protein on the surface of these and other cells to gain entry. However, the presence of CD4+ molecules alone proves to be not enough to allow viral entry into other cell types such as monocytes and dendritic cells. Therefore, a second doorway is needed for the virus to gain access to infect cells. This led to the discovery of the chemokine receptor as essential coreceptors for HIV-1. There are different types of these coreceptors for different cell types that HIV variants can use for infection of cells. Two main chemokine receptors have been identified to play a major role in HIV entry, CCR5 and CXCR4 (or fusin).
HIV tropism
HIV-1 is one of the most polymorphic viruses known and exists as a swarm of genetically related variants or quasispecies. The polymorphic nature of HIV-1 can be directly attributed to its error prone reverse transcriptase and complexity of its cDNA formation. Together with other host factors, the evolution of the viral genome underlies all of the changes in the biological characteristics of HIV-1 including cytopathic ability, immune evasion, coreceptor usage and tropism.
HIV-1 strains can be subdivided into three main groups based on their cellular tropism. They are referred to as macrophage-tropic (Mtropic) with a non-syncytium-inducing (NSI) phenotype or T-cell line tropic (T-tropic) with a syncytium-inducing (SI) phenotype or dualtropic HIV-1 strains. M-tropic NSI variants infect peripheral blood mononuclear cells (PBMC), monocytes, macrophages and T lymphocytes but not T-cell lines and are present throughout all stages of infection/disease.6-8 Ttropic SI isolates preferentially infect T lymphocytes and T-cell lines but not monocytes or macrophages and emerge in late stages of infection and is associated with progression to AIDS.9 On the other hand dual-tropic HIV-1 variants infect both monocytes/macrophage and T-cell lines, and therefore have mixed viral population of NSI/SI phenotypes.
The prevalence of SI strains usually correlates with rapid decline in CD4+ T cell count and the disease progression rate will be 3-5 fold faster than NSI strains. It is unclear whether the virus converts from an NSI to an SI strain, or an SI viral population emerges. It is worth noting that about 50% of people who die of AIDS still have a predominant NSI strain of HIV.
Defining the viral phenotype can be done by examining its ability to infect and produce syncytia in the MT2 cell line, i.e. a classification can be made of SI versus NSI. It has been shown that the viral phenotype NSI/SI could also be defined genetically through the charge of, and amino acid substitutions, in the V3 loop of the HIV gp120 envelop gene.10-12 Since the discovery of the chemokine receptors as HIV co-receptors, it has been possible to understand these phenomena at a molecular level.
HIV coreceptors
The discovery of chemokine receptors as essential coreceptors required for HIV entry has to a large extent rationalised the basis of cellular tropism and better defined HIV entry into different cells. CCR5 is present on a broad range of cells that can be infected by HIV, including T cells, monocytes and macrophages. Different HIV strains may be encountered in the body of the patients which can be classified into three variants: M-, T- and dual-tropic. Mtropic HIV variants can infect monocytes, macrophages and T lymphocytes through using CCR5 (R5), but not T cell lines as these express primarily CXCR4. T-tropic variants, which use CXCR4 (X4) as their principal coreceptor readily, infect T-cell lines and T lymphocytes but not macrophages (Figure 2).13
Figure 2.
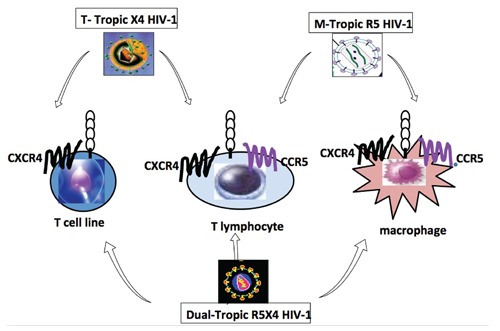
Coreceptor usage determines viral entry into different cell types and uncovered the mystery of cellular tropism. Macrophages and primary T lymphocytes express CCR5 and CXCR4 where T cell lines express only CXCR4. Macrophage (M)-tropic HIV-1 strains infect macrophages and lymphocyte using CCR5, while T-cell line (T)-tropic strains infect lymphocytes and T cell lines (but not macrophages) by using CXCR4. All three cell types are infectable by the dual-tropic strains by using either coreceptors for entry. T lymphocytes are infectable by all strains of HIV.
Dual-tropic strains of HIV-1 utilise CCR5 and CXCR4 (R5X4) to enter macrophages and T celllines, respectively, but they can also utilise combinations of major and minor coreceptors. Furthermore, there is coexpression of chemokine receptors, especially CCR5 and CXCR4, on most cell types, including blood and tissue macrophages, dendritic cells and T lymphocytes. 14 Some primary, but not laboratoryadapted X4, T-tropic isolates can also enter macrophages via CXCR4 (dual-tropic X4 strains).15 Major and minor coreceptors are also coexpressed, and CCR3, in addition to CCR5, can be utilised by M-tropic strains in fetal microglial cells,16 although there is no consistent agreement as to the relative importance of CCR3 and CCR5 in adult microglial cells.
Thus the current classification of cellular tropism of HIV-1 relies on the differential expression of CCR5 and CXCR4 in monocytes/macrophages and T-cell lines.6,7 CCR5 is important for NSI M-tropic strains (R5NSI) of HIV, most commonly observed in early stage of infection. On the other hand, CXCR4 mostly is associated with SI strains (X4SI) which are more pathogenic and they appear in some individuals with more aggressive disease. The dual-tropic HIV-1 variants infect both monocytes/macrophage and T-cell lines either through CCR5 or CXCR4 (R5X4) respectively (Figure 2). We have reported a case of homozygote for CCR5 delat32/delta32 with ability of the virus to enter various cell types including T lymphocytes and macrophages using solely CXCR4.17
Other chemokine receptors, principally CCR3 and CCR2b, function as minor HIV coreceptors. 18 Coreceptors have also shown to mediate the entry of simian immunodeficiency virus and some M-tropic HIV-1 and HIV-2 strains, including Bonzo/STRL33, Bob/GPR15, US28, CCR8, CX3CR1/V28, and CCR9 (APJ).18 Another coreceptor, GPR1, mediates the entry of SIV but not HIV-1.
CCR5 polymorphism
Naturally occurring mutations of CCR5 influence susceptibility to HIV infection. A 32- nucleotide deletion in CCR5 (CCRdelta32) was found to be common in Caucasians, with heterozygosity being present in 20% of the population and homozygosity in 1%. CD4+ T lymphocytes and macrophages from individuals homozygous for this mutant do not express functional CCR5 on the surface (CCR5 delta32/delta32) and cannot usually be infected with NSI R5 strains of HIV-1.19 Individuals with heterozygous genotype progress more slowly to AIDS and death than individuals with the wild-type CCRwt/wt genotype and are overrepresented among long-term nonprogressors.20 Only a few homozygotes infected with HIV-1 have been reported.17,21-23 The characterization of an HIV strain infecting one of these individuals has been studied by us,17 and others.24,25 The viral quasispecies in these individuals were homogeneous: T-tropic, syncytium inducing, utilized only CXCR4, and replicated well in T cells but not in macrophages. In our study, the HIV strain isolated from one individual was fully characterized for viral phenotype and genotype, macrophage tropism, and replication kinetics in MT2 cell lines and in primary macrophages and T lymphocytes from siblings who were CCR5delta32/delta32 or CCR5wt/wt. The isolated virus demonstrated dual-tropism for infection of MT2 cell lines and primary macrophages. The virus was shown to utilize predominantly CXCR4 for entry into T lymphocytes and macrophages using the HOS.CD4 cell infection assay, direct envelope protein fusion, and inhibition by anti-CXCR4 monoclonal antibodies (12G5), SDF1, and AMD3100. In addition, sequencing of the full HIV genome directly from the patient’s PBMCs revealed 21 nucleotides insertion in the V1 region of gp120. The virus envelope sequence segregated apart from both the T-cell-line-adapted tropic strains NL43 and SF2 and M-tropic strain JRFL or YU2 by phylogenetic tree analysis.
Macrophage tropism and disease progression
Macrophages act as major reservoirs for HIV-1 in tissues of the body. HIV infected macrophages may be found in brain, lungs, lymph nodes, skin, bone marrow and blood of seropositive individuals.26 They are the main source of productive infection in brain, and HIV infected macrophages in lymph nodes produce large amounts of viral particles. Infection with HIV-1 persists for weeks or months after commencement of highly active retroviral therapy (HAART). Monocytes/macrophages are believed to serve as vehicles for dissemination of HIV between different tissues of the body.26 Macrophages may play a key role in regulating the intensity and progression of disease in HIV infection, with their secretory products being implicated in the pathogenesis of AIDS dementia complex.27
The variability in susceptibility and productivity of infection shown here between neonatal and adult cells may be due to differences in the expression of chemokine receptors such as CCR5, CXCR4, CCR3 and CD4. However, CD4 expression has no contribution to the differential tropism and replication demonstrated between both cell types (Naif HM et al., 2002; unpublished data), which may highlight the utilization of CCR3 in neonatal cells but not in adult. Whether this indicates a functional role of CCR3 and/or has implications in vertical transmission, it requires further investigation. CXCR4 is expressed reasonably well on monocytes and macrophages,6 and can be used by some SI and dual-tropic primary isolates.15,17,28
Progression of acute HIV-1 infection to AIDS is accompanied by immune dysregulation and susceptibility to OIs.4 Accompanying these events, a change in HIV-1 dynamics involves an increased representation of HIV-1 positive macrophages hosts, in addition to CD4+ T cells. Although in the context of HAART, OIs are minimized as immune reconstitution occurs, HAART is neither universally available nor fully successful, requiring continued dissection of the mechanisms underlying augmented viral replication. The impact of OIs on macrophages susceptibility to HIV infection and replication revealed multiple contributing permissive factors. These include upregulation of viral coreceptors, activation of NF and production of TNF-alpha leading to induction of HIV replication in infected macrophages.29 In addition; apoptosis of T cells is also shown to act as a potent antiviral strategy to purge infected lymphoid cells. This approach fails to counter the persistence of HIV-1 infection in tissue macrophages and reservoirs monocytes.
Role of R5 and X4 HIV-1 variants in disease progression
Many distinct strains of HIV-1 have been isolated, and their variability in biological, serological, and molecular features indicate the virus is highly heterologous. These differences, again become apparent in the level of virus entry, replication kinetics, production the modulation of CD4 expression, cytopathicity, latency and inducibility, genetic evolution, viral tropism and coreceptor usage.7,8,30,31
R5 M-tropic strains are dominant in the early stages of infection, and more importantly they persist to advanced stages of disease. In a progressive study we have shown the importance of macrophages, the persistence and role of R5 M-tropic HIV-1 strains in disease progression. 7 These R5 strains from the advanced stage of disease are biologically fit and more infectious than those derived from the early time of infection (Figure 1). They are able to infect productively monocytes and macrophages. On the other hand, the R5 variants appear to be restricted in replication, particularly in undifferentiated monocytes at the level of viral entry.7,30 Thus, R5 from patients with AIDS seem to have an enhanced cytopathic activity and higher affinity for binding to CD4 and/or CCR5. We have shown that X4 variants are absent in approximately 60% of HIV-1 infected patients at the very advanced stage of disease (Figure 1A). It has been shown that HIV-1 infects macrophages and microglia in the central nervous system and causes HIV-associated dementia in 10% of patients with AIDS.32 This raises the possibility of interactions and/or contributions of multiple factors exerted in maintaining R5 persistence, such as the effect of viral evolution, specific host genetic factors and immune system pressure. In addition we have also shown that infection of monocyte and macrophage by HIV-1 is regulated by various factors.33 These factors, in brief, include cell maturation and differentiation, 6 sensitivity to beta-chemokines,34 and cytokines.35,36
Years after chronic infection is established, CXCR4 utilizing strains emerge in approximately 40% of infected individuals. The importance of the emergence of X4 strains in late stages of infection has always been directed towards their role in in immunodeficiency and rapid progression to AIDS. This is due to the seemingly obvious association between their enhanced cytopathicity and replicative ability linked to CD4+ T-cell depletion in late stages of infection. However, the mechanism by which X4 viruses are associated with accelerated disease progression has never been properly elucidated. The acceleration of HIV-1 disease progression has been attributed to the expanded spectrum of CXCR4+ precursor cells susceptible to infection by X4 strains. It has also been postulated that the decline of the host immune system associated with clinical AIDS may allow X4 viruses to evolve and replicate freely in late-stage infection.37 Furthermore, a critical question must be asked: how are X4 strains maintained at such high levels during late stages of infection when one of the main cell targets, CD4+T cells, are significantly depleted? At the same time macrophages as the other major target cells and 60% were infected by X4 strains and only 10% were shown to be resistant (Naif HM et al. 2002; unpublished data). This is still a major paradox in HIV pathogenesis and raises a few questions. What is the source of these X4 variants? How do they emerge from a relatively homogeneous R5 viral population after transmission? What factors (viral and host factors) contribute to their emergence?
Chemokines and cytokines
Multiple factors that associated with inflammation and immune activation appear to contribute to the permissiveness of mononuclear phagocytes (and other cell types) to HIV infection, particularly at the advanced course of disease. Environmental factors such as different cytokines and chemokine exhibited different profiles which may influence disease progression and HIV pathogenesis.38 It has been shown that viral replication can be stimulated by vaccination, cytokine induction, beta chemokines, endotoxins and opportunistic infections by various pathogens.
Th1 and Th2 cytokines play an important role in the pathogenesis of HIV infection and development of AIDS. In individuals infected with HIV, the normal Th1 response to viral infection is shifted to Th2 response.39
Many cytokines such as TNF-alpha and -beta, IL-1, IL-2, IL-3, IL-12, MCSF, GMCSF and IL-6 can enhance the replication of HIV and also play a key role in AIDS associated malignancies. 36,40 In contrast, other cytokines such as IFN-alpha and -beta inhibit HIV replication whereas TGF-beta, IL-4, IL-10, IL-13 and IFN-gamma have dichotomous effects on HIV replication, being stimulatory or inhibitory depending on cell types and experimental conditions. 35,36 IL-16 effect on HIV replication where most of the study have been done in cell-lines, it exerted an inhibitory effect on HIV-1 replication. 41 In our study using macrophages, IL-16 inhibited both R5 and X4 strains of HIV-1 replication and this inhibition was independent to its effect on chemokine receptors CCR5 and CXCR4 as well as CD4.42,43
The interactions among chemokine receptors, chemokines, and HIV are likely to be critical to the pathogenesis of HIV disease. The interactions among chemokine receptors, chemokines, and HIV are likely to be critical to the pathogenesis of HIV disease.43 The ligands for CCR5 (RANTES, MIP-1alpha, and MIP-1beta) and CXC chemokine receptor 4 (stromal-derived factor-1, SDF1) have been demonstrated to inhibit HIV entry into CD4+ T cells and PBMCs, as well as monocytic and CD4+ T cell lines.19,44 Given these inhibitory effects, increased production of these proteins has been suggested to be a protective host immune response against HIV infection and disease progression.44 The effects of beta-chemokines on HIV replication in monocytes and macrophages, using different experimental conditions was conducted by our group to analyze the effects of timing of exposure to betachemokines on HIV replication in monocytes and MDM in vitro. Beta-chemokine exposure produced dichotomous effects on HIV replication in this system.
Primary SI strains resistant to betachemokines. 45 They in fact upregulate T-tropic strains in CD4 T-cells but sensitive to SDF1.41 Recently dysregulation of expression of cytokines and chemokines in HIV has been demonstrated.46
References
- 1.Fauci AS.25 years of HIV. Nature. 2008;453: 289-90 [DOI] [PubMed] [Google Scholar]
- 2.Cunningham AL, Li S, Juarez J, et al. The level of HIV infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol. 2000; 68:311-7 [PubMed] [Google Scholar]
- 3.Moir S, Chun TW, Fauci AS.Pathogenic mechanisms of HIV disease. Annu Rev Pathol. 2011; 6:223-48 [DOI] [PubMed] [Google Scholar]
- 4.El-Atrouni W, Berbari E, Temesgen Z.HIV-associated opportunistic infections. Bacterial infections. J Med Liban. 2006; 54:80-3 [PubMed] [Google Scholar]
- 5.Chun TW, Fauci AS.HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS. 2012; 26:1261-8 [DOI] [PubMed] [Google Scholar]
- 6.Naif HM, Li S, Alali M, et al. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998; 72:830-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Juarez J, Alali M, et al. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999; 73:9741-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naif HM, Li S, Alali M, et al. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocytederived macrophages by using twins. J Virol. 1999; 73:4866-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glushakova S, Grivel JC, Fitzgerald W, et al. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998; 4:346-9 [DOI] [PubMed] [Google Scholar]
- 10.Fouchier R, Groenink M, Koostra N, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp20 molecule. J Virol. 1992; 66:3183-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariyoshi K, Harwood E, Chiengsong-Popov R, et al. Is clearance of HIV-1 viraemia at seroconversion mediated by neutralising antibodies? Lancet. 1992; 340:1257-8 [DOI] [PubMed] [Google Scholar]
- 12.Mild M, Kvist A, Esbjörnsson J, et al. Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection. Infect Genet Evol. 2010; 10:356-64 [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Broder CC, Kennedy PE, et al. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996; 272:872-7 [DOI] [PubMed] [Google Scholar]
- 14.Turville SG, Arthos J, Donald KM, et al. HIV gp120 receptors on human dendritic cells. Blood 2001;98:2482-8 [DOI] [PubMed] [Google Scholar]
- 15.Yi Y, Isaacs SN, Williams DA, et al. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J Virol. 1999; 73:7117-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Chen Y, Farzan M, et al. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997; 385:645-9 [DOI] [PubMed] [Google Scholar]
- 17.Naif HM, Cunningham AL, Alali M, et al. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Delta32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J Virol. 2002; 76:3114-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, He T, Huang Y, et al. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998; 72:9307-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alkhatib G, Combardiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, IP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996; 272:1955. [DOI] [PubMed] [Google Scholar]
- 20.Stewart GJ, Ashton LJ, Biti RA, et al. Increased frequency of CCR-5 delta 32 heterozygotes among long-term non-progressors withHIV-1 infection. The Australian Long-Term Non-Progressor Study Group. AIDS. 1997; 11:1833-8 [DOI] [PubMed] [Google Scholar]
- 21.Balotta C, Bagnarelli P, Violin M, et al. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS. 1997;11:F67-71 [DOI] [PubMed] [Google Scholar]
- 22.Michael NL, Nelson JA, KewalRamani VN, et al. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 delta32. J Virol. 1998; 72:6040-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biti R, Ffrench R, Young J, et al. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997; 3:252-3 [DOI] [PubMed] [Google Scholar]
- 24.Simmons G, Reeves JD, McKnight A, et al. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998; 72:8453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verani A, Pesenti E, Polo S, et al. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998; 161:2084-8 [PubMed] [Google Scholar]
- 26.Ciborowski P, Gendelman HE.Human immunodeficiency virus-mononuclear phagocyte interactions: emerging avenues of biomarker discovery, modes of viral persistence and disease pathogenesis. Curr HIV Res. 2006; 4:279-91 [DOI] [PubMed] [Google Scholar]
- 27.Ryan LA, Cotter RL, Zink WE, et al. Macrophages, chemokines and neuronal injury in HIV-1-associated dementia. Cell Mol Biol. 2002; 48:137-50 [PubMed] [Google Scholar]
- 28.Princen K, Hatse S, Vermeire K, et al. Inhibition of human immunodeficiency virus replication by a dual CCR5/CXCR4 antagonist. J Virol. 2004; 78:12996-3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choe W, Volsky DJ, Potash MJ.Activation of NF-kappa B by R5 and X4 human immunodeficiency virus type 1 induces macrophage inflammatory protein 1alpha and tumor necrosis factor alpha in macrophages. J Virol. 2002; 76:5274-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naif HM, Li S, Alali M, et al. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocytederived macrophages by using twins. J Virol. 1999; 73:4866-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavarelli M, Scarlatti G.Phenotype variation in human immunodeficiency virus type 1 transmission and disease progression. Dis Markers. 2009; 27:121-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valcour V, Sithinamsuwan P, Letendre S, et al. Pathogenesis of HIV in the central nervous system. Curr HIV/AIDS Rep. 2011; 8:54-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen OJ, Fauci AS.Host factors that affect sexual transmission of HIV. Int J Infect Dis. 1998; 2:182-5 [DOI] [PubMed] [Google Scholar]
- 34.Kelly MD, Naif HM, Adams SL, et al. Dichotomous effects of beta-chemokines on HIV replication in monocytes and monocyte-derived macrophages. J Immunol. 1998; 160:3091-5 [PubMed] [Google Scholar]
- 35.Naif HM, Chang J, Ho-Shon M, et al. Inhibition of human immunodeficiency virus replication in differentiating monocytes by interleukin 10 occurs in parallel with inhibition of cellular RNA expression. AIDS Res Hum Retroviruses. 1996; 12:1237-45 [DOI] [PubMed] [Google Scholar]
- 36.Naif HM, Li S, Ho-Shon M, et al. The state of maturation of monocytes into macrophages determines the effects of IL-4 and IL-13 on HIV replication. J Immunol. 1997; 158:501-1 [PubMed] [Google Scholar]
- 37.Mild M, Kvist A, Esbjörnsson J, et al. Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection. Infect Genet Evol. 2010; 10:356-64 [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee A, Rathore A, Vidyan S, et al. Chemokines and chemokine receptors in susceptibility to HIV-1 infection and progression to AIDS. Dis Markers. 2012;32: 143-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein SA, Dobmeyer JM, Dobmeyer TS, et al. Demonstration of the Th1 to Th2 cytokine shift during the course of HIV-1 infection using cytoplasmic cytokine detection on single cell level by flow cytometry. AIDS. 1997; 11:1111-8 [DOI] [PubMed] [Google Scholar]
- 40.Kinter AL, Godbout EJ, McNally JP, et al. The common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 induce the expression of programmed death-1 and its ligands. J Immunol. 2008; 181:6738-46 [DOI] [PubMed] [Google Scholar]
- 41.Zhou P, Goldstein S, Devadas K, et al. Human CD4+ cells transfected with IL-16 cDNA are resistant to HIV-1 infection: inhibition of mRNA expression Nat Med. 1997; 3:659-64 [DOI] [PubMed] [Google Scholar]
- 42.Nasr N.IL-16 modulation of HIV-1 replication is independent of expression of CD4 and coreceptors. Degree Diss., University of Sydney, Australia; 2002 [Google Scholar]
- 43.Lusso P.HIV and the chemokine system: 10 years later. EMBO J. 2006; 25:447-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cocchi F, DeVico A, Garzino-Demo A, et al. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD81 T cells. Science. 1995;270:1811. [DOI] [PubMed] [Google Scholar]
- 45.Jansson M, Popovic M, Karlsson A, et al. Sensitivity to inhibition by betachemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996; 93:15382-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenneth OS, Hem CJ, Richard KD, et al. Cytokine and chemokine expression profiles in HIV-1 infected patients with ocular surface squamous neoplasia from Botswana. Cancer Clin Oncology. 2012;1: 10-24 [Google Scholar]


